Abstract
Context:
Peroxisome proliferator-activated receptor gamma (PPARG) is a susceptibility locus for type 2 diabetes mellitus (T2DM). Although cross-sectional associations have been reported, primarily for Pro12Ala, few longitudinal studies in nondiabetic populations have been conducted.
Objective:
This study aimed to examine whether and to what extent variation in PPARG is associated with longitudinal changes in anthropometric and metabolic traits in Mexican Americans at risk for T2DM.
Setting and Design:
Subjects were participants of BetaGene, a family-based study of obesity, insulin resistance, and β-cell function, who completed a baseline and follow-up study visit (n = 378; mean followup, 4.6 ± 1.5 y). Phenotypes included body fat assessed by dual-energy x-ray absorptiometry; insulin sensitivity (SI), acute insulin response, and β-cell function (disposition index; DI) were estimated from iv glucose tolerance tests with Minimal Model analysis. Eighteen tag single nucleotide polymorphisms (SNPs) capturing variation in a 156-kb region surrounding PPARG were tested for association with changes in longitudinal traits. P-values were Bonferroni-corrected for multiple testing.
Results:
Six SNPs (rs2972164, rs11128598, rs17793951, rs1151996, rs1175541, rs3856806) were significantly associated with rate of change in SI after adjustment for age, sex, and body fat percentage, but not with changes in adiposity. rs17793951 also had a significant effect on change in DI over time. Association between rs1175541 and change in SI varied by changes in adiposity such that only carriers of the minor allele who reduced body fat over followup improved SI. rs1306470 (captured Pro12Ala, r2 = 0.9) was not associated with rates of change in any traits and its effects were not modified by changes in adiposity.
Conclusions:
Variation in PPARG, but not Pro12Ala, contributes to declining SI and concomitant deterioration in β-cell function in Mexican Americans at risk for T2DM.
Peroxisome proliferator-activated Receptor gamma (PPARG) has long been recognized as a susceptibility locus for type 2 diabetes mellitus (T2DM). PPARG is expressed in adipocytes and encodes a nuclear transcription factor with a major role in adipocyte differentiation and function (1). In addition to associations with diabetes risk, variation in PPARG has also been shown to be associated with body mass index (BMI) (2–9), waist circumference (3, 4), waist-hip ratio (5), body fat distribution (4, 5, 8), fasting insulin (8–10), and insulin sensitivity (7, 9, 10) in nondiabetic individuals. However, other studies have failed to replicate these associations (11–16). Moreover, most of the studies that investigated these relationships were based on cross-sectional designs and/or focused primarily on the effects of Pro12Ala.
Among the studies that have examined longitudinal quantitative trait associations with PPARG (17–23), most examined Pro12Ala in relation to changes in body weight over time in the context of an intervention trial aimed at weight loss among obese and/or glucose-intolerant subjects. Two of these studies reported an association between the Ala allele and weight loss (19, 22), and one with weight gain (17), among individuals randomly assigned to placebo. In a population-based study of nondiabetic individuals in the United States, Fornage et al (20) observed opposite effects of the Ala allele for two racial groups; presence of an Ala allele was associated with a significant 15-year decline in adiposity among African Americans but an increase among whites. Significant decreases in fasting insulin and insulin resistance associated with Ala were only observed among African Americans (20). In a study of nondiabetic French individuals, Jaziri et al (23) reported that carriers of the Ala allele had significantly lower fasting insulin and insulin resistance after 6 years of followup compared with those with the Pro/Pro genotype. The inconsistent longitudinal findings reported to date within both homogenous and racial/ethnically diverse groups and scant data in the Mexican-American population underscore the need for investigation of the longitudinal effects of variation in PPARG in Mexican Americans at risk for T2DM.
The present study examined the effects of variation in PPARG on longitudinal changes in anthropometric and metabolic traits in a Mexican-American cohort enriched with individuals at high risk for T2DM. We also assessed whether the observed associations between the genetic variants and change in metabolic traits over time varied by changes in adiposity over time.
Materials and Methods
Subject recruitment
Participants of this study were from BetaGene, a family-based study of obesity, insulin resistance, and β-cell dysfunction in Mexican Americans. Details regarding recruitment have been previously described (24). Briefly, probands qualified for participation if they 1) were of Mexican ancestry (both parents and at least 3/4 of grandparents Mexican or of Mexican descent, 2) had a confirmed diagnosis of gestational diabetes mellitus (GDM) or normal glucose levels (no GDM) during a pregnancy within the 5 years of study enrollment, and 3) had no evidence of β-cell autoimmunity by glutamic acid decarboxylase-65 testing. GDM and non-GDM probands were identified from the patient populations of Los Angeles County/University of Southern California Medical Center, Kaiser Permanente Southern California, and obstetrical/gynecological clinics yat local southern California hospitals, and were frequency matched on age, BMI, and parity categories. Family members of GDM probands were recruited and included siblings, cousins, spouses, offspring, parents, and connecting aunts and uncles. Spouses and offspring of non-GDM probands were also invited to participate. Protocols for BetaGene were approved by the Institutional Review Boards of each institution, and all participants provided written informed consent prior to study enrollment.
Phenotyping for BetaGene was performed on two separate visits to the General Clinical Research Center. The first visit consisted of a physical examination, DNA collection, administration of food frequency and physical activity questionnaires, and a 2-hour 75-g oral glucose tolerance test (OGTT) with blood samples obtained before and at 30-minute intervals after glucose ingestion, as previously described (24). Fasting blood samples were also collected for lipid measurements. Participants with fasting glucose less than 126 mg/dL (< 7 mmol/l) were invited for a second visit, which consisted of a dual-energy x-ray absorptiometry scan for body composition and an insulin-modified iv glucose tolerance test (IVGTT) as previously described (24). All participants in the proband generation (n = 1247; 72% female; mean ± SD age, 34.7 ± 8.2 y) underwent the full phenotyping protocol, whereas parents, uncles, aunts, spouses, and offspring had only a physical examination, fasting glucose measurement, and DNA collection.
