Abstract
GOLPH3 is the first example of an oncogene that functions in secretory trafficking at the Golgi. The discovery of GOLPH3’s roles in both cancer and Golgi trafficking raises questions about how GOLPH3 and the Golgi contribute to cancer. Our recent investigation of the regulation of GOLPH3 revealed a surprising response by the Golgi upon DNA damage that is mediated by DNA-PK and GOLPH3. These results provide new insight into the DNA damage response with important implications for understanding the cellular response to standard cancer therapeutic agents.
Keywords: DNA damage, Golgi, GOLPH3, cancer, phosphatidylinositol-4-phosphate
GOLPH3 is an Oncogene
GOLPH3 is an oncogene that functions in secretory trafficking at the Golgi (1–5). Through genome-wide analysis of human cancers, Lynda Chin and colleagues found high frequency of amplification of GOLPH3 in several solid tumor types including 56% of lung cancers, 38% of ovarian cancers, 32% of breast cancers, 33% of pancreatic cancers, 37% of prostate cancers, 32% of melanomas, and 24% of colon carcinomas (5). They then went on to show that GOLPH3 is, in fact, an oncogene, capable of cooperating with other oncogenes to cause transformation in both cell culture and xenograft mouse models. In particular, they observed that overexpression of GOLPH3 could cooperate with B-RAF(V600E) in TERT-immortalized human melanocytes to allow growth in semi-solid media, and with HRAS(G12V) in Ink4a/Arf-deficient primary mouse embryonic fibroblasts to cause focus formation. They also observed that overexpression of GOLPH3 dramatically increased mouse xenograft tumor growth for WM239A melanoma, A549 lung adenocarcinoma, and 1205LU melanoma cell lines. Systematic data from The Cancer Genome Atlas also detect amplification of GOLPH3 in cancers, albeit at lower frequency (e.g., 9.6% of lung adenocarcinomas) (6). The differences in frequency may reflect differences in methods, issues associated with high-throughput screening approaches, or relate to known inconsistencies in the cancer genome datasets (7, 8).
Since the initial publication, over twenty studies have validated GOLPH3 as an oncogene, demonstrating its ability to cause transformation, observing frequent overexpression in a variety of cancers, and showing a correlation between high levels of expression and poor patient prognosis. Evidence of transformation by overexpression of GOLPH3 has been reported in MDA-MB-231 and MCF7 breast cancer cell lines (9–11), and U251 and U87 glioblastoma cell lines (12, 13). Frequent overexpression of GOLPH3 and correlation with poor prognosis have been reported in multiple tumor types including 58–72% of non-small cell lung cancers (14, 15), 52% of breast cancers (11), 70% of prostate cancers (16), 73% of pancreatic ductal adenocarcinomas (17), 65% of hepatocellular carcinomas (18, 19), 55–58% of gastric cancers (20, 21), 53% of renal cell carcinomas (22), 41–53% of glioblastomas (12, 23, 24), 49% of esophageal squamous carcinomas (25), and 45% of ovarian carcinomas (26, 27). Overexpression of GOLPH3 occurs frequently in rhabdomyosarcoma, and knockdown of GOLPH3 in rhabdomyosarcoma cell lines impairs cell proliferation (28). Abnormal expression of microRNA-126 has been associated with increased proliferation, migration, and invasion of esophageal squamous cell carcinoma in a manner that depends on the ability of the microRNA to increase expression of GOLPH3 (29). Taken together, the data argue that overexpression of GOLPH3 is a common feature of many solid tumors that helps drive oncogenic transformation and generally portends poor prognosis.
The Golgi PI4P/GOLPH3/MYO18A/F-Actin Pathway
Working from a different angle, our laboratory discovered GOLPH3 as a novel effector of phosphatidylinositol-4-phosphate (PI4P), playing a critical role in Golgi to plasma membrane trafficking (1). PI4P was known to be highly enriched at the trans-Golgi (30) and to be somehow required for Golgi to plasma membrane trafficking across species (31–33). We found that GOLPH3 binds tightly and specifically to PI4P, resulting in robust localization of GOLPH3 to the trans-Golgi from yeast to humans (1). We further showed that GOLPH3 interacts tightly with an unconventional myosin, MYO18A, recruiting it to the Golgi. MYO18A binds to F-actin and the complex applies a tensile force that pulls on the Golgi membrane. This tensile force deforms the Golgi membrane to participate in the process of vesicle budding for vesicles trafficking from the Golgi to the plasma membrane (1–3).
