Abstract
Context:
A common polymorphism in the gene encoding the activating deiodinase (Thr92Ala-D2) is known to be associated with quality of life in millions of patients with hypothyroidism and with several organ-specific conditions. This polymorphism results in a single amino acid change within the D2 molecule where its susceptibility to ubiquitination and proteasomal degradation is regulated.
Objective:
To define the molecular mechanisms underlying associated conditions in carriers of the Thr92Ala-D2 polymorphism.
Design, Setting, Patients:
Microarray analyses of 19 postmortem human cerebral cortex samples were performed to establish a foundation for molecular studies via a cell model of HEK-293 cells stably expressing Thr92 or Ala92 D2.
Results:
The cerebral cortex of Thr92Ala-D2 carriers exhibits a transcriptional fingerprint that includes sets of genes involved in CNS diseases, ubiquitin, mitochondrial dysfunction (chromosomal genes encoding mitochondrial proteins), inflammation, apoptosis, DNA repair, and growth factor signaling. Similar findings were made in Ala92-D2-expressing HEK-293 cells and in both cases there was no evidence that thyroid hormone signaling was affected ie, the expression level of T3-responsive genes was unchanged, but that several other genes were differentially regulated. The combined microarray analyses (brain/cells) led to the development of an 81-gene classifier that correctly predicts the genotype of homozygous brain samples. In contrast to Thr92-D2, Ala92-D2 exhibits longer half-life and was consistently found in the Golgi. A number of Golgi-related genes were down-regulated in Ala92-D2-expressing cells, but were normalized after 24-h-treatment with the antioxidant N-acetylecysteine.
Conclusions:
Ala92-D2 accumulates in the Golgi, where its presence and/or ensuing oxidative stress disrupts basic cellular functions and increases pre-apoptosis. These findings are reminiscent to disease mechanisms observed in other neurodegenerative disorders such as Huntington's disease, and could contribute to the unresolved neurocognitive symptoms of affected carriers.
Hypothyroidism is found in about 4.6% of the U.S. population age 12 and older (1). The current standard of care for these patients is treatment with daily tablets of the long-lived prothyroid hormone (TH), levothyroxine (L-T4). T4 is subsequently activated to T3 outside of the thyroid parenchyma via the deiodinases, ie, D1 and D2. Unfortunately therapy with L-T4 alone does not resolve symptoms in all hypothyroid patients, with approximately 12% of the patients remaining symptomatic despite normalization of serum TSH and TH levels (2, 3). Impaired cognition, fatigue, and difficulty losing weight are the main residual symptoms of these patients, for which we lack understanding and have no mechanistic explanation.
A prevalent Thr92Ala-D2 polymorphism [between 12% and 36% of the population are homozygotes (4)] has been identified that results in a single amino change at position 92 within an 18 amino acid loop that controls D2 ubiquitination for proteasomal destruction (5, 6). Hypothyroid individuals carrying this polymorphism were found to have a preference for a therapy that includes T3 vs monotherapy with L-T4 alone (7), suggesting defective Ala92-D2 catalysis. In addition, the Thr92AlaD2 polymorphism has been associated with conditions aside from symptomatic hypothyroidism such as mental retardation (8), low IQ (9), and bipolar disorder (10); this supports the hypothesis that Ala92-D2-expressing is disruptive aside from impaired T4 activation.
Here we used a multifaceted strategy to define the molecular foundation of the clinical syndromes associated with the Thr92AlaD2 polymorphism. There are unique modifications in the cellular transcriptome identified in human brains homozygous for the polymorphism that are independent of TH signaling. These transcriptional changes included upregulation of processes related to the mitochondria, Golgi apparatus/ER transport, oxidative stress and apoptosis, suggesting a molecular basis underlying cerebral symptomatology in affected individuals. A cellular model revealed that Ala92-D2 protein exhibits a longer half-life and, as opposed to Thr92-D2, can be found in the Golgi apparatus. Cells expressing Ala92-D2 also exhibited alteration in expression of Golgi markers, a finding that absolved with antioxidant treatment. Notably, in both the human brain and cell models there is molecular and physiological evidence of dysregulation in EGF receptor signaling, a pathway known to be altered in oxidative stress (11) and play an important role in cognitive development (12) and function (13–16).
Materials and Methods
Human brain samples
The University of Miami (UM) Brain Endowment Bank provided genomic DNA and brain tissue samples from postmortem human donors; protocols at UM were IRB-approved. Cause of death was limited to accident or sudden cardiac death without medical intervention or prolonged agonal state. Postmortem interval at specimen collection was < 24 h, brain pH (quality measure) was > 6.0. Genomic DNA from 95 brain samples was genotyped for the Thr92AlaD2 polymorphism by sequence analysis according to previously published methods (17). Brain samples from 19 patients without known thyroid or neurologic disease (six from homozygous Thr92-D2, seven heterozygotes, and six homozygotes for Ala92-D2) were matched by age (ANOVA P = .46), sex (male), race (Caucasian), and BMI (ANOVA P = .66) and chosen for further studies. Homogenous samples were dissected from frozen coronal blocks based on surface and cytoarchitectural landmarks from Brodmann's Area 38 (temporal cortex) by neuroanatomist and stored at −80°C.
Microarray studies of human brain
RNA was extracted (RNeasy Lipid Tissue Mini Kit, Qiagen) and cDNA generated (First Strand cDNA Synthesis Kit, Roche). RNA from all 19 human brain samples from each of the three genotypes (Ala92-D2 homozygotes, Hets, and Thr92-D2 homozygotes) was analyzed by microarray at the Joslin Diabetes Center Genomics Core Laboratory. Gene expression was evaluated using Genechip Human Gene 2.0 ST arrays (Affymetrix), which utilizes a whole-transcript design to assess > 30 000 coding genes. Gene expression data was preprocessed using Affymetrix Expression Console. Differential expression analysis was performed in the Affymetrix Transcriptome Analysis Console to identify individual genes demonstrating enrichment in three comparisons: Ala92-D2 homozygotes vs Thr92-D2 homozygotes (Supplemental Table 2), Ala92-D2 homozygotes vs Hets, and Hets vs Thr92-D2 homozygotes. Expression values (signal) of individual genes were log2 transformed. One-way ANOVA was used to calculate P values for each fold change (linear); multitesting correction was then performed using the Benjamini-Hochberg Step-Up FDR-controlling procedure for all the expressed genes. Genes were considered statistically significant with an ANOVA P value < .05. A custom heat map displaying expression values of the 25 with the highest and lowest fold change was generated (HeatMapViewer, GenePattern, Broad Institute).
Gene ontology analysis was used to determine differences in gene sets between phenotypes [Gene Set Enrichment Analysis (GSEA), Broad Institute]. Expression values for all genes from all 19 brain samples were used; no filter was applied to eliminate genes with low expression. GSEA included calculation of enrichment scores (ES), estimation of significance level of ES (nominal p-value), and adjustment for multiple hypothesis testing including the normalized enrichment score (NES) and false discovery rate (FDR). Gene sets with an FDR ≤ 25% were limited (3) and given the goal of hypothesis generation, a nominal P value of < 1% was chosen to indicate significance. Core enrichment of individual genes within these gene sets was defined as those genes contributing to the leading-edge subset. All individual genes demonstrating core enrichment within the enriched gene sets were further considered by investigators (investigator analysis); this consisted of searches in publically available databases (PubMed, UniProt). A heatmap of the top 50 ranking genes from each genotype was generated by GSEA. GSEA was also applied to an investigator-generated custom gene set of known T3-responsive genes (18, 19).
Pathway analysis was performed with Ingenuity Pathway Analysis (IPA). The signal intensity values from the entire microarray dataset (all genes, all 19 samples) were used to identify canonical pathways altered between the three genotypes. When viewing the EGF Signaling pathway alone (P < .001, ratio 50/56 genes), individual genes demonstrating alteration in our dataset were highlighted in an IPA-generated pathway diagram.
Class prediction was performed (WeightedVoting, GenePattern) via two approaches. First the algorithm created a 10-gene classifier that was able to correctly predicted the genotypes of 12/12 samples; 2 of these 10 genes were SNORD16 and TRAPPC4. Then, the weighted voting prediction algorithm was used with an investigator-derived list of 79 genes as a classifier, where 11/12 sample genotypes were correctly predicted. Lastly, the 79-gene classifier was supplemented with the addition of SNORD16 and TRAPPC4 and applied to the microarray data from the Thr92-D2 and Ala92-D2 homozygotes.
