Abstract
An important goal of synthetic biology is the rational design and predictable implementation of synthetic gene circuits using standardized and interchangeable parts. However, engineering of complex circuits in mammalian cells is currently limited by the availability of well-characterized and orthogonal transcriptional repressors. Here, we introduce a library of 26 reversible transcription activator-like effector repressors (TALERs) that bind newly designed hybrid promoters and exert transcriptional repression through steric hindrance of key transcriptional initiation elements. We demonstrate that using the input-output transfer curves of our TALERs enables accurate prediction of the behavior of modularly assembled TALER cascade and switch circuits. We also show that TALER switches employing feedback regulation exhibit improved accuracy for microRNA-based HeLa cancer cell classification versus HEK293 cells. Our TALER library is a valuable toolkit for modular engineering of synthetic circuits, enabling programmable manipulation of mammalian cells and helping elucidate design principles of coupled transcriptional and microRNA-mediated post-transcriptional regulation.
Introduction
Synthetic gene circuits are designed to implement desired cellular functions in living cells through sensing, integration and processing of molecular information and are assembled by functionally connecting genetic regulatory devices. A variety of synthetic circuits have been developed to implement customized and programmed functions in cells, including dynamic behaviors1-3, switches and memory4-8, cell-cell communication9-11, adaptation12, 13, cell polarization14, digital and analog computation15-21, and complex biosynthetic pathways22. Most of these circuits have been constructed by costly and inefficient “trial-and-error” methods with limited parts. To simplify design and optimization of large and complex synthetic circuits for sophisticated manipulation of living cells, a large library of well-characterized synthetic components and a set of appropriate computational models and simulation methods are needed.
Engineering synthetic transcriptional activators and repressors to support scalable circuit design is an important objective in the field of mammalian synthetic biology. A common strategy for the construction of synthetic mammalian/eukaryotic transcriptional repressors is to fuse a transcriptional repression domain to an engineered DNA binding protein such as zinc finger protein23, 24, transcription activator like effector (TALE) protein25-27 and more recently deactivated Cas9 (dCas9) nuclease in an RNA-guided CRISPR (clustered regularly interspaced short palindromic repeats) system28, 29. However, transcriptional repression domains, such as Krüppel-associated box (KRAB) repression domain or mSin interaction domain (SID4) usually cause epigenetic modifications along the target promoter region, resulting in slow temporal responses because transcription reactivation can be delayed significantly5, 25, 30. Thus, such transcriptional repressor fusions are not suitable for constructing synthetic gene circuits that require reversible control of gene expression. Another mode of transcriptional repression is achieved through steric hindrance with no functional repression domains. Variable repression efficiency has been shown in the context of mammalian gene regulation12, 18, 28, 29, 31.
Here we introduce a framework for engineering reversible TALE transcriptional repressors (TALERs) that suppress gene expression through steric hindrance of key transcriptional elements within their cognate promoters (Fig. 1a). We developed a mathematical model solely based on our experimental measurements of individual TALER input-output transfer curves, and used this model to quantitatively predict the fold induction of TALER cascades and the steady state behavior of TALER switches constructed with two cross repressed TALER proteins. Finally, we implemented TALER sensory switches controlled by cell-type specific microRNAs and assay their behavior in a mixed cell population, resulting in distinct outputs in two types of co-cultured cancer cells with greatly improved classification precision. Therefore, our library of reversible TALER devices can serve as standard genetic parts for modular assembly of synthetic circuits with predictable and programmable behavior in mammalian cells for biotechnology and biomedical applications. Creation of synthetic networks using these TALER devices will also be valuable for understanding systematic correlations between network motifs with solely transcriptional regulation and motifs with coupled transcriptional and microRNA-mediated post-transcriptional regulation in mammalian cells.
Figure 1. Design and construction of TALE repressors for mammalian cells.
(a) Schematic representation of TALE repressors with repeat di-variable residue (RVD) domains. The repeat domains are shown in orange blocks with RVDs in magenta and the binding specificity of RVDs is shown below. The last domain is a 15 amino acid half repeat domain depicted in a grey box. The green block represents the requirement of a leading repeat that binds T. The blue block represents a nucleus localization signal (NLS). (b) Design of a fluorescent reporter assay for measuring TALER repression fold and orthogonality. Tx: binding site for TALER x. Blue arrow represents a minimal CMV promoter. Lines with arrows indicate up regulation and lines with bars indicate down regulation. (c) Functional assay of TALERs in HEK293 cells. Selected RVD domains and binding sequences are shown on the left. Blue bars represent repression fold and red boxes represent repression percentage. Each bar/box shows mean ± SD from three independent flow cytometry experiments. (d) TALER orthogonality matrix. Each box in matrix represents fluorescent reporter expression measured in co-transfection of indicated TALER protein and promoter.