The BetaGene follow-up study (BetgaGene II) was designed to recall 400 participants from the proband generation of BetaGene for assessment of longitudinal changes in anthropometric and metabolic traits (25). Subjects with a fasting glucose level of at least 7.0 mmol/L were ineligible for further follow-up testing. Otherwise, both clinic visits described above were repeated on a subset of GDM probands, their siblings and cousins, and non-GDM probands (n = 390; 74% female; mean ± SD age, 39.5 ± 8.4 y; mean ± SD follow-up time, 4.6 ± 1.5 y). Due to follow-up study eligibility criteria, those who returned for followup had slightly lower median fasting and 2-hour glucose levels than those who did not participate (P = .09 and P = .04, respectively); no other significant differences between these two groups were observed (data not shown). We report results from 378 of the 390 BetaGene II participants, for whom complete genotype and phenotype data described below were available.
Assays
Plasma glucose was measured on an autoanalyzer using the glucose oxidase method (YSI Model 2300; Yellow Springs Instruments, Yellow Springs, OH). Insulin was measured by two- site immunoenzymometric assay (TOSOH Biosciences, San Francisco, CA) that has less than 0.1% cross reactivity with proinsulin and intermediate-split products.
Molecular analysis
Genotyping for BetaGene was performed on the Illumina OmniExpress platform. We selected 207 SNPs residing in PPARG, as well as eight SNPs in the regions 5-kb upstream and downstream of the gene for the current study. Genotype data were tested for non-Mendelian inheritance with MENDEL v.12.0 (26). We tested for deviation from Hardy-Weinberg equilibrium and estimated allele frequencies by maximum likelihood using SOLAR v.6.6.2 (27). Pair-wise linkage disequilibrium (LD) and haplotype block structure for the SNPs in PPARG were assessed using Haploview v.4.1 (28). Haplotype block structure was assessed using the method of Gabriel et al (29). After excluding SNPs for which the assay failed (n = 3), that failed (n = 10), were monomorphic (n = 30), or had minor allele frequencies (MAFs) less than 5% (n = 75), tag SNPs were selected from among the remaining 97 SNPs using TAGGER, with the default setting of pairwise r2 ≥ 0.8, as implemented in Haploview (28). We identified proxies for two common SNPs of interest that were not genotyped on the OmniExpress chip (rs10865710 and rs7649970) from the 1000 Genomes MXL panel (30).
For all BetaGene participants, we estimated proportions of admixture using a genome-wide set of 117 347 LD-pruned markers available on the OmniExpress array and the program ADMIXTURE v.1.21 (31). We included data for 591 individuals from the HapMap CEU, CHB, YRI, and MEX populations as proxies for European, Asian, African and Mexican ancestral populations, respectively. Assuming four subpopulations was appropriate for these data and yielded the lowest cross-validation error (data not shown). Proportions of admixture for BetaGene, compared with the HapMap CEU, CHB, YRI, and MEX populations are shown in Supplemental Figure 1.
Measures of insulin response to glucose
We calculated two measures of insulin response to glucose: the difference between the OGTT 30′ and fasting plasma insulin (30′ΔInsulin) and the incremental area under the insulin curve during the first 10 minutes of the IVGTT (acute insulin response; AIR). IVGTT glucose and insulin data were analyzed using the minimal model (MINMOD Millennium v.5.18) to derive insulin sensitivity (SI) (32). β-cell function was measured by disposition index (DI = SI × AIR), a measure of β-cell compensation for insulin resistance (33).
Data analysis
Demographic characteristics for the BetaGene II cohort are described by median and corresponding 25th and 75th percentiles. Descriptive analyses were performed using SAS software v.9.2 (SAS Institute Inc, Cary, NC). Baseline quantitative traits with skewed distributions were statistically transformed to approximate univariate normality for cross-sectional analyses. The rates of change in AIR, SI, and DI were computed as the difference between the log of the follow-up value and the log of the baseline value divided by total follow-up time; the rate of change in percent body fat was calculated as the difference between the follow-up value and baseline value, divided by total follow-up time. The resulting distributions for the rate of change in each trait were approximately normal. We tested each tag SNP for association with both baseline and the rate of change in anthropometric and metabolic traits using likelihood ratio tests within a variance components framework implemented in SOLAR (27), to account for familial correlation. An additive genetic model was assumed for SNPs with MAF greater than 20%, and a dominant model otherwise. All models were adjusted for age, sex, and baseline BMI or body fat percentage (where appropriate). To determine whether results would be further affected by population substructure and/or changes in adiposity over time, we also examined models that were additionally adjusted for estimated proportions of admixture and/or change in body fat percentage.
P-values were Bonferroni corrected for 18 tag SNPs to account for multiple testing, although because of the modest level of correlation underlying many of the SNPs examined and the assumption of independence inherent in the applying a Bonferroni correction, corrected P-values are likely overly conservative. Age-, sex-, and percent body fat–adjusted geometric means and SEs, computed by the Delta Method (34), for SI at baseline and mean followup, are presented by genotype for all tag SNPs. To determine whether the effect of the six SNPs associated with rate of change in SI varied by changes in body fat percentage over time, we tested the interaction between each of the six SNPs and the rate of change in body fat percentage using a 1-df likelihood ratio test. P-values for the tests of interaction were Bonferroni-corrected for the six interaction tests performed.
We estimated a priori power to assess our ability to detect association between SNPs in PPARG and longitudinal change in SI. For the lowest frequency SNP, which had a MAF = 11.0% (eg, Pro12Ala, or its proxy rs13064760) and was modeled under a dominant genetic model with genotype-specific sample sizes of 299 and 79 participants, respectively, we had 80% power to detect a difference of 0.08 in the mean rate of change in SI (× 10−3/min per pmol/L per year) between the two groups, assuming a 100% coefficient of variation in each group, two-sided test of no difference and α = 0.0028 (Bonferroni corrected for 18 SNPs). Under these assumptions, we had 80% power to detect a difference of 0.06 in the mean rate of change in SI for a SNP with MAF = 20.0%.
Results
The sample was composed of 378 BetaGene II participants, of whom 74.3% were female, and the median (25th, 75th percentile) baseline age was 34.6 (29.4, 40.4) years. Median BMI and body fat percentage were 28.7 kg/m2 and 36.1%, respectively. At baseline, median age, anthropometric, and metabolic profiles were generally similar for GDM probands and their family members, although GDM probands tended to be at the higher end and their cousins at the lower end of the range of values for each characteristic (Table 1). Non-GDM probands were similar to GDM probands in median age, BMI, and body fat percentage, by virtue of the fact that these two groups were initially frequency matched at baseline. However, non-GDM probands tended to have lower median OGTT-measured glucose and insulin, higher SI and DI, and lower low-density lipoproteins and triglyceride levels than GDM probands (Table 1). Overall, study participants did not show statistically significant changes in any anthropometric traits between baseline and follow-up visits. However, deterioration in metabolic profile over time was evident; at followup, participants had significantly higher median OGTT-measured 2-hour glucose and insulin, lower SI and DI, and higher triglyceride levels than they did at baseline (data not shown).