Generation of stable cell lines and cell culture
HEK-293 cells (American Type Culture Collection) were cultured in 150-mm dishes with DMEM (Life Technologies) supplemented with 10% fetal bovine serum (FBS). Unless noted otherwise, all experiments were performed with 10% FBS-containing media. In particular, HEK-293 cells were chosen for this experiment as they express low levels of the deiodinases (20) and TR (21) and because they have certain properties consistent with neuronal lineage (22, 23).
The 6×His-CysD2-YFP (D2) vector was created fusing enhanced yellow fluorescent protein (EYFP) in frame to the C terminus of 133Cys/266Cys (CysD2) mutant human D2. The D2-EYFP cassette was inserted in-frame between EcoRI-NotI of pcDNA™4/ HisMax C (Invitrogen). The Thr92Ala (T92A) mutant of 133Cys/266Cys was generated by overlap extension PCR. HEK-293 cell lines stably expressing wild type Thr92-D2HY or polymorphic Ala92-D2HY were established by transfecting 2.5 μg of His-D2-YFP vector using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's instructions. D2HY-expressing clones of HEK-293 cells were selected 48 h after transfection by antibiotic resistance (Zeocin, 300 μg/mL) for 2 weeks. The control yellow fluorescent protein (YFP)-expressing vector has been described elsewhere (24); these D2HY-expressing cells exhibit similar properties compared to cells with native D2 (24). Some cells were grown to confluence and then treated with 1 mM N-acetylecysteine (NAC, Sigma) or 1 M trimethylamine-N-oxide (TMAO, Sigma) for 24 h. In addition, some cells were treated with brefeldin A (BFA, Tocris), dissolved in abs ethanol for 30 minutes at 0.5 ug/mL and then processed for electron microscopy (EM).
Flow cytometry
BD FACSCalibur flow cytometer was used for these studies. For apoptosis, Annexin V-PE apoptosis detection kit (eBioscience) was used for annexin V cell surface staining per manufacturer's protocol. For cell cycle analysis, cells were collected, washed, and resuspended. −20°C absolute ethanol was added in dropwise manner to the cell while vortexing. HEK-293 cells were fixed for 1 h at 4°C. After washing twice, 40 μg/mL propidium iodide and 1 μg/mL RNase were added and incubated 3 h at 4°C and subsequently analyzed by FACS. All of the FACS data were obtained after correct compensation setting using single labeled control. For cell size analysis, forward scatter was used as a measure of relative size.
Coculture of Thr92-D2HY and Ala92-D2HY-expressing cells with T3-responsive HeLa cells
As a positive control, HeLa cells were grown to confluence in solutions of 0 nM T3 or 100 nM T3 (19) and harvested for RT-qPCR of known T3-responsive genes, BCL3 and SPOT14; both genes were significantly increased in the 100 nM confirming T3-responsiveness in our system. Then, Thr92-D2HY and Ala92-D2HY HEK cells were grown to confluence in the upper chamber of a Transwell permeable support (Corning) above, but not in physical contact with, highly T3-responsive HeLa cells (American Type Culture Collection) in 10% FBS. HeLa cells were harvested, RNA extracted and processed for RT-qPCR of T3-responsive genes.
Immunofluorescence
Cells were plated on poly-D-lysine-coated chamber slides and fixation, imaging and co-localization were performed as previously described (25). D2 was imaged with 1:500 αYFP (Rockland immunologicals) and 1:1000 α-nuclear lamin (Cell Signaling). For the Golgi immunofluorescence, cells were grown on culture slide until confluent. After fixation in 4% paraformaldehyde (Electron Microscopy Science) for 30 minutes, cells were permeabilized by 0.5% Triton X-100 and subsequently blocked by Fish Skin Gelatin (Biotum). GM130 antibody (CellSignaling technology) and antiGFP antibody (Rockland immunologicals) were added at 1 ug/mL concentration at 4°C overnight. Secondary antibodies (Life Technologies) were incubated for 1 h and made into a slide using SloFade Gold mounting medium (Life Technologies). Images were acquired using a Nikon eclipse Ti microscope with C1 confocal system. Images were subsequently analyzed by NISelement AR or ImageJ software.
For immunofluorescence after CHX treatment, each chamber was fixed as previously described (26) and then incubated in a mixture of mouse Na+/K+ATPase antiserum (M7-PB-E9) at 1:250 dilution and rabbit GFP antiserum at 1:10000 dilution in PBS containing 2% normal horse serum and 0.2% sodium azide (antiserum diluent) for 2 days at 4°C. After rinses in PBS, the cells were incubated in biotinylated donkey antimouse IgG for 2 h (1:500; Jackson Immunoresearch Lab) followed by treatment in the avidin-biotin-peroxidase complex (ABC Elite; 1:1000; Vector Laboratories) in 0.05 M Tris buffer for 1 h at room temperature. The signal of Na+/K+ATPase was amplified with biotinylated tyramide for 10 minutes using the TSA amplification kit (Perkin Elmer Life and Analytical Sciences) according to the manufacturer's instruction. The cells were incubated in Streptavidin Cy5 (1:250; Jackson Immunoresearch Lab) and Alexa 555 conjugated antirabbit IgG (1:500, Invitrogen) for 2 h at room temperature. Sections were mounted on glass slides and coverslipped with Vechtashield Mounting medium (Vector). Imaging was performed on a Zeiss LSM 780 Confocal Microscope.
Ultrastructural studies
2 × 105 Thr92-D2HY or Ala92-D2HY stably expressing HEK-293 cells were plated on 2-well Permanox plastic chamber slides (Lab-Tek). The next day, cells were treated with 100 uM CHX or DMSO as a vehicle for 2 h and fixed in the chamber slides with the mixture of 3% PFA and 1% glutaraldehyde in 0.1 M phosphate buffer (PB) pH7.4 (PB) at 37°C for 1 h. After three washes for 2 minutes with 0.01 M PBS pH7.4, the cells were cryoprotected in 30% sucrose in PBS for 30 minutes at room temperature and then, quickly frozen over liquid nitrogen. The cells were washed again 3× with 0.01 M PBS and then treated with 2% normal horse serum (NHS; in PBS) for 20 minutes. Pretreated cells were covered with rabbit anti-GFP serum (1:10 000) diluted in 2% NHS in PBS for 2 days at 4°C. After rinsing in PBS and in 0.1% cold water fish gelatin +1% bovine serum albumin (BSA) in PBS, the cells were incubated in donkey antirabbit IgG conjugated with 0.8 nm colloidal gold (Electron Microscopy Sciences) diluted at 1:100 in PBS containing 0.1% cold water fish gelatin and 1% BSA overnight at 4°C. After rinsing in PBS and fixed with 1.25% glutaraldehyde in 0.1 M PB at room temperature for 10 minutes. After rinsing in PBS and in Aurion buffer for 20 minutes, gold particles were silver intensified using the Aurion R-Gent SE-LM Kit (Amersham-Pharmacia Biotech UK, Buckinghamshire, UK). The cells were washed 3× for 2 minutes in 0.2 M sodium citrate, pH7.5 followed by washes in 0.1 M PB. Cells were treated with 1% osmium tetroxide in 0.1 M PB for 20 minutes at 4°C, followed by treatment with 2% uranyl acetate in 70% ethanol for 10 minutes. After dehydration in an ascending series of ethanol and acetonitrile, the cells were embedded in Durcupan ACM epoxy resin (Fluka) in a gelatin capsule, and polymerized at 56°C for 2 days. Serial 60–70 nm thick utlrasections were cut with Leica ultracut UCT ultramicrotome (Leica Microsystems). The ultrathin sections were mounted onto Formvar-coated single slot grids, contrasted with 2% lead citrate and examined with a Jeol-100 C transmission electron microscope (EM).
Western blot quantitation
Western blot was performed as 30 μg of total protein was resolved on a 4–12% SDS-PAGE gel. The samples were transferred to PVDF transfer membrane (Immobolin-FL, Millipore), incubated with various antibodies overnight at 4°C and subsequently quantitated by using the LiCOR Odyssey instrument with Odyssey Image Studio software simultaneously (D2 vs actin or tubulin) using different infrared channels.