Results
Construction of TALER library and promoter optimization
To test whether an engineered TALER protein without a repression domain can inhibit transcription, we designed a new TALER promoter architecture that is transcriptionally inactivated upon TALER binding (Fig. 1b). The promoter contains five upstream activation binding sites (5×UAS) and a minimal CMV promoter flanked by two TALER binding sites. Trans-activator Gal4VP16 expressed from a constitutive promoter associates with UAS binding sites and activates the expression of a red fluorescent reporter gene, mKate2. In the presence of a TALER protein, the repressor binds its recognition sites at both the 5’ and 3’ of the minimal CMV promoter and blocks transcription. We chose 26 DNA sequences ranging from 12-bp to 24-bp previously shown to have strong affinity to their cognate TALE proteins in different contexts and no full matches within human promoter sequences except TALER24 (Supplementary Results, Supplementary Table 1)32, 33. By transient co-transfection in human embryonic kidney cell line (HEK293), we found that 23 out of our 26 TALER promoter/repressor pairs exhibit greater than 90% repression efficiency and 16 TALERs repress their cognate promoters more than 100 fold over the control (Fig. 1c). We then examined the orthogonality of the top ten strongest TALERs by measuring expression levels of each mKate2 reporter separately in the presence of each of the ten different TALER proteins. All of these TALERs exhibited strong repression toward their cognate promoters and had little effect on all other TALER promoters (Fig. 1d). For example, the repression efficiency of TALER 1, 9, 10, 12, 14 and 21 toward their cognate promoters is more than 100-fold stronger than against the other non-cognate promoters. To verify that binding sequences of the TALERs have minimal potential interference with endogenous transcriptome, we performed RNA sequencing analysis on cells transfected with these five strongest TALERs and cells transfected with a control circuit. While we found a few differentially expressed genes (False detection rate < 20% after Benjamini Hochberg correction, Supplementary Fig. 1), potential TALER targets (Supplementary Table 4) were not observed to have statistically significant differences in gene expression. Therefore, we chose these TALER proteins as building blocks for constructing the synthetic cascade and switch circuits, as discussed below.
Before construction of these circuits, we sought to further improve repression efficiency. First, we replaced either the 5’ TALER1 binding site or the 3’ TALER1 binding site with a non-cognate TALER binding site that serves as a mock sequence (Supplementary Fig. 2a). The results showed that the 3’ TALER1 binding site is essential for strong repression activity, while the 5’ TALER1 binding site alone has a much weaker effect. When both binding sites are present, higher repression fold was observed. We next examined whether the distance between two TALER binding sites affects the repression efficiency by inserting a spacer between the minimal CMV promoter and the 3’ TALER binding site. A series of promoters were engineered with distances ranging from 72-bp to 100-bp between the two binding sites (Supplementary Fig. 2b). Unlike the tetrameric LacI that shows periodic repression efficiency due to the phasing difference between two LacO sites34, we found that TALER does not display phasing behavior and that TALER binding sites closer to the minimal CMV promoter result in stronger repression. Subsequently, we constructed a few TALER promoters (pTxBS4) with three TALER binding sites positioned downstream of the minimal CMV promoter (Supplementary Fig. 2c). As desired, previously weak TALERs exhibited stronger repression with this optimization, although additional binding sites caused a modest reduction in fold repression for a previously efficient TALER (Supplementary Fig. 2c).
Next, we created a cell line that harbor a synthetic circuit with six genes and four regulatory units using our site-specific recombination35 (Supplementary Fig. 2d). EYFP fluorescence intensity is used to estimate expression of TALER14 protein upon doxycycline (Dox) induction and mKate2 intensity reflects regulated expression of the cognate TALER14 promoter. By alternatively adding and removing Dox in cell culture every eight days, we demonstrated that the TALER14 device exhibits reversible gene regulation during a period of 24 days (Supplementary Fig. 2d).