Interference with GOLPH3 or MYO18A strongly impairs Golgi to plasma membrane trafficking as shown by many different assays, including measurement of vesicular stomatitis virus G glycoprotein trafficking, measurement of total secretory flux by metabolic pulse-chase analysis, live-cell imaging of vesicle exit from the Golgi, and measurement of hepatitis C virus secretion (1–3). We found that the familiar appearance of the Golgi, an extended ribbon observed by light microscopy and flattened stacks observed by electron microscopy, is a consequence of the tensile force applied through the GOLPH3/MYO18A-dependant linkage of the Golgi membrane to the F-actin cytoskeleton. Thus, the shape of the Golgi is a consequence of the mechanism of trafficking. Furthermore, Golgi morphology serves as a functional readout of the GOLPH3 pathway and Golgi trafficking.
DNA Damage Response and the Golgi
The effect of DNA damage on the Golgi had not been studied previously. While we were studying the regulation of GOLPH3, we found that it is phosphorylated on T143, which we noted to be in the context of Q at the +1 position, forming a TQ motif (4, 34). TQ/SQ is the preferred substrate motif for the DNA damage-activated protein kinases ATM, ATR, and DNA-PK (35, 36). This raised the question of whether there might be a direct signaling pathway from the DNA damage response to GOLPH3 and the Golgi.
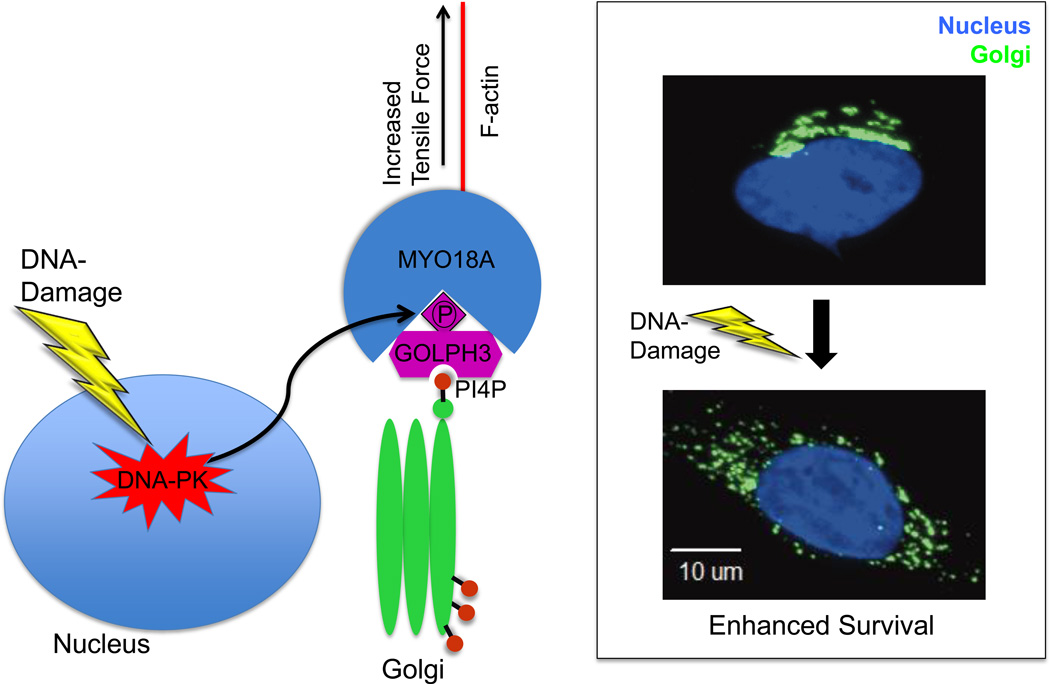
We found that exposure of mammalian cells to agents that cause DNA double-stranded breaks (ionizing radiation, camptothecin, or doxorubicin) leads to a significant alteration of Golgi morphology from the normal perinuclear ribbon to a fragmented, punctate Golgi that is dispersed throughout the cytoplasm (see Figure) (4). We found that this dramatic alteration of Golgi morphology is a common feature of the DNA damage response in mammalian cells, including a broad range of cell lines as well as primary mouse and human cells.