Table 1.
Characteristics of the BetaGene Longitudinal Cohort (n = 378) at Baseline
| Characteristic | GDM Probands (n = 79) | Siblings (n = 184) | Cousins (n = 80) | Non-GDM Probands (n = 35) |
|---|---|---|---|---|
| Female, n (%) | 79 (100.0) | 114 (62.0) | 53 (66.3) | 35 (100.0) |
| Age, y | 35.2 (32.3, 39.9) | 35.3 (29.0, 42.4) | 32.5 (24.1, 38.7) | 34.0 (31.4, 37.3) |
| Anthropometrics | ||||
| BMI, kg/m2 | 30.1 (26.3, 32.8) | 28.8 (26.2, 33.0) | 27.0 (22.7, 30.5) | 28.2 (24.3, 32.7) |
| Body fat percent | 38.5 (36.3, 42.7) | 34.3 (26.1, 40.4) | 32.5 (23.8, 37.8) | 38.4 (33.4, 40.5) |
| Trunk fat, kg | 14.5 (12.7, 18.2) | 12.4 (9.8, 17.5) | 10.6 (7.7, 15.1) | 11.9 (8.6, 17.2) |
| Waist circumference, cm | 95.0 (89.5, 103.0) | 94.0 (86.0, 103.0) | 90.0 (78.0, 99.0) | 88.0 (82.0, 96.5) |
| OGTT | ||||
| Fasting glucose, mmol/L | 5.2 (4.8, 5.5) | 5.1 (4.7, 5.4) | 5.0 (4.7, 5.3) | 4.7 (4.4, 4.9) |
| 2-h glucose, mmol/L | 8.0 (6.8, 9.6) | 7.3 (6.1, 8.5) | 6.4 (5.3, 7.7) | 5.7 (4.9, 6.8) |
| 30′ Δ insulin, pmol/L | 300 (162, 510) | 339 (204, 558) | 324 (201, 507) | 306 (192, 564) |
| Fasting insulin, pmol/L | 54 (36, 84) | 42 (24, 72) | 36 (18, 60) | 30 (18, 48) |
| 2-h insulin, pmol/L | 468 (276, 798) | 348 (228, 570) | 273 (117, 402) | 264 (180, 378) |
| IVGTT | ||||
| SI, × 10−3/min per pmol/L | 2.46 (1.66, 3.72) | 2.54 (1.69, 3.57) | 3.15 (2.22, 4.69) | 3.46 (2.24, 4.75) |
| AIR, pmol/L × 10 min | 1682 (958, 3580) | 2925 (1688, 4695) | 2867 (1560, 4559) | 3460 (2080, 4970) |
| DI | 4966 (2746, 6934) | 7528 (4628, 11 341) | 7903 (5664, 12 056) | 11, 379 (7297, 14 228) |
| Lipids | ||||
| Cholesterol, mg/dL | 168 (152, 196) | 173 (156, 199) | 165 (149, 183) | 151 (141, 172) |
| HDL, mg/dL | 46 (40, 52) | 45 (38, 50) | 49 (41, 57) | 50 (42, 61) |
| LDL, mg/dL | 100 (79, 121) | 106 (87, 127) | 97 (77, 111) | 85 (71, 102) |
| Triglycerides, mg/dL | 100 (76, 158) | 110 (68, 161) | 78 (46, 114) | 67 (41, 101) |
Abbreviations: HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Unless otherwise stated, values are median (25th, 75th percentile).
Estimated pairwise LD and haplotype block structure for the 97 SNPs included in this analysis are shown in Supplemental Figure 2. Among these 97 SNPs, we identified 18 tag SNPs (Table 2 and Supplemental Figure 3). Tag SNP MAFs ranged from 11.0–43.3%. C34G (Pro12Ala; rs1801282) was tagged by rs13064760 (r2 = 0.9). C1431T (rs3856806) was captured as a tag SNP. C-681G (rs10865710) and C-689T (rs7649970) were tagged by rs11128598 (r2 = 1.0) and rs17036328 (r2 = 1.0), respectively.
Table 2.
Characteristics of 18 Tag SNPs in PPARGa
| SNP | Position | Location | Major Minor Allele | MAF |
|---|---|---|---|---|
| rs13076933 | 12327431 | upstream | T G | 13.5% |
| rs2972164 | 12334416 | intron | C T | 30.6% |
| rs11128598b | 12353326 | intron | T C | 41.3% |
| rs17036281 | 12354411 | intron | T G | 28.2% |
| rs13064760c | 12369401 | intron | C T | 11.0% |