Isolation of subcellular membrane fractions
Cells were collected, resuspended in PBS solution with Complete-mini EDTA-free protease inhibitor (Roche) at 4°C. Cells were homogenized by ten passages through a ball homogenizer (Isobiotec) at 10 μm clearance. Homogenates were centrifuged for 10 minutes at 800 × g. The resulting nuclear (N') pellet was washed twice with ice-cold PBS by centrifugation, while the supernatant fraction, containing cytosolic membranes and the cytosol, was designated as lysate and used for D2 activity assay. For western blot, nuclear and lysate samples were prepared using nonionic detergent Triton X-100. 293 cells were treated with 0.5% Triton X-100 at 4°C and centrifuged at 800 × g for 10 minutes to isolate nuclear (N) fraction. The supernatant after centrifugation, which contained Triton X-100 soluble membranes and cytosol, was designated as lysate.
D2 activity assay
D2 activity was assayed in cell lysates and nuclear fractions using 150 μg cell sonicated protein incubated in PE-EDTA pH7.35 buffer at 37°C in the presence of 1 uM 125I-T4 and 20 uM DTT for 2 h (19). The assay was terminated by the addition of TCA and horse serum and 125I released quantified in a gamma counter. Background activity was measured for each sample in the presence of 100 uM 125I-T4. Results are expressed as fmols 125I/min/mg protein.
Microarray studies of cultured cells
RNA was extracted from confluent cells from each group (RNeasy kit, Qiagen). Microarray data from samples from three plates each of the three cell types (YFP, Thr92-D2HY, and Ala92-D2HY) were collected and analyzed as above. In addition, hierarchical clustering analysis was performed (HierarchicalClustering, GenePattern) using the pairwise average-linkage clustering method and Pearson correlation column distance measure to generate a dendogram. Creation and application of the class predictor (classifier) was performed (KNNXValidation, GenePattern) where the k-nearest-neighbors (KNN) class prediction algorithm was used to run class prediction iteratively against the dataset from all three cell genotypes. The PredictionResultsViewer and FeatureSummaryViewer modules were then used to obtain values for the absolute error rate (incorrect cases/total cases) and ROC error rate [(receiver operating characteristic (ROC); fraction of true positives vs the fraction of false positives] and to view the genes comprising the classifier. To compare overlap between gene expression from the brain and cell microarray GSEAs, individual genes demonstrating core enrichment within the enriched gene sets were entered into VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html), where Venn diagrams were used to identify common genes.
RT-qPCR
RT-qPCR (StepOnePlus Real-Time PCR Detection System, Applied Biosystems) using Taqman reagents (Applied Biosystems) was performed on human brain samples with the following conditions: 2 minutes at 50°C followed by 10 minutes at 95°C, 15 s at 95°C, 1 minute at 60°C × 40. Standard curves consisted of five points of serially diluted cDNA from all samples. Cyclophilin A (CycloA) was used as an internal control gene and there was no difference in CycloA expression between the three genotypes. The coefficient of correlation (r2) was > 0.98 for all curves; amplication efficiency ranged from 90% to 110%. Results expressed as the ratio of target mRNA to CycloA mRNA. For HEK-293 cell PCR,18s was used for housekeeping.
In HeLa cells, BCL3 and SPOT14 were measured by RT-qPCR using SYBR green Fastmix (Quanta Bioscience) with the following conditions: 20 s at 95°C, 3 s at 95°C, and 30 s at 60°C × 40, and 15 s at 95°C followed by 1 minute at 60°C and 15 s at 95°C. Standard curve, r2 and amplification efficiency were as above.
Statistics
All data were analyzed using PRISM software (GraphPad). Unless otherwise indicated, data represent mean ± SEM, comparisons between two groups were analyzed with a two-tailed Student's t-test and comparisons between more than two groups were carried out by ordinary one-way ANOVA. A P value < .05 was considered significant.
Analysis of the human brain RT-qPCR data was done using Agilent Genespring GX version 12.6. The probeset data was log2 transformed and grouped according to Thr92-D2 homozygote, Het, and Ala92-D2 homozygous status. Each individual gene was compared using fold change between the three groups pairwise, selecting those genes that had an absolute fold change ≥ 1.5 in at least one of the three pairwise group comparisons. Each gene other than the mutant DIO2 was also correlated with DIO2 using Pearson's correlation. For clustering analysis, the data was further normalized to means of normal samples. This normalized gene expression data was clustered genewise using an unsupervised hierarchical clustering algorithm, with Euclidean distance metric and Ward's linkage rule.
Results
Analysis of the human brain transcriptome in patients carrying the Thr92-D2 polymorphism
The effects of the D2 polymorphism in humans were analyzed in the cerebral cortex of recently deceased adult accident/sudden death victims without known neurological or thyroid disease. Genotyping on brain samples from 95 donors revealed 24 homozygous for the Thr92AlaD2 polymorphism, 52 heterozygous and 25 normal patients; these frequencies are in the Hardy-Weinberg equilibrium. Brain samples from 19 male Caucasian donors (six samples from homozygote Thr92-D2, seven heterozygotes and six homozygotes for Ala92-D2) were matched by age and BMI and chosen for further studies (Supplemental Table 1). Homogenous samples from Brodmann's Area 38 of the temporal cortex were evaluated for gene expression. Given the diverse array of phenotypes exhibited by carriers of the polymorphism and the findings of normal catalytic activity of the D2 enzyme (27, 28), we hypothesized that this alteration could result in noncatalytic cellular consequences. Thus an unbiased whole-transcript microarray approach was used to assess the potential impact of the polymorphism and generate hypotheses regarding the mechanism of dysfunction in Ala92-D2 expression.
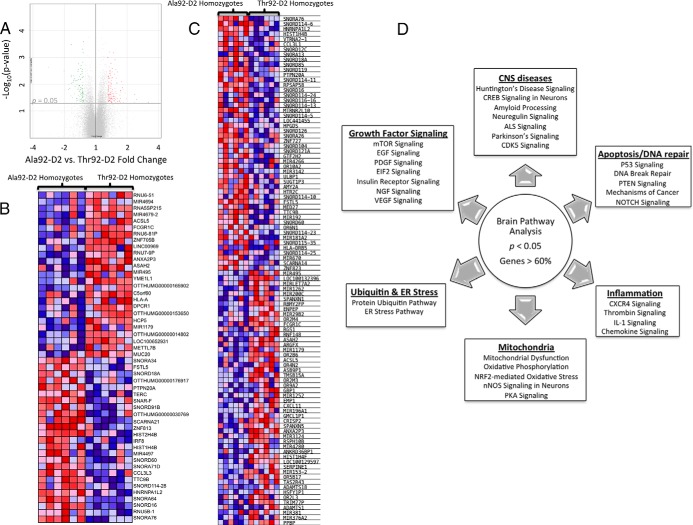
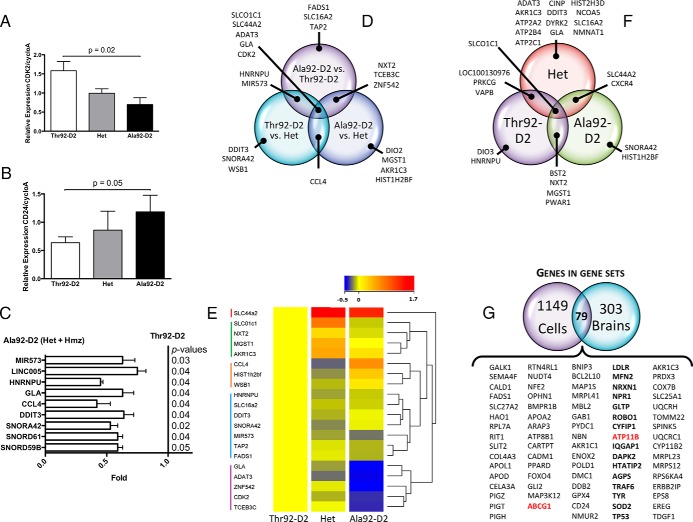
First, differentially expressed individual genes were identified by comparing the data from three genotypes: Ala92-D2 homozygotes vs Thr92-D2 homozygotes, Ala92-D2 homozygotes vs heterozygotes (Het), and Hets vs Thr92-D2 homozygotes. Differences in gene expression between the Ala92-D2 homozygote vs Thr92-D2 homozygote comparison were considered at a P value < .05 (Figure 1A). As a result, 839 named genes were found to be differentially expressed, where 110 of these were altered at a fold change > 1.3 or < −1.3 (Supplemental Table 2). A heat map incorporating the 25 genes with the highest or lowest expression values by fold change between these two genotypes was built (Figure 1B); it is notable that despite statistical significance between the two genotypes, the expression of some genes is heterogeneous reflecting their multifactorial regulation.