Figure.
Mechanism of DNA damage induced dispersal of the Golgi. In response to DNA damage, DNA-PK is activated and phosphorylates GOLPH3 on the T143 TQ motif. This phosphorylation results in increased GOLPH3 interaction with MYO18A and consequently an increased tensile force on the Golgi, leading to Golgi vesiculation, fragmentation, and dispersal. The DNA-PK/GOLPH3/MYO18A pathway is required for cell survival following DNA damage, and overexpression of GOLPH3 confers resistance to killing by DNA damaging agents.
We mapped the signaling pathway responsible for DNA damage induced Golgi dispersal (see Figure) (4). We found that DNA-PK (but not ATM or ATR) directly phosphorylates GOLPH3 on the T143 TQ site, and this is required for DNA damage induced Golgi dispersal. We further showed that this phosphorylation of GOLPH3 leads to enhanced interaction between GOLPH3 and MYO18A, resulting in an increased tensile force, and thus increased vesiculation of the Golgi. This vesiculation ultimately causes Golgi fragmentation and dispersal.
The end result of DNA damage induced Golgi dispersal is impaired protein trafficking from the Golgi to the plasma membrane. However, we speculate that the immediate consequence may be an initial burst of anterograde trafficking, which eventually leads to Golgi fragmentation, and subsequently results in impaired Golgi function and reduced trafficking. Future experiments with better temporal resolution will be needed to test this possibility.
While the exact mechanism remains uncertain, it is clear that the DNA-PK/GOLPH3/MYO18A pathway is required for cell survival following DNA damage. Depletion of GOLPH3 or MYO18A results in a significant increase in cellular apoptosis in response to DNA damage. Likewise, depletion of DNA-PKcs (catalytic subunit), GOLPH3, or MYO18A reduces the ability of the cells to survive and proliferate after DNA damage. Furthermore, overexpression of GOLPH3 enhances cell survival in response to DNA damage. This enhanced cell survival due to GOLPH3 overexpression requires both the ability of GOLPH3 to localize to the Golgi and phosphorylation of GOLPH3 on the DNA-PK site. We think it is likely that there exist one or more proteins involved in either survival signaling or death signaling that depend on trafficking from the Golgi to the plasma membrane for their function, and that altered trafficking following DNA damage, by altering the function of these proteins, produces a survival benefit. However, the critical cargoes have yet to be identified.
Logic of the DNA Damage Response
To step back, at the most basic level the DNA damage response has evolved in cells to detect DNA lesions, signal a cellular response, and repair the damaged DNA. The main signaling components in the DNA damage response are the PI3K-like protein kinases ATM, ATR, and DNA-PKcs. The protein kinases ATM and ATR are responsible for the classical DNA damage signaling response, including CHK1/2-dependent cell cycle arrest, activation of DNA repair, and transcriptional regulation of proteins in the DNA damage response (37). ATM and ATR are the more ancient DNA damage response kinases, conserved back to yeast. In a single-celled organism like yeast, every cell attempts to repair the DNA damage to try to survive. By contrast, in higher multicellular organisms each cell must somehow decide whether to repair the DNA damage to allow cellular survival or instead to trigger apoptosis for the benefit of the organism. DNA-PK evolved later, appearing in the genomes of higher multicellular organisms. DNA-PK is a multimeric complex consisting of a catalytic subunit, DNA-PKcs, and the regulatory subunits, Ku70 and Ku80. It plays a critical role in the DNA damage response, particularly in the recognition and repair of double-stranded breaks (38). Cells and mice that lack DNA-PK exhibit increased cell death in response to DNA damage. Interestingly, DNA-PK has a number of diverse and unusual cellular functions.