| rs17793951 | 12370737 | intron | A G | 16.8% |
| rs17036328d | 12390484 | intron | T C | 12.7% |
| rs4135268 | 12437237 | intron | C G | 11.0% |
| rs4135275 | 12443844 | intron | A G | 23.4% |
| rs1151996 | 12445807 | intron | T G | 20.4% |
| rs1151998 | 12446492 | intron | T C | 43.3% |
| rs1175541 | 12465488 | intron | C A | 18.9% |
| rs13099634 | 12468463 | intron | C T | 33.8% |
| rs1152001 | 12471862 | intron | T C | 17.7% |
| rs1152002 | 12471871 | intron | G A | 29.4% |
| rs3856806e | 12475557 | exon 6 | C T | 12.0% |
| rs4135300 | 12475994 | 3′ UTR | C T | 11.4% |
| rs9833097 | 12478817 | downstream | G A | 28.5% |
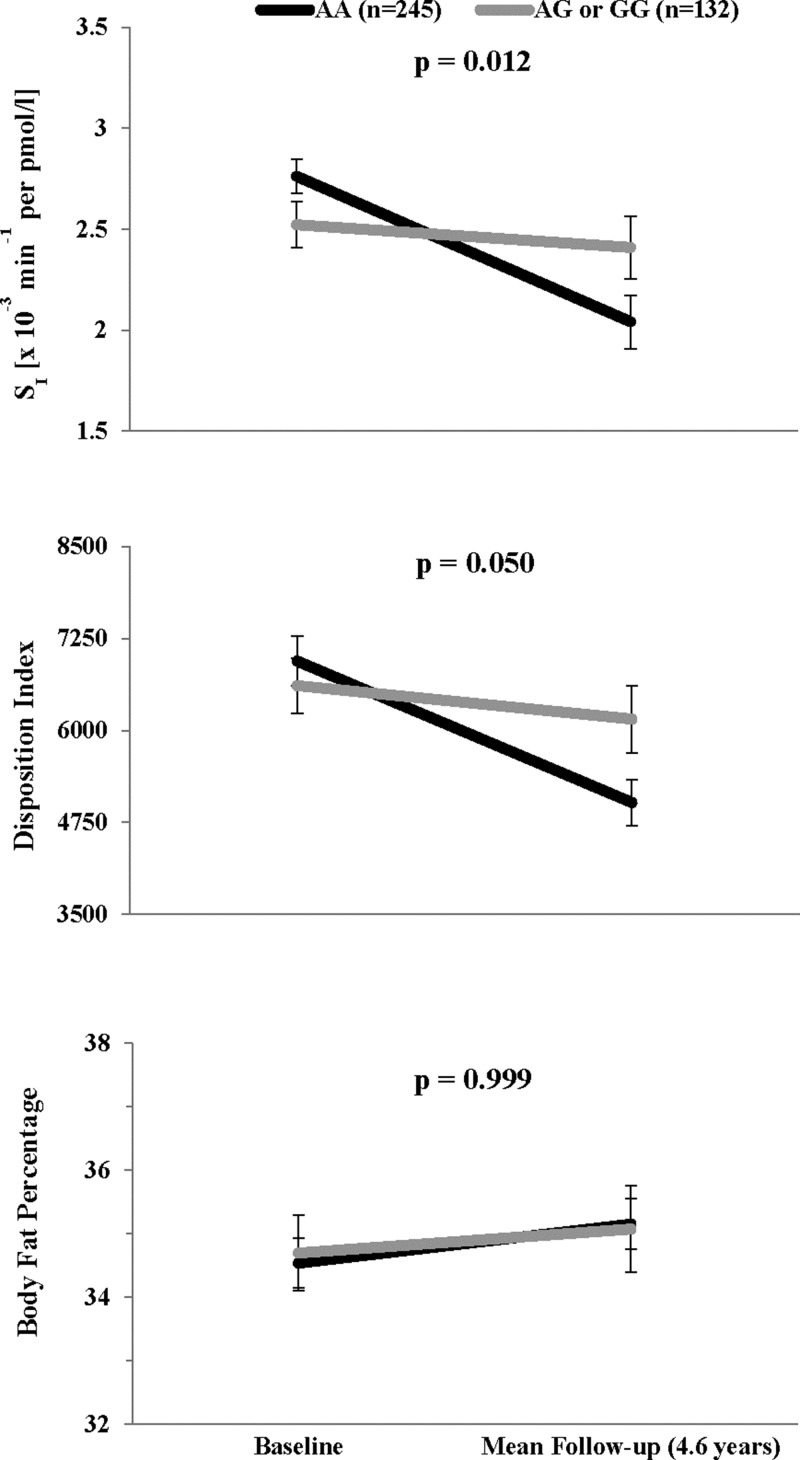
After adjustment for age and sex and correction for multiple testing, none of the 18 tag SNPs were significantly associated with any of the anthropometric or metabolic traits at baseline (all corrected P ≥ .20). Moreover, none were associated with any cross-sectional traits at followup (all corrected P ≥ .10). Cross-sectional associations with metabolic traits remained similar after further adjustment for BMI or body fat percentage. None of the SNPs were significantly associated with the rate of change in anthropometric traits; rs2972164 was only marginally associated with the rate of change in BMI (corrected P = .08). However, six SNPs: rs2972164, rs11128598, rs17793951, rs1151996, rs1175541 and rs3856806, were significantly associated with the rate of change in SI in models adjusted for age, sex, and baseline body fat percentage (corrected P = .006, .037, .012, .004, .016, and .037 respectively). Age-, sex- and percent body fat–adjusted mean values of SI at baseline and follow-up are shown by genotype for each of the 18 tag SNPs in Table 3. In general, the minor alleles of rs2972164, rs17793951, rs1151996, and rs1175541 were associated with slower rates of decline in SI, whereas those for rs11128598 and rs3856806 were associated with faster rates of decline. Only rs17793951 was associated with the rate of change in DI (corrected P = .05). Adjusted mean values of SI, DI, and body fat percentage at baseline and followup are shown for rs17793951 in Figure 1. Individuals carrying a G allele at this locus manifested little change in SI or DI over time, whereas those homozygous for the A allele decreased SI with concomitant decreases in DI, although there were no differences in the rate of change in body fat percentage by genotype (Figure 1). Results were similar after adjusting models for baseline BMI instead of body fat percentage, or further adjusting models for changes in adiposity (eg, BMI or body fat percentage) over time. Additional adjustment for population substructure did not alter these findings. Analysis of female participants only, most of the cohort, yielded similar estimates although some of these results became nonsignificant after multiple testing correction due to reduced sample size.
Table 3.