Figure 1.
Microarray analysis of human brain from patients homozygous and heterozygous for the Thr92AlaD2 polymorphism. A, Volcano plot of expression data in the comparison of Ala92–D2 (n = 6) and Thr92–D2 (n = 6) homozygotes; highlighted genes are significant at a P value < .05 and a fold change > 1.3 (red) or < −1.3 (green). Generated in Affymetrix Transcriptome Analysis Console; (B) heat map of the 25 most enriched and 25 most down-regulated genes in human brain samples from Ala92–D2 and Thr92–D2 homozygotes by fold change from differential expression analysis; expression values are represented as colors (red: increased expression; blue decreased expression) with the degree of color saturation indicating degree of expression; distinct transcriptional trends are observed; (C) Heat map of the top 50 features exhibited by polymorphic homozygotes vs normal patients as calculated in the gene set enrichment analysis software; (D) Canonical pathways identified in analysis of all brain samples (n = 19) where individual pathways are grouped by overlap in function, each of these pathways was altered at P < .05 and with a ratio of genes affected within the pathway > 60%.
In order to evaluate the physiologic context of the transcriptional footprint left by Ala92-D2-expression, the microarray data were then analyzed by GSEA (29, 30). When performed in the Ala92-D2 vs Thr92-D2 samples, GSEA revealed enrichment of 18 gene sets at a nominal P value < .05 (3 of which were additionally enriched at an FDR < 0.25) and downregulation of 10 gene sets at a nominal P value < .05 (Supplemental Table 3). Within these gene sets a total of 382 genes demonstrated core enrichment; a heat map of the 50 top genes for each phenotype was generated (Figure 1C). Investigator analysis of transcripts identified by GSEA showed that genes enriched in Ala92-D2 homozygotes were primarily related to apoptosis, oxidation-reduction, mitochondrial function, carbohydrate metabolism, ribosome components, inflammation, lipid/fatty acid metabolism, cytoskeleton structure, Golgi/ER transport, DNA repair, and transcription regulation. Genes downregulated in Ala92-D2 homozygotes were related to cell growth, cell cycle control, phospholipid maintenance, EGFR signaling, phosphatidylinositol, and lipoprotein metabolism.
To test whether carriers of the Ala92-D2 genotype exhibited changes in TH signaling we examined microarray data for the expression level of a set of typical T3-responsive genes (Supplemental Table 4) (18, 19) and found no significant differences by GSEA (Supplemental Figure 1A). Next, expression data were processed through pathway analysis software to identify cellular pathways that altered at the P < .05 level when the three genotypes are assessed. Many of the top pathways exhibiting significant changes in at least 60% of their genes shared overlapping functional significance (Figure 1D); Huntington's Disease signaling was the top canonical pathway identified (ratio of genes affected 180/225, P value < .0001).
A cell model to assess the impact of Thr92-D2 expression
Findings from the human brain microarray supported an association between gene expression pattern and genotype and thus served as grounds for further investigation using a cell model. HEK-293 cells were chosen as suitable candidates for the model as they exhibit many features of neuronal lineage (22, 23) and are known for lacking in deiodinase expression (20); these cells also lack in TR, which eliminates cell changes due to altered TH signaling (21). Thus, we used a previously characterized HEK-293 cell model that stably expresses a His/YFP-doubled tagged Sec133Cys-Thr92D2 (Thr92-D2HY) (24). In this setting, Thr92-D2HY exhibited similar cellular properties as endogenously expressed D2 and the host cells were not adversely affected when compared to cells stably expressing YFP (24). Here we observed that stable expression of Ala92-D2HY protein also did not result in gross cellular phenotypic alterations (Figure 2, A and B). Ala92-D2HY sorted predominantly to the ER and no differences were observed in cell size as assessed by flow cytometry (Supplemental Figure 2, A and B), cell cycle (Supplemental Figure 2C) or cell duplication time (Supplemental Figure 2D) when compared with cells stably expressing Thr92-D2HY.
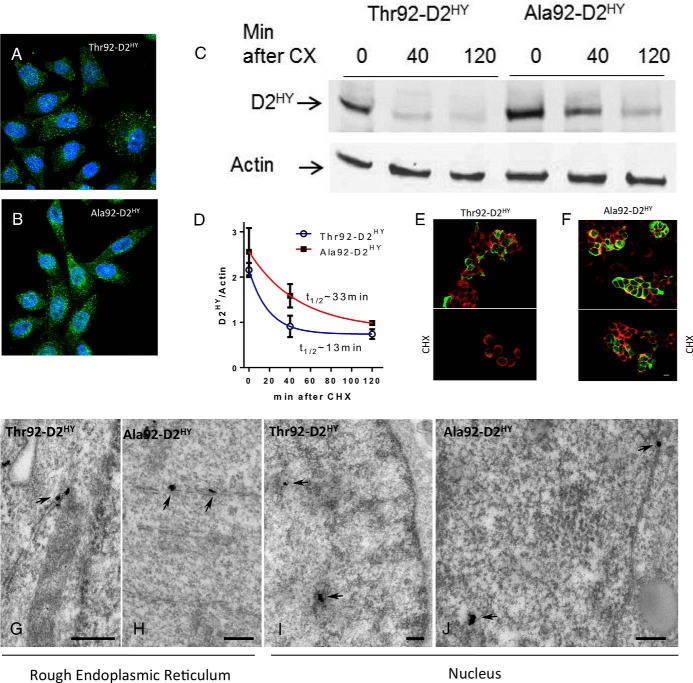
Figure 2.
Thr92–D2HY and Ala92–D2HY are both found in the nucleus but Ala92–D2HY has a longer half-life. A, B, Immunofluorescence of HEK-293 cells stably expressing D2HY proteins; D2 was stained with αYFP (green) and is found throughout the ER; nuclei were stained with DAPI (blue); (C) the indicated cells were incubated for 40–120 minutes with 100 uM CHX and subsequently harvested and processed for western analysis using αYFP and αActin; (D) quantification of the bands shown in C; data were analyzed by nonlinear regression assuming that D2 decays exponentially after addition of CHX; all data are the mean ± SEM of three entries per data-point; (E–F) immunofluorescent detection of plasma membrane by the Na+/K+-ATPase (red) and αYFP of D2HY fusion proteins (green) in the same cells as A-B before (above) and after (below) treatment with CHX for 120 minutes; (G) EM of HEK-293 cells stably expressing D2HY proteins as in A-B; silver grains denoting D2HY can be visualized in the rough endoplasmic reticulum (arrows) in both Thr92–D2HY and (H) Ala92–D2HY-expressing cells; (I) the nucleus also contains Thr92–D2HY and (J) Ala92–D2HY proteins; scale bars = 0.25 um.
Both Thr92-D2HY and Ala92-D2HY are found in the nucleus and ER but only Ala92-D2HY has a longer half-life and can be found in the Golgi apparatus
The Thr for Ala substitution in the D2 molecule is in the instability loop (5, 6), possibly affecting D2 turnover rate and other functions of Ala92-D2HY-expressing cells. Thus, we first looked at the rate of disappearance of the Ala92-D2HY protein following exposure to its natural substrate T4 that accelerates D2 ubiquitination and proteasomal degradation (31). Notably, whereas there was progressive loss of Thr92-D2HY after T4 was added to the medium, the abundance of the Ala92-D2HY protein remained largely unaffected by T4, suggesting impaired ubiquitination/degradation (Supplemental Figure 3, A and B). Indeed, we used cycloheximide (CHX) to arrest protein synthesis and found that Ala92-D2HY had a longer half-life (∼33 vs ∼13 minutes) when compared to Thr92-D2HY protein (Figure 2, C and D). This was confirmed with immunofluorescent localization where more Ala92-D2HY protein is seen after CHX treatment (Figure 2, E and F).