One clearly unique cellular function of DNA-PK is the ability to phosphorylate and activate AKT, observed by Hemmings and colleagues (39). Upon DNA damage DNA-PK phosphorylates AKT on S473, activating its kinase activity. Activation of AKT results in downstream signaling that is well known to enhance cellular survival (40), and thus this is one mechanism by which DNA-PK can generate a survival signal.
DNA-PK also has been shown to play an intriguing role in metabolic gene regulation (41). In response to feeding and insulin signaling, DNA-PK is recruited to transcriptional promoters, where it phosphorylates upstream stimulatory factor (USF-1). The phosphorylation of USF-1 leads to the recruitment of additional transcription factors, including the histone acetyltransferase p300 associated factor (P/CAF), which acetylates USF-1, leading to activation of gene expression. While Sul and colleagues show this unique role of DNA-PK occurs in response to insulin stimulation, it seems likely to be true in response to IGF-1, and perhaps other receptor tyrosine kinases, as well. Notably, the genes that are activated include some that might confer a cellular survival benefit, potentially providing another link from DNA-PK to survival signaling.
Our recent data demonstrate that DNA-PK plays an essential role in regulating the Golgi in response to DNA damage, via the GOLPH3/MYO18A pathway (4). This pathway is required for normal cell survival in response to DNA damage and when enhanced can confer resistance to killing by DNA damaging agents. Here again, we observe a role for DNA-PK in generating a survival signal. Thus, we propose that perhaps DNA-PK evolved in higher multicellular organisms to weigh in on the cellular decision to survive or die following DNA damage. DNA-PK has multiple mechanisms to bias cells toward survival following DNA damage.
Regulation of the Golgi
Previously popular models of Golgi function have failed to provide insight into regulation of the Golgi in response to changes in the status of the cell. This is unexpected, since regulation is the overarching theme in biology. Recent data now demonstrate interesting regulation of the Golgi via the PI4P/GOLPH3/MYO18A pathway. Mayinger and colleagues found that growth factor signaling leads to increased Golgi to plasma membrane trafficking (42). The mechanism involves translocation of the SAC1 PI4P-4-phosphatase away from the Golgi to the endoplasmic reticulum, resulting in a rise in Golgi PI4P levels. The DNA damage response provides another, dramatic example of regulation of the Golgi. In this case, the regulation in response to DNA damage occurs by modulating the strength of the interaction between GOLPH3 and MYO18A (4).
GOLPH3L is a paralog of GOLPH3 that appears late in evolution, restricted to vertebrates, with expression restricted to highly secretory cell types (3). We found that GOLPH3L acts as a dominant-negative inhibitor of the PI4P/GOLPH3/MYO18A pathway. It does so because it is able to bind to PI4P, but is unable to bind to MYO18A. Our data indicate that GOLPH3L acts as a throttle to damp-down secretory trafficking in highly secretory cells. Thus, the PI4P/GOLPH3/MYO18A pathway is emerging as a convergent hub for regulation of the Golgi.
The Role of GOLPH3 and the Golgi in Cancer
The output of the Golgi, chiefly trafficking of proteins to the plasma membrane either to remain there or to be released into the extracellular space, likely has pleiotropic effects on cellular function. Thus, we might expect dysregulation of the Golgi to play a role in common human disease. The fact that GOLPH3 is an oncogene fulfills that prediction, although the mechanism remains poorly understood.
The role of GOLPH3 in the Golgi DNA damage response may provide some insight into the role of GOLPH3 in cancer. We observe that overexpression of GOLPH3, by driving the Golgi DNA damage response, confers resistance to killing by DNA damaging therapeutic agents. Overexpression of GOLPH3 in a variety of clinical cancers has been shown to predict poor prognosis. While poor prognosis can be interpreted in many ways, it definitely means that these patients’ cancers have progressed despite standard therapy. Standard therapy relies heavily on DNA damaging chemotherapeutics or radiation. And thus, the fact that overexpression of GOLPH3 can confer resistance to killing by DNA damaging agents may explain its role in determining poor prognosis. Future studies will be needed to determine if overexpression of GOLPH3 is a useful marker for predicting responsiveness to particular therapeutics, and whether inhibition of the GOLPH3 pathway will potentiate standard therapies to provide therapeutic benefit.