Modular construction of TALER cascade circuits
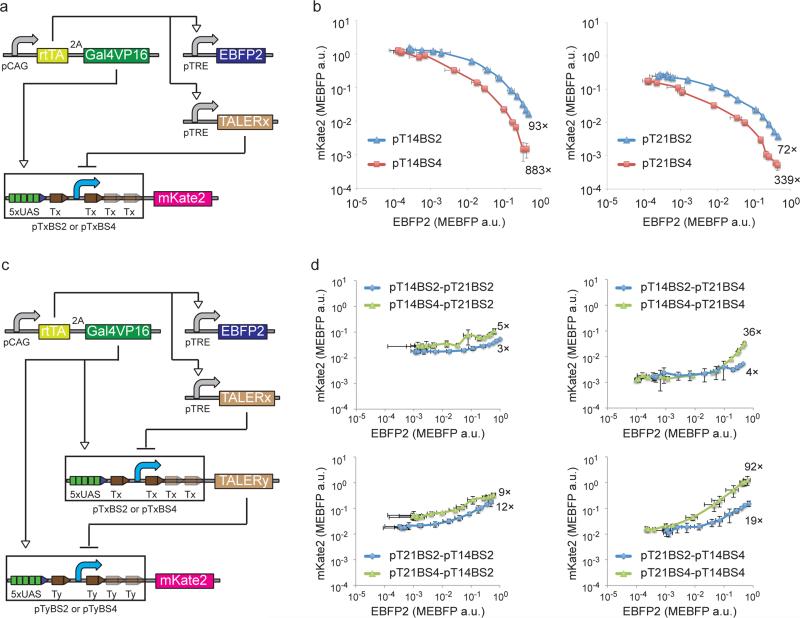
After creation and validation of strong and orthogonal TALERs, we proceeded to characterize the dose responses of our TALER devices with either two flanking binding sites (pTxBS2) or four binding sites (pTxBS4) (Fig. 2a). We co-transfected the circuit-encoding plasmids into HEK293 cells and measured EBFP2 and mKate2 levels in the presence of a series of Dox concentrations. We also established a color model that allows us to convert between EBFP2 and mKate2 measurements (Supplementary Fig. 3). This data, termed transfer curve (Fig. 2b), provided detailed characterization of TALER promoter response to different TALER protein concentrations, as approximated by EBFP2 fluorescence. The transfer curves revealed that three binding sites downstream of the minimal CMV promoter improved TALER 14 and 21 repression efficiency (Fig. 2b).
Figure 2. Design and construction of TALER cascade.
(a) Schematic representation of a circuit for measuring transfer curves. pTxBS2 and pTxBS4 are TALER promoters with two binding sites or four binding sites, respectively. (b) Circuit outputs for characterizing transfer curves indicated in (a) at varying Dox concentrations. Each data point shows mean ± SD from three independent replicates. (c) Schematic representation of a cascade circuit. For simplicity, pTxBS2 and pTxBS4 are depicted in the same diagram with two downstream binding sites in pTxBS4 promoter colored with reduced intensity. (d) Measured output from cascade circuit indicated in (c) at varying Dox concentrations. Each cascade is named by concatenating the name of the first TALER, ‘-‘, and the name of the second TALER. Each data point shows mean ± SD from three independent replicates.
We then investigated the development of composite circuits using TALERs as building modules. We concatenated two TALER protein/promoter pairs by placing one repressor gene under the control of another TALER promoter, thus making a TALER cascade (Fig. 2c), where the output should follow the input. For all eight possible cascades assembled by using TALER14 and TALER21 protein/promoter pairs with either two (pTxBS2) or four binding sites (pTxBS4), the output levels of the TALER cascades increased with increasing concentrations of Dox (Fig. 2d). The fold change between the minimum and maximum output levels of these cascades ranged from 3-fold up to 92-fold. We also examined noise propagation in the cascade circuits and observed that these two-stage cascades attenuate noise found in their single-stage TALER constituents in most cases (Supplementary Fig. 4). To test our ability to predict composition of multiple regulatory devices, we developed a computational model for each cascade by functionally composing two TALER transfer curves used in these cascades (Supplementary Fig. 5a and 5b). The observed and predicted cascade fold change correlated with an R squared of 0.81 (Supplementary Fig. 5c). These results support our ability to build and quantitatively predict the output induction fold of modularly assembled TALER cascades, although further efforts may be useful for improving the precision of the predictions.