A more detailed analysis of the immunofluorescence microscopy images shows that D2HY protein can also be found closely associated with the nucleus (Figure 2, A and B). In fact, when stained for nuclear lamin (Supplemental Figure 3, E and F), colocalization of D2HY proteins was demonstrated, but no differences were observed between Thr92-D2 and Ala92-D2 proteins (Pearson's coefficient: 0.48 ± 0.02 vs 0.49 ± 0.03; n = 10). The presence of D2 in the ER and nucleus was also detected using EM, but no differences were observed between Thr92-D2 and Ala92-D2 proteins in these cellular compartments (Figure 2, G and J). Next, we isolated the nuclear fraction and detected Thr92-D2 and Ala92-D2 proteins by western analysis, both types of D2 disappearing with CHX treatment (Supplemental Figure 3C). Notably, nuclear D2 is catalytically active where its activity is not influenced by the polymorphism (Supplemental Figure 3D).
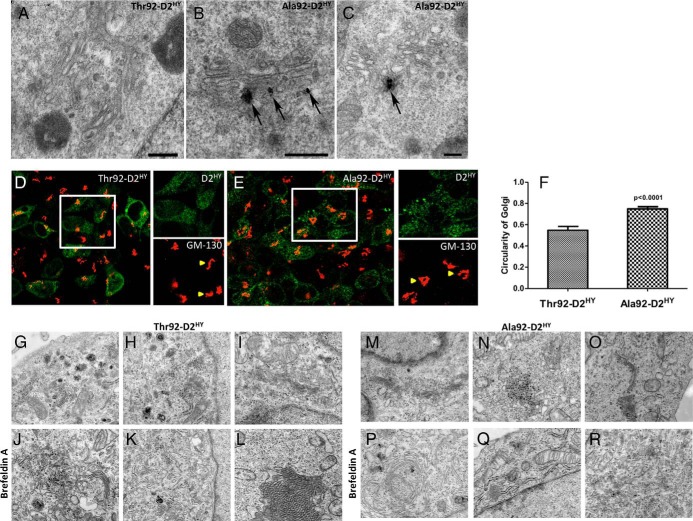
A remarkable new finding of the present studies is the observation with EM that only Ala92-D2HY could be identified in the Golgi apparatus (Figure 3, A-C). Even when cells are depleted of D2 by treatment with CHX, D2 was still consistently observed in the membranes of the Golgi apparatus (Supplemental Figure 4). Structural differences in Golgi as assessed by immunofluorescence were apparent under these conditions with Golgi in Ala92-D2HY-expressing cells exhibiting a circular configuration as opposed to its normal ribbon morphology (Figure 3, D-F). These findings were also documented by EM but only when the Golgi apparatus in Ala92-D2HY-expressing cells was further stressed with BFA treatment (Figure 3, G-I, M-O).
Figure 3.
Ala92–D2HY, but not Thr92–D2HY, can be found in the Golgi. A, Electron microscopy of Thr92–D2HY expressing cells where a typical Golgi apparatus is devoid of D2 protein; (B, C) same as A except that Ala92–D2HY expressing cells were studied; in this case, silver grains denoting the presence of Ala92–D2HY protein are observed associate to the Golgi apparatus (arrow); scale bars = 0.25 um; (D) Immunofluorescence staining of Thr92–D2HY and (E) Ala92–D2HY (green) expressing cells with cis-Golgi marker GM-130 (red). White box is enlarged in green (D2) and red (GM-130) channels. Yellow arrows show Thr92–D2HY and Ala92–D2HY specific cis-Golgi staining features, where Ala92–D2HY Golgi demonstrates a circular morphology compared to the ribbon configuration in the Thr92–D2HY-expressing cells; (F) The circularity index of the cis-Golgi complex in individual D2HY-expressing cells was measured in ImageJ where the cis-Golgi structure in Ala92–D2HY cells had higher circularity values than from Thr92–D2HY cells; (G-I, M-O) Untreated cells display the typical appearance of Golgi-complex independently from the type of D2 expressed in the cells. The cisternae of the Golgi complex were organized in parallel, slightly curved and surrounded by small Golgi vesicles; (J-L) In cells expressing the Thr92–D2HY, BFA treatment (0.5 μg/mL) resulted in a disorganization of the Golgi apparatus with scattered, dilated and short cisternae. However, some organized Golgi can be observed; (P-R) In BFA-treated Ala92D2HY-expressing cells, circular Golgi complexes were present that were otherwise unidentified in Thr92–D2HY expressing cells.
Defining a transcriptional fingerprint of Ala92-D2HY expression in cultured cells
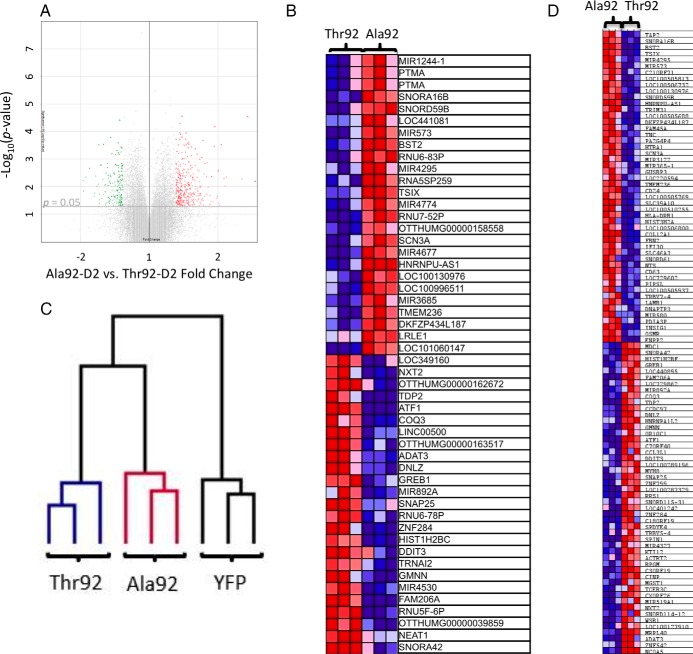
Given that fundamental biological differences exist between Thr92-D2HY and Ala92-D2HY that could underlie the transcriptional patterns observed in affected human brains, we next turned to an unbiased microarray approach to assess the impact of Ala92-D2HY expression in our cell model. Differential expression analysis was performed to identify individual genes altered in the three cell lines, Ala92- D2HY vs Thr92- D2HY, Ala92- D2HY vs YFP, Thr92 D2HY vs YFP. A volcano plot for Ala92- D2HY- vs. Thr92- D2HY-expressing cells allowed for visualization of the differences in gene expression at a P value < .05 (Figure 4A). This resulted in 3155 named genes differentially expressed, where 263 of these were altered at a fold change > 1.3 or < −1.3 (Supplemental Table 5). The 50 most affected genes by the Ala92-D2HY expression were plotted in a heat map (Figure 4B).
Figure 4.
Microarray analysis of HEK-293 cells stably expressing Ala92–D2HY or Thr92–D2HY proteins. A, Volcano plot of expression data in the comparison of Ala92–D2HY (n = 3) with Thr92–D2HY -expressing cells (n = 3); highlighted genes are significant at a P value < .05 and a fold change > 1.3 (red) or < −1.3 (green); (B) cultured cells differing only in their stable expression of Ala92–D2HY vs Thr92–D2 HY have unique transcriptional features where heat mapping of the 25 most enriched and 25 most downregulated genes as identified by fold change calculated in differential expression analysis are shown; (C) dendogram created via hierarchical clustering analysis reveals that mRNA expression patterns of Thr92–D2, Ala92–D2, and YFP-expressing cells cluster with samples of the same phenotype; Ala92–D2 cells exhibit a transcriptional expression pattern more similar to Thr92–D2-expressing cells than to YFP controls; (D) heat map of the top 50 genes up- and down-regulated genes as ranked by gene set enrichment analysis software.
We then performed a hierarchical clustering analysis (32). The resulting dendogram utilized the expression profiles of 6643 genes within these phenotypic contexts and indicated that the gene expression pattern appropriately clustered the three samples in groups with their respective phenotypes and that Ala92-D2 and Thr92-D2 clusters exhibited a more similar gene expression pattern than the YFP controls (Figure 4C). Also, we used class prediction analysis of the microarray dataset to create and evaluate a genetic classifier using the KNN class prediction method (Supplemental Table 6). Utilization of this classifier on the cell microarray data correctly classifies 9/9 samples (0 absolute error, 0 ROC error, confidence of prediction of each sample 0.667).