Meana SI at Baseline and at Mean Follow-up Time (4.6 y) by Genotype, According to Additive or Dominant Genetic Models as Appropriate, for 18 tag SNPs in PPARG
| SNP | SI × 10−3/min per pmol/L |
||||
|---|---|---|---|---|---|
| Baseline | seb | Mean Followup (4.6 y) | seb | P-Valuec | |
| rs13076933 | .185 | ||||
| TT | 2.73 | 0.08 | 2.08 | 0.13 | |
| TG/GG | 2.56 | 0.13 | 2.41 | 0.17 | |
| rs2972164 | .006 | ||||
| CC | 2.76 | 0.10 | 2.00 | 0.16 | |
| CT | 2.65 | 0.10 | 2.26 | 0.13 | |
| TT | 2.39 | 0.18 | 2.59 | 0.24 | |
| rs11128598 | .037 | ||||
| TT | 2.67 | 0.13 | 2.38 | 0.13 | |
| TC | 2.62 | 0.09 | 2.12 | 0.14 | |
| CC | 2.92 | 0.18 | 1.86 | 0.26 | |
| rs17036281 | 1.0 | ||||
| TT | 2.63 | 0.10 | 2.20 | 0.11 | |
| TG | 2.63 | 0.10 | 2.14 | 0.14 | |
| GG | 3.33 | 0.23 | 2.01 | 0.50 | |
| rs13064760 | 1.0 | ||||
| CC | 2.69 | 0.08 | 2.22 | 0.10 | |
| CT/TT | 2.65 | 0.12 | 1.95 | 0.22 | |
| rs17793951 | .012 | ||||
| AA | 2.76 | 0.09 | 2.04 | 0.13 | |
| AG/GG | 2.52 | 0.11 | 2.41 | 0.15 | |
| rs17036328 | 1.0 | ||||
| TT | 2.68 | 0.08 | 2.23 | 0.11 | |
| TC/CC | 2.67 | 0.12 | 1.96 | 0.19 | |
| rs4135268 | 1.0 | ||||
| CC | 2.69 | 0.08 | 2.19 | 0.11 | |
| CG/GG | 2.66 | 0.18 | 2.06 | 0.19 | |
| rs4135275 | 1.0 | ||||
| AA | 2.64 | 0.08 | 2.09 | 0.13 | |
| AG | 2.75 | 0.12 | 2.34 | 0.15 | |
| GG | 2.70 | 0.30 | 1.89 | 0.36 | |
| rs1151996 | .004 | ||||
| TT | 2.83 | 0.09 | 2.04 | 0.14 | |
| TG | 2.49 | 0.11 | 2.35 | 0.15 | |
| GG | 2.34 | 0.20 | 2.39 | 0.26 | |
| rs1151998 | 1.0 | ||||
| TT | 2.60 | 0.11 | 2.23 | 0.15 | |
| TC | 2.68 | 0.10 | 2.16 | 0.15 | |
| CC | 2.83 | 0.17 | 2.06 | 0.20 | |
| rs1175541 | .016 | ||||
| CC | 2.83 | 0.09 | 2.08 | 0.14 | |
| CA/AA | 2.45 | 0.10 | 2.30 | 0.13 | |
| rs13099634 | 1.0 | ||||
| CC | 2.55 | 0.10 | 2.14 | 0.11 | |
| CT | 2.74 | 0.10 | 2.22 | 0.17 | |
| TT | 3.04 | 0.22 | 2.05 | 0.29 | |
| rs1152001 | .118 | ||||
| TT | 2.68 | 0.08 | 2.30 | 0.11 | |
| TC/CC | 2.68 | 0.11 | 1.89 | 0.17 | |
| rs1152002 | .100 | ||||
| GG | 2.91 | 0.10 | 2.11 | 0.16 | |
| GA | 2.46 | 0.09 | 2.17 | 0.12 | |
| AA | 2.69 | 0.25 | 2.43 | 0.23 | |
| rs3856806 | .037 | ||||
| CC | 2.66 | 0.08 | 2.27 | 0.11 | |
| CT/TT | 2.75 | 0.14 | 1.82 | 0.21 | |
| rs4135300 | 1.0 | ||||
| CC | 2.71 | 0.08 | 2.20 | 0.11 | |
| CT/TT | 2.59 | 0.18 | 2.01 | 0.19 | |
| rs9833097 | 1.0 | ||||
| GG | 2.68 | 0.10 | 2.23 | 0.12 | |
| GA | 2.63 | 0.10 | 2.12 | 0.14 | |
| AA | 2.97 | 0.21 | 1.88 | 0.38 | |
Values shown are geometric means, adjusted for age, sex, and baseline body fat percentage.
ses computed by the δ Method (38).
P-value for the association between the rate of change in SI and SNP, under an additive or dominant genetic model as appropriate, adjusted for age, sex, and baseline body fat percentage. P-values are Bonferroni-corrected for the 18 tag SNPs tested.
Figure 1.
Association between rs17793951 and changes in SI, DI, and body fat percentage over followup. Black lines indicate genotype AA; gray lines indicate genotypes AG or GG. Values shown for SI and DI at baseline and at mean follow-up time (4.6 y) are geomtric means with SEs computed by delta method, adjusted for age, sex, and body fat percentage. Values shown for body fat percentage are arithmetic means and SEs, adjusted for age and sex. P-values are Bonferroni corrected for 18 tag SNPs.
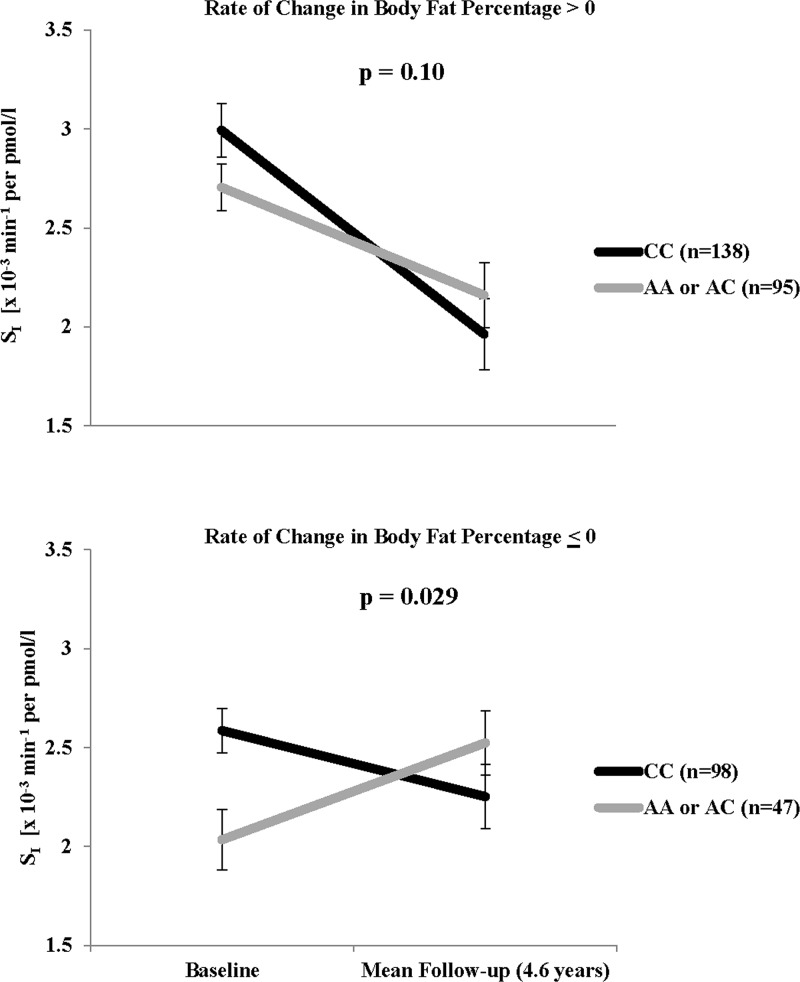
Among the six SNPs associated with rate of change in SI, only the association with rs1175541 varied significantly with rate of change in body fat percentage (corrected interaction P = .013). Adjusted mean values by rs1175541 genotypes, stratified by rate of change in body fat percentage > 0 (increased body fat over time) and ≤ 0 (no change or reduced body fat over time) are shown in Figure 2. Overall, 233 participants (62% of cohort) increased body fat during followup. Among these individuals, SI decreased over time regardless of genotype (corrected P = .10). However, among individuals whose body fat percentage remained the same or decreased over time, SI increased for those carrying the minor allele, but continued on a decreasing trend in those homozygous for the major allele (corrected P = .029).