TALER sensory switches controlled by synthetic shRNAs
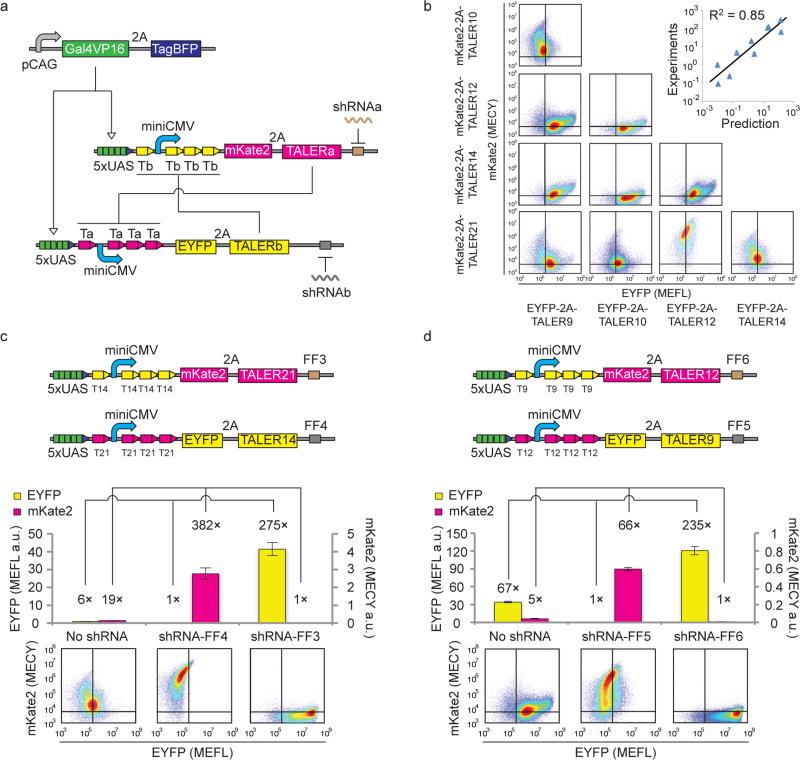
Genetic switches that can sense important cellular states are important for cellular decision-making in mammalian cells36, 37. Synthetic gene switches have been created previously using two cross-inhibiting transcriptional repressors, and the state of these switches was toggled by alleviating the repression of one transcriptional repressor with small external molecules4, 5. With strong TALER repressors in hand, we sought to construct TALER switches using two cross-repressed TALER devices and rewire microRNAs/shRNAs to control the state of the switch (Fig. 3a). For building blocks, we chose TALER 9, 10, 12, 14 and 21 and the corresponding promoters with four binding sites, since they were characterized above to be efficient and orthogonal repressors. A bicistronic construct with a self-cleaving 2A linker38 was used to express each TALER protein along with either mKate2 or EYFP fluorescent reporter. Input-output transfer curves were measured using a Dox-inducible system similar to the one described above (Supplementary Fig. 5d, 5e, and 5f), and a color model was established for conversion between EYFP and mKate2 measurements (Supplementary Fig. 3).
Figure 3. Design and construction of TALER sensory switches.
(a) Schematics of TALER switching circuits. Each TALER level is monitored by 2A-linked mKate2 or EYFP fluorescence. shRNAs are represented by wiggly lines, and shRNA targets are shown as blocks in brown or grey color. (b) Indicated TALER switches were co-transfected into HEK293 cells. Representative flow cytometry scatter plots measured 48 h post-transfection are shown in a matrix layout, where each row shows the same mKate2-linked TALER used in transfection experiments and each column represents the same EYFP-linked TALER used in transfection experiments. Correlation of the ratio of EYFP to mKate2 is shown in the top-right graph. Each data point shows mean ± SD from three independent replicates. (c) and (d) Setting the states of TALER21-TALER14 switches (c) or TALER21-TALER9 switches (d) by shRNAs. The upper panels represent schematics of TALER switches. Each bar shows mean ± SD of EYFP or mKate2 from three independent replicates. The lower panel represents representative flow cytometry scatter plots measured 48 h post-transfection.