Acknowledgements
The authors thank Drs. Farber-Katz, Dippold, and Ng, and other members of the Field lab, past and present, who have contributed to this work.
Grant Support
Dr. Buschman thanks the American Cancer Society for a Lee National Denim Day Postdoctoral Fellowship Award PF-t3-367-01-CDD. Dr. Rahajeng gratefully acknowledges support by a Ruth L. Kirchstein National Research Service Award NIH/NCI T32 CA009523. Dr. Field thanks the Era of Hope Scholar Award from the Breast Cancer Research Program of the Department of Defense W81XWH-10-1-0822, the NIH Director’s New Innovator Award DP2-OD004265, and the Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed by the authors.
References
- 1.Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishe B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng MM, Dippold HC, Buschman MD, Noakes CJ, Field SJ. GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Mol Biol Cell. 2013;24:796–808. doi: 10.1091/mbc.E12-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, et al. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156:413–427. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson AM, Yates T, Li Y, Trotter EW, Fawdar S, Chapman P, et al. Discrepancies in Cancer Genomic Sequencing Highlight Opportunities for Driver Mutation Discovery. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadalayil L, Rafiq S, Rose-Zerilli MJ, Pengelly RJ, Parker H, Oscier D, et al. Exome sequence read depth methods for identifying copy number changes. Brief Bioinform. 2014 doi: 10.1093/bib/bbu027. [DOI] [PubMed] [Google Scholar]
- 9.Salem AF, Whitaker-Menezes D, Lin Z, Martinez-Outschoorn UE, Tanowitz HB, Al-Zoubi MS, et al. Two-compartment tumor metabolism: autophagy in the tumor microenvironment and oxidative mitochondrial metabolism (OXPHOS) in cancer cells. Cell Cycle. 2012;11:2545–2556. doi: 10.4161/cc.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tokuda E, Itoh T, Hasegawa J, Ijuin T, Takeuchi Y, Irino Y, et al. Phosphatidylinositol 4-phosphate in the Golgi apparatus regulates cell-cell adhesion and invasive cell migration in human breast cancer. Cancer Res. 2014;74:3054–3066. doi: 10.1158/0008-5472.CAN-13-2441. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R, et al. Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin Cancer Res. 2012;18:4059–4069. doi: 10.1158/1078-0432.CCR-11-3156. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Ding Z, Mo J, Sang B, Shi Q, Hu J, et al. GOLPH3 promotes glioblastoma cell migration and invasion via the mTOR-YB1 pathway in vitro. Mol Carcinog. 2014 doi: 10.1002/mc.22197. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Xue P, Yang M, Shi H, Lu D, Wang Z, et al. Protein kinase D2 promotes the proliferation of glioma cells by regulating Golgi phosphoprotein 3. Cancer Lett. 2014;355:121–129. doi: 10.1016/j.canlet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Ma M, Han B. GOLPH3 high expression predicts poor prognosis in patients with resected non-small cell lung cancer: an immunohistochemical analysis. Tumour Biol. 2014 doi: 10.1007/s13277-014-2357-3. [DOI] [PubMed] [Google Scholar]
- 15.Lu M, Tian Y, Yue WM, Li L, Li SH, Qi L, et al. GOLPH3, a Good Prognostic Indicator in Early-stage NSCLC Related to Tumor Angiogenesis. Asian Pac J Cancer Prev. 2014;15:5793–5798. doi: 10.7314/apjcp.2014.15.14.5793. [DOI] [PubMed] [Google Scholar]
- 16.Hua X, Yu L, Pan W, Huang X, Liao Z, Xian Q, et al. Increased expression of Golgi phosphoprotein-3 is associated with tumor aggressiveness and poor prognosis of prostate cancer. Diagn Pathol. 2012;7:127. doi: 10.1186/1746-1596-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang LJ, Wang KB, Liu LS, Chen LZ, Peng BG, Liang LJ, et al. Overexpression of GOLPH3 is associated with poor prognosis and clinical progression in pancreatic ductal adenocarcinoma. BMC Cancer. 2014;14:571. doi: 10.1186/1471-2407-14-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu GS, Li YQ, Yang YM, Shi W, Liao AJ, Yao YH, et al. High expression of Golgi phosphoprotein-3 is associated with poor survival in patients with hepatocellular carcinoma. Tumour Biol. 2014;35:8625–8632. doi: 10.1007/s13277-014-2105-8. [DOI] [PubMed] [Google Scholar]