Figure 2.
Change in SI over followup for rs1175541, stratified by rate of change in body fat percentage. Black lines indicate genotype CC; gray lines indicate genotypes AA or AC. Values shown at baseline and at mean follow-up time (4.6 y) are geometric means and SEs, adjusted for age and sex. P-values are Bonferroni corrected for six interaction tests.
Discussion
In this Mexican-American cohort enriched with individuals at risk for T2DM, we observed association between variation in PPARG and the rate of decline in SI over time, independent of cross-sectional or longitudinal changes in adiposity. We also found the effect of one SNP, rs1175541, on the rate of change in SI to vary by longitudinal changes in body fat percentage. To our knowledge, this is the first study to demonstrate differential effects of variation in PPARG on insulin sensitivity by changes in adiposity over time.
In a single-arm intervention study of 95 overweight and obese Japanese women assigned to 14 weeks of caloric restriction (∼1200 kcal/d), Matsuo et al (21) reported significant decreases in several anthropometric and metabolic traits such as body weight, body fat percentage, triglycerides, and fasting glucose. Matsuo et al also found that changes in adiposity over the course of the trial varied significantly by PPARG genotypes. Specifically, study participants carrying minor alleles for any one of six SNPs (rs2959272, rs1386835, rs709158, rs1175540, rs1175544, rs1797912), all of which are located in a large intron of PPARG, experienced significantly greater weight reduction than those homozygous for the major allele. The largest and most significant weight-loss association was observed with rs1175544, which was also associated with decreases in waist circumference and fat mass. In BetaGene, rs1175544 is in strong LD with rs1175541 (r2 = 0.9), whereas its MAF is only half of that observed in the Japanese study (19 vs 38%). Although rs1175541 was not significantly associated with changes in any anthropometric traits in our study, it was associated with the rate of change in SI. Furthermore, it significantly interacted with change in body fat over time, such that improvement in SI due to reduction in body fat percentage was only apparent among those carrying the minor allele. Taken together, these data suggest that rs1175541, and/or other intronic variants in LD with it, may have an important regulatory role in body composition and concomitant changes in insulin sensitivity over time.
Two variants previously shown to be associated with BMI and/or body composition in Caucasian samples, C-681G (rs10865710; tagged by rs11128598) and C1431T (rs3856806) (8, 11, 39), were not significantly associated with baseline or change in adiposity in the BetaGene longitudinal cohort. However, these variants were associated with change in SI over time. To date, studies of these variants have been conducted in other European, African American, Indian, and Asian populations (6, 9, 23, 35–39), although associations have not been replicated in nonwhite populations. Although the MAF for C1431T in BetaGene participants was similar to that typically observed among white individuals (12 vs 12–13%), the MAF for C-681G (or its proxy) was strikingly different (41 vs 25–26%). Further, these reports were based on cross-sectional analyses, making direct comparisons of SNP associations with longitudinal traits difficult. Interestingly, Wei et al (6) found that white individuals carrying the T allele of C1431T or the G allele of C-681G had a higher BMI than those homozygous for the C allele of either SNP. A few studies in nonwhite populations, whereas finding no significant association between these alleles and obesity, have observed their association with markers of insulin resistance (9), proinflammatory adipokines (39), or risk for impaired glucose tolerance and T2DM (37). In our study, the 1431T and -681G alleles (or the minor allele of the proxy SNP) were both associated with faster decline in SI, which suggests a consistent allelic effect on anthropometric and metabolic traits in different racial/ethnic groups. Interestingly, C1431T (rs3856806) has been reported as a cis eQTL associated with significant increases PPARG expression in adipose tissue of 166 female subjects from the Multiple Tissue Human Expression Resource (MuTHER) study (40). Adipose-specific cis eQTL effects for this SNP were also replicated at a nominal level in an additional 856 female subjects from MuTHER (41). Additional experimental studies suggest that C1431T resides in a region that is highly conserved, hypersensitive to DNAse, and a putative transcription factor binding site (42, 43). Taken together, the evidence suggests that this synonymous variant may also have an important PPARG regulatory function.
Similarly, Pro12Ala (rs1801282; tagged by rs13064760) and C-689T (rs7649970; tagged by rs17036328) were not associated with baseline levels of or rates of change in anthropometric or metabolic traits. In a population-based longitudinal study of nondiabetic French individuals, Jaziri et al (23) observed significant effects of Pro12Ala on 6-year change in fasting insulin and homeostasis model of assessment–insulin resistance, a surrogate measure of insulin resistance. Although associations between Pro12Ala and baseline or changes in anthropometric traits were not reported, the average BMI of these individuals was lower and in the normal range (∼24 kg/m2) (23), whereas that of our study participants was much higher (∼29 kg/m2). Conversely, in a study of nondiabetic African American and white individuals in the United States, Fornage et al (20) reported Pro12Ala associations with 15-year changes in anthropometric traits in both racial groups, but significant effects on change in homeostasis model of assessment were observed only among African Americans. Interestingly, this association was slightly attenuated and became nonsignificant after adjusting for baseline BMI (20), despite the fact that Pro12Ala was not significantly associated with BMI among African Americans in this study.