We performed nullcline analysis by plotting the input-output transfer curve of each TALER against output-input transfer curves of all other TALERs (Supplementary Fig. 5g). In a nullcline graph, line intersections should indicate equilibrium states of TALER switches. We subsequently measured the performance of several TALER switches at 48 hours post transfection. The ratio of EYFP to mKate2 obtained by experiments and the ratio predicted by nullcline analysis correlate well with an R squared of 0.85 (Fig. 3b), suggesting that experimentally derived nullcline analysis is a useful approach for guiding the design of TALER switches. We observed two general trends among the TALER switches we tested. First, as expected, when two TALERs were imbalanced, the TALER switch tended to generate output from the stronger TALER device and repress output from the weaker one. Second, when the two TALERs were balanced, the output from both of the TALERs was relatively low at 48 hour (Fig. 3b).
We next tested whether synthetic shRNAs can be used to modulate the state of the switches by placing four tandem repeats of shRNA target sites in the 3’UTR of TALER genes. TALER pairs were co-transfected without shRNA or with the corresponding shRNA. Our result validated that shRNA input can set both balanced and imbalanced switches to either state for all TALER pairs tested at 48 hours post-transfection (Fig. 3c, 3d, and Supplementary Fig. 6) except for one imbalanced TALER12-TALER21 switch comprising the weakest and strongest TALERs (Supplementary Fig. 6e).
TALER sensory switch improves cell-type classification
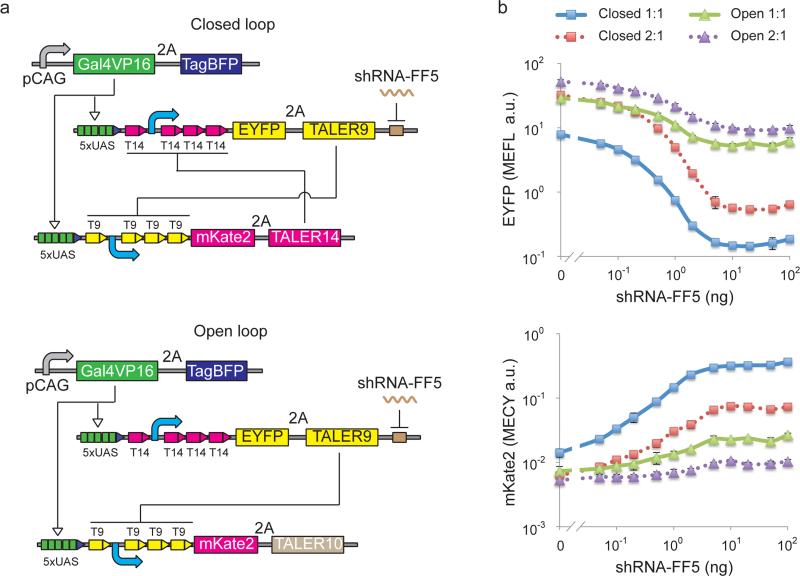
We have previously demonstrated a synthetic multi-input logic circuit that can specifically identify HeLa cervical cancer cells by sensing and performing logic computation based on a HeLa-specific microRNA expression profile18. This classifier circuit required extensive optimization to reduce leaky output expression because our initial HeLa-high microRNA sensors had lower than desired signal-to-noise ratio. In a relevant study, it has been shown that mutual inhibition can function as a robust switch for cell polarization14. We hypothesized that an imbalanced TALER switch would improve microRNA/shRNA sensing and yield a higher ON/OFF ratio. Computational simulation using TALER transfer curves suggests that improved sensitivity can be obtained from closed-loop (Online Methods and Supplementary Fig. 7). To verify the prediction experimentally, we chose TALER9 and TALER14, an imbalanced TALER pair, and constructed a closed-loop sensory switch with mutual inhibition that could be regulated by synthetic shRNA-FF5, and compared its behavior to an open-loop switch where TALER9 represses the promoter of TALER10 but not vice versa (Fig. 4a). In this experiment, the fold-change of EYFP was used to compare the sensitivity to shRNA of the closed-loop switch to that of the direct sensing circuit, and the fold-change of mKate2 was used to compare the sensitivity to shRNA of the closed-loop switch to that of the cascade circuit. When an equal DNA concentration of two TALER encoding plasmids was used in transfection experiments, we observed ~26 fold mKate2 induction and ~54 fold EYFP reduction without or with 100 ng shRNA-FF5 for the closed-loop switch, in comparison to ~4 fold mKate2 induction and ~5 fold EYFP reduction for the open-loop switch (Fig. 4b and Supplementary Fig. 8). When a 2:1 ratio of TALER9 and TALER14 was used, in comparison to the amount needed for equal DNA concentration of transfected TALERs, roughly eight times more shRNA was needed to induce mKate2 or reduce EYFP to the same levels (Fig. 4b). These results suggested that a closed-loop TALER sensory switch can significantly improve signal sensitivity and precision for microRNA sensing, and that the sensitivity to microRNA input is tunable by changing the ratio of the two cross-repressing TALERs.