In order to evaluate the physiologic context of the transcriptional footprint left by Ala92-D2HY-expression, the microarray data were analyzed by GSEA (29, 30), which revealed 97 gene sets enriched at a nominal P value of < 1% in Ala92-D2HY vs Thr92-D2HY-expressing cells (Supplemental Table 7). Subsequent investigator analysis indicated that a predominance of the genes contained in these sets played a role in NF-κB signaling, inflammation, lysosome, cell cycle control, DNA repair, and cytoskeleton maintenance. Twenty-nine of these gene sets contain genes involved in cell membrane transport or function (Supplemental Table 7). There were 63 gene sets downregulated in Ala92-D2HY-expressing cells that were involved in DNA metabolism, cell cycle maintenance, transcription regulation, apoptosis, EGFR signaling, and mitochondria (Supplemental Table 6). A GSEA-generated heat map of the top 50 genes (up and down) by their ranking criteria was obtained (Figure 4D).
Next we tested whether differences between Ala92-D2 vs Thr92-D2 could interfere with TH signaling by measuring the expression level of a custom set of typical T3-responsive genes (Supplemental Table 4) (18, 19) and, as in the brains, found no significant differences by GSEA (Supplemental Figure 1B). Differences in TH activation between Ala92-D2 vs Thr92-D2 were also evaluated using a previously characterized co-culture system (33), where T3-responsive cells were co-cultured with Thr92-D2HY- or Ala92-D2HY-expressing cells. Here HeLa cells were used as targets as they express two highly sensitive T3-responsive genes, ie, BCL3 or SPOT14 (34, 35), but no differences in the expression of either gene were observed in co-cultures with either Thr92-D2HY or Ala92-D2HY-expressing cells (Supplemental Figure 5).
Are the genes identified in the cells also altered in the human brain samples obtained from Ala92-D2 homozygous donors?
To that end we generated a list of 40 genes (40-list) identified in the cell microarray GSEA based on their representation within prominent gene ontology classes and analyzed their expression level in all brain samples by RT-qPCR. CDK2 expression inversely correlated with the Ala92-D2 allele dose (Figure 5A), whereas the opposite was observed for CD24 (Figure 5B). Next, we combined expression data of Hets and Ala92-D2 homozygotes to test whether the expression of these 40 genes was indistinctly affected by at least one copy of the AlaD2 allele. This strategy confirmed that 9 other genes identified in the cells, totaling 11 genes (11-list), were affected by the AlaD2 allele in the human brain (Figure 5C). The 40-list was further analyzed using the fold change ranking approach (36) that yielded 21 genes (7 genes in common with the 11-list) with expression level that varied > 1.5-fold between any two genotypes (Figure 5D) and clustered into 5 groups (Figure 5E).
Figure 5.
Identification of genes differentially affected by Ala92–D2 expression in both cells and human brains. Forty genes identified in the cell microarray (i) for their significant alteration in expression and (ii) representation of unique physiologic themes were chosen for additional studies in human brain. A, Relative expression of CDK2 by RT-qPCR in human brain samples from Ala92–D2 and Thr92–D2 homozygotes and heterozygous; (one-way ANOVA P = .01, two-tailed t-test Ala92–D2 vs Thr92–D2, P = .02); (B) same as A except that CD24 was studied; (one-tailed t test Ala92–D2 vs Thr92–D2, P = .05); (C) when expression data from heterozygous and homozygous samples were combined, significant differences in comparison to Thr92–D2 brain samples was observed for 9 genes; (D) 21 genes exhibited an expression level that varied > 1.5-fold between any two genotypes; these 21 genes are depicted in a Venn diagram to show genes identified in one or more pairwise genotype comparisons; (E) hierarchical clustering of these genes (aside from DIO2) after normalization to the expression level of Thr92–D2 homozygotes using Euclidean distance metric yields a dendogram showing 5 clusters; expression levels are indicated by heatmap (low: blue; high: red) created in Agilent Genespring GX 12.6; (F) D2 expression in human brain correlated significantly (Pearson correlation at P < .05) with the expression of 27 genes; Venn diagram showing overlap between genes that correlated with D2 in Thr92–D2 homozygotes, Hets and Ala92–D2 homozygotes samples; (G) 79 individual genes common to both the cell and human brain microarray gene set enrichment analyses were identified using Venn diagrams, constituting the Ala92–D2 fingerprint; these include two Golgi-specific genes (red) and 15 Golgi-related genes (bold).
Next, we asked whether the expression of any of the genes in the 40-list correlated with the level of D2 expression within each genotype. Using the Pearson correlation analysis we identified 27 genes that significantly correlated with the D2 mRNA level within samples of each of the three genotypes (Supplemental Table 8; Figure 5F). Notably, only the thyroid hormone transporter gene SLCO1C1 (OATP14) expression correlated significantly with D2 mRNA independently of genotype (Figure 5F).
Using human brain and cell microarray data to define the Ala92-D2 genetic fingerprint
Venn diagrams were used to identify common genetic features between human brain and cell microarray datasets. We first looked at individual genes enriched in the gene sets from cell- (1228 genes) and human brain- (382 genes) GSEA and identified 79 common genes (79-list) (Figure 5G). Notably, the gene set for Epidermal Growth Factor Receptor Signaling Pathway (EGFR) was down-regulated in human brain homozygous for Ala92-D2 and Ala92-D2-expressing cells (Supplemental Table 3, Supplemental Table 7; Supplemental Figure 6).
Next, a classifier of the 79-list was created and applied to the microarray data sets from the Thr92-D2 and Ala92-D2 homozygous human brain donors to predict their genotype; all of the Thr92-D2 samples were correctly classified as well as 5/6 Ala92-D2 samples. When this classifier was strengthened with the addition of two genes identified from a computer-generated weighted voting prediction model, SNORD16 and TRAPPC4 (Supplemental Table 2), the 81-gene classifier correctly predicted the genotypes of all homozygous Thr92-D2 and Ala92-D2 human brain samples (0 absolute error, 0 ROC error, confidence of prediction of each sample 0.028–0.704). We also applied the 81-gene classifier to the data set of all 19 samples to predict carrier status of at least one allele, which was correct in 18 patients (Absolute error 0.05, ROC error 0.04).
Oxidative stress plays a role in the genetic fingerprint of Ala92-D2HY-expressing cells
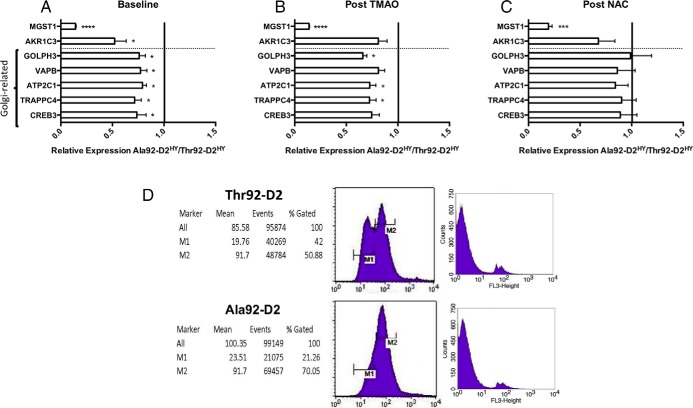
Next we used the cell model to assess findings obtained in the combined genetic analysis of brain and cell microarrays, such as Golgi function and oxidative stress (Figure 1D; Supplemental Table 3, Supplemental Table 7). We looked at the expression of seven genes affected by the Ala92-D2HY before (Figure 6A) and after treatment with the chemical chaperone TMAO (minimizes ER stress; Figure 6B) or the antioxidant NAC (Figure 6C). Here, we also included a subset of five Golgi apparatus related genes given the ectopic presence of Ala92-D2HY in this structure and the finding of three Golgi-specific genes (ATP11B, ABCG1, TRAPPC4) and 15 Golgi-related genes (Figure 5G) in the 81-gene list. Whereas treatment with TMAO for 24 h normalized expression of only one of those genes (Figure 6C), exposure to NAC had a more consistent pattern of abating differences between Ala92-D2HY and Thr92-D2HY-expressing cells in 5 of the 7 genes analyzed, all related to the Golgi apparatus (Figure 6C). Notably, the observation that Ala92-D2HY expressing cells exhibit an increase in the preapoptosis marker Annexin-V (Figure 6D) also points towards oxidative stress as well as disruption in the EGFR signaling pathway (37), similar to what was observed in EGFR−/− cortical astrocytes (38).