Few studies have examined the effects of variation in PPARG specifically in Mexican or Mexican-American populations (3, 24, 44, 45); all were cross sectional and focused exclusively on associations with Pro12Ala. One study reported significant associations between Pro12Ala and measures of adiposity (3), whereas the others failed to replicate this finding. We previously reported no main effect of Pro12Ala on cross-sectional anthropometric or metabolic traits in the full BetaGene cohort (24). Thus it is not surprising that we observed no such associations in the BetaGene II cohort. This report is the first to examine longitudinal changes in adiposity, insulin sensitivity, and β-cell function associated with variation in PPARG in Mexican Americans at risk for T2DM.
Strengths of our study also include our family-based design and ascertainment of study participants with parents and at least 3/4 grandparents of Mexican ancestry, both of which partially protect from population stratification in the study design, as well as our ability to statistically control for admixture with genome-wide data. Moreover, our longitudinal design and detailed phenotyping with dual-energy x-ray absorptiometry and IVGTTs for estimation of body fat percentage and direct assessment of insulin sensitivity and β-cell function, respectively, are advantageous for studying physiological changes over time that precede T2DM. However, our study is not without limitations. Despite the unique array of genotype and longitudinal trait data available in BetaGene participants, the sample size of the subset with follow-up phenotyping is relatively modest. Given the multiple testing penalty applied, it is possible that we were unable to detect variation in PPARG associated with very small changes in body fat or SI, particularly for low-frequency SNPs with small effects. However, we were well-powered to detect differences in the mean rate of change in these traits for the SNPs identified in this report. Finally, our study design did not afford us the opportunity to assess T2DM development among all study participants; further investigation of the direct contribution of these variants to T2DM risk in Mexican Americans is warranted.
In conclusion, we did not observe associations between any variants in PPARG and baseline levels of or changes in measures of adiposity. We did find that variation in PPARG, but not Pro12Ala, contributes to declining SI and concomitant deterioration in β-cell function, after accounting for adiposity. The beneficial metabolic effects of weight loss or reductions in body fat may also depend on variation in PPARG. These data may have important implications for the treatment of obesity and prevention of T2DM in Mexican Americans.
Acknowledgments
The authors thank the families who participated in the BetaGene Study and also acknowledge the efforts of our recruiting and technical staff.
This work was supported by National Institutes of Health Grants R01-DK-61628 and UL1RR031986.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AIR
- acute insulin response
- BMI
- body mass index
- CEU
- European ancestral population CHB. Asian ancestral population
- DI
- disposition index
- GDM
- gestational diabetes mellitus
- IVGTT
- iv glucose tolerance testl
- LD
- linkage disequilibrium
- MAF
- minor allele frequency
- MEX
- Mexican ancestral population
- MuTHER
- Multiple Tissue Human Expression Resource
- OGTT
- oral glucose tolerance test
- PPARG
- Peroxisome proliferator-activated receptor gamma
- SI
- insulin sensitivity
- SNP
- single nucleotide polymorphism
- T2DM
- type 2 diabetes mellitus YRI, African ancestral population.
References
- 1. Fajas L, Auboeuf D, Raspé E, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–18789. [DOI] [PubMed] [Google Scholar]
- 2. Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. [DOI] [PubMed] [Google Scholar]
- 3. Cole SA, Mitchell BD, Hsueh WC, et al. The Pro12Ala variant of peroxisome proliferator-activated receptor-gamma2 (PPAR-gamma2) is associated with measures of obesity in Mexican Americans. Int J Obes Relat Metab Disord. 2000;24:522–524. [DOI] [PubMed] [Google Scholar]
- 4. Robitaille J, Després JP, Pérusse L, Vohl MC. The PPAR-gamma P12A polymorphism modulates the relationship between dietary fat intake and components of the metabolic syndrome: Results from the Québec Family Study. Clin Genet. 2003;63:109–116. [DOI] [PubMed] [Google Scholar]
- 5. Kim KS, Choi SM, Shin SU, Yang HS, Yoon Y. Effects of peroxisome proliferator-activated receptor-gamma 2 Pro12Ala polymorphism on body fat distribution in female Korean subjects. Metabolism. 2004;53:1538–1543. [DOI] [PubMed] [Google Scholar]
- 6. Wei Q, Jacobs DR, Jr, Schreiner PJ, Siscovick DS, Steffes MW, Fornage M. Patterns of association between PPARgamma genetic variation and indices of adiposity and insulin action in African-Americans and whites: The CARDIA Study. J Mol Med (Berl). 2006;84:955–965. [DOI] [PubMed] [Google Scholar]
- 7. Dongxia L, Qi H, Lisong L, Jincheng G. Association of peroxisome proliferator-activated receptorgamma gene Pro12Ala and C161T polymorphisms with metabolic syndrome. Circ J. 2008;72:551–557. [DOI] [PubMed] [Google Scholar]
- 8. Bhatt SP, Misra A, Sharma M, Luthra K, Guleria R, Pandey RM, Vikram NK. Ala/Ala genotype of Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-γ2 gene is associated with obesity and insulin resistance in Asian Indians. Diabetes Technol Ther. 2012;14:828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prakash J, Srivastava N, Awasthi S, et al. Association of PPAR-γ gene polymorphisms with obesity and obesity-associated phenotypes in North Indian population. Am J Hum Biol. 2012;24:454–459. [DOI] [PubMed] [Google Scholar]
- 10. Douglas JA, Erdos MR, Watanabe RM, et al. The peroxisome proliferator-activated receptor-gamma2 Pro12A1a variant: Association with type 2 diabetes and trait differences. Diabetes. 2001;50:886–890. [DOI] [PubMed] [Google Scholar]
- 11. Mori Y, Kim-Motoyama H, Katakura T, et al. Effect of the Pro12Ala variant of the human peroxisome proliferator-activated receptor gamma 2 gene on adiposity, fat distribution, and insulin sensitivity in Japanese men. Biochem Biophys Res Commun. 1998;251:195–198. [DOI] [PubMed] [Google Scholar]
- 12. Clement K, Hercberg S, Passinge B, et al. The Pro115Gln and Pro12Ala PPAR gamma gene mutations in obesity and type 2 diabetes. Int J Obes Relat Metab Disord. 2000;24:391–393. [DOI] [PubMed] [Google Scholar]