- 19.JianXin J, Cha Y, ZhiPeng L, Jie X, Hao Z, Meiyuan C, et al. GOLP3 is a predictor of survival in patients with hepatocellular carcinoma. Clin Invest Med. 2014;37:E233. [PubMed] [Google Scholar]
- 20.Hu BS, Hu H, Zhu CY, Gu YL, Li JP. Overexpression of GOLPH3 is associated with poor clinical outcome in gastric cancer. Tumour Biol. 2013;34:515–520. doi: 10.1007/s13277-012-0576-z. [DOI] [PubMed] [Google Scholar]
- 21.Peng J, Fang Y, Tao Y, Li K, Su T, Nong Y, et al. Mechanisms of GOLPH3 Associated with the Progression of Gastric Cancer: A Preliminary Study. PLoS One. 2014;9:e107362. doi: 10.1371/journal.pone.0107362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y, Wu G, Liao Y, Xiao G, Ma X, Zou X, et al. GOLPH3 is a novel marker of poor prognosis and a potential therapeutic target in human renal cell carcinoma. Br J Cancer. 2014;110:2250–2260. doi: 10.1038/bjc.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XY, Liu W, Chen SF, Zhang LQ, Li XG, Wang LX. Expression of the Golgi phosphoprotein-3 gene in human gliomas: a pilot study. J Neurooncol. 2011;105:159–163. doi: 10.1007/s11060-011-0573-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Xu T, Qin R, Yan Y, Chen C, Chen Y, et al. Overexpression of Golgi phosphoprotein-3 (GOLPH3) in glioblastoma multiforme is associated with worse prognosis. J Neurooncol. 2012;110:195–203. doi: 10.1007/s11060-012-0970-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang JH, Chen XT, Wen ZS, Zheng M, Deng JM, Wang MZ, et al. High expression of GOLPH3 in esophageal squamous cell carcinoma correlates with poor prognosis. PLoS One. 2012;7:e45622. doi: 10.1371/journal.pone.0045622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Wang X, Wu Y, Sun B, Lv H, Rong F, et al. Overexpression of GOLPH3 protein is associated with worse prognosis in patients with epithelial ovarian cancer. Tumour Biol. 2014 doi: 10.1007/s13277-014-2411-1. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y, Ren Y, Zhang X, Lin L, Liu Y, Rong F, et al. High GOLPH3 expression is associated with a more aggressive behavior of epithelial ovarian carcinoma. Virchows Arch. 2014;464:443–452. doi: 10.1007/s00428-014-1536-3. [DOI] [PubMed] [Google Scholar]
- 28.Kunigou O, Nagao H, Kawabata N, Ishidou Y, Nagano S, Maeda S, et al. Role of GOLPH3 and GOLPH3L in the proliferation of human rhabdomyosarcoma. Oncol Rep. 2011;26:1337–1342. doi: 10.3892/or.2011.1413. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Meng F, Ma J, Yu Y, Hua X, Qin J, et al. Insulin receptor substrate-1 and Golgi phosphoprotein 3 are downstream targets of miR126 in esophageal squamous cell carcinoma. Oncol Rep. 2014;32:1225–1233. doi: 10.3892/or.2014.3327. [DOI] [PubMed] [Google Scholar]
- 30.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, et al. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 31.Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- 32.Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 33.Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, et al. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 34.Foiani M, Bartek J. Golgi feels DNA's pain. Cell. 2014;156:392–393. doi: 10.1016/j.cell.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 36.O'Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, et al. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- 37.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 40.Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 41.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–1072. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knodler A, Nicolson T, et al. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]