Figure 6.
Normalization of gene expression profile in Ala92–D2 expressing-cells by antioxidant and the chemical chaperone NAC. A, Expression levels of genes identified in cell microarray were confirmed by RT-qPCR in Thr92–D2HY and Ala92–D2HY-expressing cells where selected genes exhibited lower expression levels in Ala92–D2HY; (B) treatment with chemical chaperone, trimethylamine oxide (TMAO), normalizes two genes; (B) after 24 h of treatment with N-acetylcysteine (NAC), the expression levels of the Golgi-related genes normalized with respect to their expression levels in Thr92–D2HY-expressing cells; (two-tailed t-tests *P ≤ .05, *** P ≤ .001, ****P ≤ .0001, all data are the mean ± SEM of 3 entries per data-point); (D) The early stage of apoptosis was examined by annexin V/propidium iodide staining of intact cells; cells were gated per YFP signal, annexin V-PE/propidium iodide were FL2/FL3. Left two panels are annexin V-PE staining, where M1 represents nonapoptotic cells and M2 represents apoptotic cells; right two panels are propidium iodide staining to differentiate late-apoptotic from necrotic cells.
Discussion
A striking observation in the human brain and cell models is that expression of Ala92-D2 interferes with basic cellular processes in a pattern that establishes an 81-gene fingerprint of transcriptional alterations related to CNS diseases, ubiquitin and ER stress, mitochondrial dysfunction, inflammation, apoptosis, DNA repair and growth factor signaling. It is conceivable that the change in the single amino acid produces a protein that upsets basic ER functions triggering ER stress and changes in ubiquitination genes; the Ala92-D2 ultimately escapes to the Golgi apparatus and disrupts its normal ribbon morphology. Thus mitochondrial dysfunction, inflammation and apoptosis are likely consequences of primary disruption in ER and Golgi. The prolonged half-life exhibited by Ala92-D2HY could also be a consequence of its presence in the Golgi, sheltered from the typical proteasomal machinery present in its native ER location. It is notable that treatment with an antioxidant agent normalized the expression of multiple genes affected by Ala92-D2HY, indicating that oxidative stress plays a role in defining the genetic fingerprint. Thus, it is plausible that this fingerprint underlies the different clinical phenotypes associated with homozygosity for Ala92-D2.
Another striking observation in the present studies was that both native and polymorphic D2 were found associated with the nuclear compartment in the HEK-293 cells. Of course this could be the result of D2 overexpression in this cell model and final determination awaits the development of high-quality D2 antisera to study endogenous D2 expression. If confirmed in vivo, this would explain the decades-old kinetic and physiological data that D2-generated T3 contributes much to the occupation of TR (39, 40). In the brain of course, D2 is expressed in glial cells while TR is expressed predominantly in neurons and thus a higher level of TR occupancy is most likely the result of accelerated local T3 production and signaling through a paracrine mechanism (33). In any event, that no differences in nuclear D2 were observed between Thr92-D2HY and Ala92-D2HY proteins support the notion that TH signaling is not grossly affected by this polymorphism (Supplemental Figure 3, C and D).
A strength of the present study is the complementary usage of microarray analysis in brain and cells, which led to the refinement of the genetic fingerprint initially discovered in the brains. The resulting 81-gene fingerprint was highly efficacious as a genotype classifier in these samples. Along these lines, the observation that Huntington's disease was the top pathway identified in the gene analysis of the human brains indicates similarities between the two diseases might exist. Indeed, Huntington's is a CNS degenerative disease that causes chorea and loss of cognition that is caused by Huntingtin, an ER-associated protein that normally translocates to and from the cell nucleus (41). Carriers of the mutant huntingtin gene express an abnormally long version of the huntingtin protein that is hydrolyzed into smaller peptides that accumulate and disrupt the normal function of neurons. This leads to ER and oxidative stress, and eventually to apoptosis of neurons in the striatum, explaining the signs and symptoms of Huntington's disease (42, 43). While it is too early to draw any definitive conclusions, the parallels between the Huntington's disease mechanism and the observations in our Ala92-D2HY cell model are intriguing and could ultimately indicate that carriers for the D2 variant may be predisposed to neurodegenerative processes. The gene analyses also point towards alternative disease mechanisms, such as disruption of the EGF signaling pathway, which is known for playing a role in cognitive development (12) and impairment in diseases including Parkinson's (14, 15), Alzheimer's (16), and schizophrenia (13).
It is clear that the clinical phenotypes associated the Ala92-D2 polymorphism are not limited to hypothyroid patients, but can be traced back to tissues/organs that express D2 (44). However, it is unclear why hypothyroid carriers of the Ala92-D2 polymorphism would prefer combination therapy with L-T4 and L-T3 (7) given that TH signaling is not affected by Ala92-D2. While much additional investigation is still needed, it is conceivable that compensatory mechanisms developed in carriers of the Ala92-D2 polymorphism lose effectiveness in the setting of treatment with L-T4, when patients exhibit an elevation in serum T4/T3 ratio (45, 46). On this note, D2 mRNA levels in the brain correlate with the TH transporter OATP14 (Figure 5F). Therefore, the present findings do not provide a direct rationale for combination therapy with L-T4 and L-T3.
In conclusion, we have uncovered a unique 81-gene fingerprint left in the brain and in cells expressing Ala92-D2. This is caused by cellular accumulation of Ala92-D2 that can be found ectopically in the Golgi apparatus, and is associated with ER and oxidative stress, as well as preapoptosis. There are striking similarities between these findings and changes observed in other well-characterized brain degenerative diseases. Future research should clarify whether the disease mechanisms proposed in the present investigation contribute to the phenotype of Ala92-D2 carriers.
Acknowledgments
We thank Selmar Leeuwenburgh for assistance with genotyping and Katelyn Hughes for assistance with performance of microarray studies.
This work was supported by grants from the National Institute of Diabetes and Digestive Kidney Diseases (DK58538), the Hungarian Brain Research Program, and the Lendület grant of the Hungarian Academy of Sciences. The University of Miami Brain Endowment Bank is funded in part with Federal funds from the NIMH, NINDS, and NICHD, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN271201300028C.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CHX
- cycloheximide
- EM
- electron microscope
- FBS
- fetal bovine serum
- FDR
- false discovery rate
- IPA
- ingenuity pathway analysis
- KNN
- k-nearest-neighbors
- L-T4
- levothyroxine
- NES
- normalized enrichment score
- PB
- phosphate buffer
- ROC
- receiver operating characteristic
- TH
- prothyroid hormone
- UM
- University of Miami
- YFP
- yellow fluorescent protein.