- 13. Meirhaeghe A, Fajas L, Helbecque N, et al. Impact of the Peroxisome Proliferator Activated Receptor gamma2 Pro12Ala polymorphism on adiposity, lipids and non-insulin-dependent diabetes mellitus. Int J Obes Relat Metab Disord. 2000;24:195–199. [DOI] [PubMed] [Google Scholar]
- 14. Evans D, Wolf AM, Nellessen U, et al. Association between polymorphisms in candidate genes and morbid obesity. Int J Obes Relat Metab Disord. 2001;25(Suppl 1):S19–S21. [DOI] [PubMed] [Google Scholar]
- 15. Frederiksen L, Brodbaek K, Fenger M, et al. Comment: Studies of the Pro12Ala polymorphism of the PPAR-gamma gene in the Danish MONICA cohort: Homozygosity of the Ala allele confers a decreased risk of the insulin resistance syndrome. J Clin Endocrinol Metab. 2002;87:3989–3992. [DOI] [PubMed] [Google Scholar]
- 16. Nelson TL, Fingerlin TE, Moss L, Barmada MM, Ferrell RE, Norris JM. The PPARgamma Pro12Ala polymorphism is not associated with body mass index or waist circumference among Hispanics from Colorado. Ann Nutr Metab. 2007;51:252–257. [DOI] [PubMed] [Google Scholar]
- 17. Lindi V, Sivenius K, Niskanen L, Laakso M, Uusitupa MI. Effect of the Pro12Ala polymorphism of the PPAR-gamma2 gene on long-term weight change in Finnish non-diabetic subjects. Diabetologia. 2001;44:925–926. [DOI] [PubMed] [Google Scholar]
- 18. Nicklas BJ, van Rossum EF, Berman DM, Ryan AS, Dennis KE, Shuldiner AR. Genetic variation in the peroxisome proliferator-activated receptor-gamma2 gene (Pro12Ala) affects metabolic responses to weight loss and subsequent weight regain. Diabetes. 2001;50:2172–2176. [DOI] [PubMed] [Google Scholar]
- 19. Franks PW, Jablonski KA, Delahanty L, et al. The Pro12Ala variant at the peroxisome proliferator-activated receptor gamma gene and change in obesity-related traits in the Diabetes Prevention Program. Diabetologia. 2007;50:2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fornage M, Jacobs DR, Steffes MW, et al. Inverse effects of the PPAR(gamma)2 Pro12Ala polymorphism on measures of adiposity over 15 years in African Americans and whites. The CARDIA study. Metabolism. 2005;54:910–917. [DOI] [PubMed] [Google Scholar]
- 21. Matsuo T, Nakata Y, Katayama Y, et al. PPARG genotype accounts for part of individual variation in body weight reduction in response to calorie restriction. Obesity (Silver Spring). 2009;17:1924–1931. [DOI] [PubMed] [Google Scholar]
- 22. Delahanty LM, Pan Q, Jablonski KA, et al. Genetic predictors of weight loss and weight regain after intensive lifestyle modification, metformin treatment, or standard care in the Diabetes Prevention Program. Diabetes Care. 2012;35:363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaziri R, Lobbens S, Aubert R, et al. The PPARG Pro12Ala polymorphism is associated with a decreased risk of developing hyperglycemia over 6 years and combines with the effect of the APM1 G-11391A single nucleotide polymorphism: The Data From an Epidemiological Study on the Insulin Resistance Syndrome (DESIR) study. Diabetes. 2006;55:1157–1162. [DOI] [PubMed] [Google Scholar]
- 24. Black MH, Fingerlin TE, Allayee H, et al. Evidence of interaction between PPARG2 and HNF4A contributing to variation in insulin sensitivity in Mexican Americans. Diabetes. 2008;57:1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiang AH, Takayanagi M, Black MH, et al. Longitudinal changes in insulin sensitivity and beta cell function between women with and without a history of gestational diabetes mellitus. Diabetologia. 2013;56:2753–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lange K, Papp JC, Sinsheimer JS, Sripracha R, Zhou H, Sobel EM. Mendel: The Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29:1568–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 29. Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 30. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN. MINMOD Millennium: A computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther. 2003;5:1003–1015. [DOI] [PubMed] [Google Scholar]
- 33. Bergman RN. Lilly lecture 1989. Toward physiological understanding of glucose tolerance. Minimal-model approach. Diabetes. 1989;38:1512–1527. [DOI] [PubMed] [Google Scholar]
- 34. Rosner B. 1993. Fundamentals of biostatistics. In: Biometrical Journal. 5th ed Boston: Brooks/Cole, Cengage Learning; 150–150. [Google Scholar]
- 35. Doney A, Fischer B, Frew D, et al. Haplotype analysis of the PPARgamma Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genet. 2002;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meirhaeghe A, Fajas L, Gouilleux F, et al. A functional polymorphism in a STAT5B site of the human PPAR gamma 3 gene promoter affects height and lipid metabolism in a French population. Arterioscler Thromb Vasc Biol. 2003;23:289–294. [DOI] [PubMed] [Google Scholar]
- 37. Tai ES, Corella D, Deurenberg-Yap M, et al. Differential effects of the C1431T and Pro12Ala PPARgamma gene variants on plasma lipids and diabetes risk in an Asian population. J Lipid Res. 2004;45:674–685. [DOI] [PubMed] [Google Scholar]
- 38. Meirhaeghe A, Cottel D, Amouyel P, Dallongeville J. Association between peroxisome proliferator-activated receptor gamma haplotypes and the metabolic syndrome in French men and women. Diabetes. 2005;54:3043–3048. [DOI] [PubMed] [Google Scholar]
- 39. Haseeb A, Iliyas M, Chakrabarti S, et al. Single-nucleotide polymorphisms in peroxisome proliferator-activated receptor gamma and their association with plasma levels of resistin and the metabolic syndrome in a South Indian population. J Biosci. 2009;34:405–414. [DOI] [PubMed] [Google Scholar]
- 40. Nica AC, Parts L, Glass D, et al. The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLoS Genet. 2011;7:e1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grundberg E, Small KS, Hedman ÅK, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karolchik D, Barber GP, Casper J, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2013;41:D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duran-Gonzalez J, Ortiz I, Gonzales E, et al. Association study of candidate gene polymorphisms and obesity in a young Mexican-American population from South Texas. Arch Med Res. 2011;42:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martínez-Gómez LE, Cruz M, Martínez-Nava GA, et al. A replication study of the IRS1, CAPN10, TCF7L2, and PPARG gene polymorphisms associated with type 2 diabetes in two different populations of Mexico. Ann Hum Genet. 2011;75:612–620. [DOI] [PubMed] [Google Scholar]