References
- 1. Golden SH, Robinson KA, Saldanha I, Anton B, Ladenson PW. Clinical review: Prevalence and incidence of endocrine and metabolic disorders in the United States: a comprehensive review. J Clin Endocrinol Metab. 2009;94(6):1853–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Canaris GJ, Manowitz NR, Mayor G, Ridgway EC. The Colorado thyroid disease prevalence study. Arch Intern Med. 2000;160(4):526–534. [DOI] [PubMed] [Google Scholar]
- 3. Wiersinga WM, Duntas L, Fadeyev V, Nygaard B, Vanderpump MP. 2012 ETA Guidelines: The Use of L-T4 + L-T3 in the Treatment of Hypothyroidism. Eur Thyroid J. 2012;1(2):55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dora JM, Machado WE, Rheinheimer J, Crispim D, Maia AL. Association of the type 2 deiodinase Thr92Ala polymorphism with type 2 diabetes: case-control study and meta-analysis. Eur J Endocrinol. 2010;163(3):427–434. [DOI] [PubMed] [Google Scholar]
- 5. Dentice M, Bandyopadhyay A, Gereben B, et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol. 2005;7(7):698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callebaut I, Curcio-Morelli C, Mornon JP, et al. The iodothyronine selenodeiodinases are thioredoxin-fold family proteins containing a glycoside hydrolase clan GH-A-like structure. J Biol Chem. 2003;278(38):36887–36896. [DOI] [PubMed] [Google Scholar]
- 7. Panicker V, Saravanan P, Vaidya B, et al. Common variation in the DIO2 gene predicts baseline psychological well-being and response to combination thyroxine plus triiodothyronine therapy in hypothyroid patients. J Clin Endocrinol Metab. 2009;94(5):1623–1629. [DOI] [PubMed] [Google Scholar]
- 8. Guo TW, Zhang FC, Yang MS, et al. Positive association of the DIO2 (deiodinase type 2) gene with mental retardation in the iodine-deficient areas of China. J Med Genet. 2004;41(8):585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor P, Okosieme O, Sayers A, et al. Effect of low thyroid hormone bioavailability on childhood cognitive development: data from the Avon Longitudinal Study of Parents and Children birth cohort. The Lancet. 2014;383:S100. [Google Scholar]
- 10. He B, Li J, Wang G, et al. Association of genetic polymorphisms in the type II deiodinase gene with bipolar disorder in a subset of Chinese population. Prog Neuropsychopharmacol Biol Psychiatry. 2009;33(6):986–990. [DOI] [PubMed] [Google Scholar]
- 11. Wang X, McCullough KD, Franke TF, Holbrook NJ. Epidermal growth factor receptor-dependent Akt activation by oxidative stress enhances cell survival. J Biol Chem. 2000;275(19):14624–14631. [DOI] [PubMed] [Google Scholar]
- 12. Futamura T, Kakita A, Tohmi M, Sotoyama H, Takahashi H, Nawa H. Neonatal perturbation of neurotrophic signaling results in abnormal sensorimotor gating and social interaction in adults: implication for epidermal growth factor in cognitive development. Mol Psychiatry. 2003;8(1):19–29. [DOI] [PubMed] [Google Scholar]
- 13. Kao WT, Wang Y, Kleinman JE, et al. Common genetic variation in Neuregulin 3 (NRG3) influences risk for schizophrenia and impacts NRG3 expression in human brain. Proc Natl Acad Sci U S A. 2010;107(35):15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pellecchia MT, Santangelo G, Picillo M, et al. Serum epidermal growth factor predicts cognitive functions in early, drug-naive Parkinson's disease patients. J Neurol. 2013;260(2):438–444. [DOI] [PubMed] [Google Scholar]
- 15. Chen-Plotkin AS, Hu WT, Siderowf A, et al. Plasma epidermal growth factor levels predict cognitive decline in Parkinson disease. Ann Neurol. 2011;69(4):655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marksteiner J, Kemmler G, Weiss EM, et al. Five out of 16 plasma signaling proteins are enhanced in plasma of patients with mild cognitive impairment and Alzheimer's disease. Neurobiol Aging. 2011;32(3):539–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zevenbergen C, Klootwijk W, Peeters RP, et al. Functional Analysis of Novel Genetic Variation in the Thyroid Hormone Activating Type 2 Deiodinase. J Clin Endocrinol Metab. 2014:99:E2429–2436. [DOI] [PubMed] [Google Scholar]
- 18. Hernandez A, Morte B, Belinchón MM, Ceballos A, Bernal J. Critical role of types 2 and 3 deiodinases in the negative regulation of gene expression by T3 in the mouse cerebral cortex. Endocrinology. 2012;153(6):2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bianco AC, Anderson G, Forrest D, et al. American thyroid association guide to investigating thyroid hormone economy and action in rodent and cell models. Thyroid. 2014;24(1):88–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeöld A, Doleschall M, Haffner MC, et al. Characterization of the nuclear factor-kappa B responsiveness of the human dio2 gene. Endocrinology. 2006;147(9):4419–4429. [DOI] [PubMed] [Google Scholar]
- 21. Zhang CY, Kim S, Harney JW, Larsen PR. Further characterization of thyroid hormone response elements in the human type 1 iodothyronine deiodinase gene. Endocrinology. 1998;139:1156–1163. [DOI] [PubMed] [Google Scholar]
- 22. Thomas P, Smart TG. HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods. 2005;51(3):187–200. [DOI] [PubMed] [Google Scholar]
- 23. Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16(8):869–871. [DOI] [PubMed] [Google Scholar]
- 24. Arrojo E Drigo R, Egri P, Jo S, Gereben B, Bianco AC. The type II deiodinase is retrotranslocated to the cytoplasm and proteasomes via p97/Atx3 complex. Mol Endocrinol. 2013;27(12):2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jo S, Kalló I, Bardóczi Z, et al. Neuronal hypoxia induces hsp40-mediated nuclear import of type 3 deiodinase as an adaptive mechanism to reduce cellular metabolism. J Neurosci. 2012;32(25):8491–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kallo I, Mohacsik P, Vida B, et al. A novel pathway regulates thyroid hormone availability in rat and human hypothalamic neurosecretory neurons. PLoS One. 2012;7(6):e37860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Canani LH, Capp C, Dora JM, et al. The type 2 deiodinase A/G (Thr92Ala) polymorphism is associated with decreased enzyme velocity and increased insulin resistance in patients with type 2 diabetes mellitus. J Clin Endocrinol Metab. 2005;90(6):3472–3478. [DOI] [PubMed] [Google Scholar]
- 28. Peeters RP, van Toor H, Klootwijk W, et al. Polymorphisms in thyroid hormone pathway genes are associated with plasma TSH and iodothyronine levels in healthy subjects. J Clin Endocrinol Metab. 2003;88(6):2880–2888. [DOI] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. [DOI] [PubMed] [Google Scholar]
- 31. Sagar GD, Gereben B, Callebaut I, et al. Ubiquitination-induced conformational change within the deiodinase dimer is a switch regulating enzyme activity. Mol Cell Biol. 2007;27(13):4774–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. [DOI] [PubMed] [Google Scholar]
- 33. Freitas BC, Gereben B, Castillo M, et al. Paracrine signaling by glial cell-derived triiodothyronine activates neuronal gene expression in the rodent brain and human cells. J Clin Invest. 2010;120(6):2206–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma D, Fondell JD. Temporal formation of distinct thyroid hormone receptor coactivator complexes in HeLa cells. Mol Endocrinol. 2000;14(12):2001–2009. [DOI] [PubMed] [Google Scholar]
- 35. Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci U S A. 2002;99(12):7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MAQC Consortium, Shi L, Reid LH, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang N, Zhang JH, Qiu F, Tashiro S, Onodera S, Ikejima T. Inhibition of EGFR signaling augments oridonin-induced apoptosis in human laryngeal cancer cells via enhancing oxidative stress coincident with activation of both the intrinsic and extrinsic apoptotic pathways. Cancer Lett. 2010;294(2):147–58. [DOI] [PubMed] [Google Scholar]
- 38. Wagner B, Natarajan A, Grünaug S, Kroismayr R, Wagner EF, Sibilia M. Neuronal survival depends on EGFR signaling in cortical but not midbrain astrocytes. EMBO J. 2006;25(4):752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gereben B, Zavacki AM, Ribich S, et al. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev. 2008;29(7):898–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Silva JE, Dick TE, Larsen PR. The contribution of local tissue thyroxine monodeiodination to the nuclear 3,5,3'-triiodothyronine in pituitary, liver, and kidney of euthyroid rats. Endocrinology. 1978;103(4):1196–1207. [DOI] [PubMed] [Google Scholar]
- 41. Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16(21):2600–2615. [DOI] [PubMed] [Google Scholar]
- 42. Labbadia J, Morimoto RI. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38(8):378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vidal R, Caballero B, Couve A, Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington's disease. Curr Mol Med. 2011;11(1):1–12. [DOI] [PubMed] [Google Scholar]
- 44. Bianco AC, Casula S. Thyroid Hormone Replacement Therapy: Three 'Simple' Questions, Complex Answers. Eur Thyroid J. 2012;1:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gullo D, Latina A, Frasca F, Le Moli R, Pellegriti G, Vigneri R. Levothyroxine monotherapy cannot guarantee euthyroidism in all athyreotic patients. PLoS One. 2011;6(8):e22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stock JM, Surks MI, Oppenheimer JH. Replacement dosage of L-thyroxine in hypothyroidism. A re-evaluation. N Engl J Med. 1974;290(10):529–533. [DOI] [PubMed] [Google Scholar]