Figure 4. Response of TALER sensory switches to shRNA inputs.
(a) Schematic representation of a closed-loop TALER switch with mutual inhibition and the open-loop counterpart without mutual inhibition. (b) TALER switch behavior in response to varying amounts of shRNA inputs. Ratio of TALER9 to TALER14 (or TALER10) used in transfection experiments is indicated as 1:1 or 2:1. Each data point shows mean ± SD of fluorescent reporter from three independent replicates. Closed: closed-loop circuit; Open: open-loop circuit.
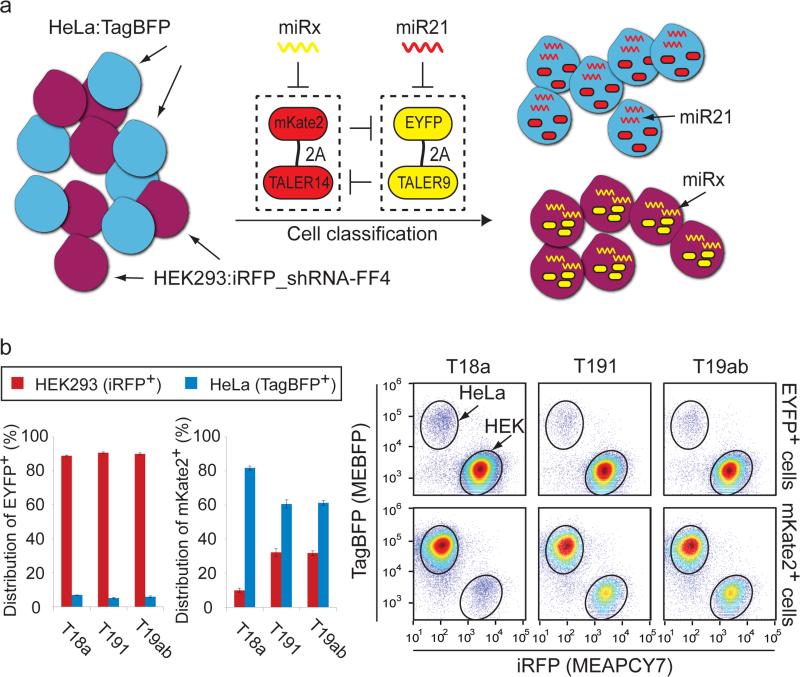
Next, we sought to connect endogenous microRNAs to control TALER switches. We engineered a HeLa cell line (HeLa:TagBFP) that expresses a blue fluorescent protein TagBFP and a HEK293 cell line (HEK293:iRFP_shRNA-FF4) expressing a near-infrared fluorescent protein iRFP39 and synthetic shRNA-FF4 (Supplementary Fig. 9a). HeLa-specific miR2118 and HEK293:iRFP_shRNA-FF4 specific shRNA-FF4 were used to control the TALER9-TALER14 switch by placing four tandem repeats of miR21 and shRNA-FF4 target sites in the 3’-UTR of EYFP-2A-TALER9 and mKate2-2ATALER14 constructs, respectively (Fig. 5a). The TALER9-TALER14 switch with different ratios was transfected into a mixture of HeLa:TagBFP and HEK293:iRFP_shRNA-FF4 cells. We found that for all ratios, the switch generated high EYFP and essentially no mKate2 in HEK293:iRFP_shRNA-FF4 cells, whereas the switch expressed high mKate2 and almost no EYFP in HeLa:TagBFP cells (Supplementary Fig. 9b), suggesting that the switch can function with a wide range of individual TALER concentrations. The most accurate cell identification was observed when the switch with a 1:1 ratio was transfected into a HEK293/HeLa cell mixture (Supplementary Fig. 9c).
Figure 5. Connecting microRNAs to regulate TALER sensory switches enables cell-type classification.
(a) Experimental outline. HEK293 cells (purple circle) that express a synthetic shRNA-FF4 and iRFP along with HeLa cells (blue circle) that express TagBFP were transfected with TALER9-TALER14 switches in response to indicated microRNAs or shRNA-FF4. (b) TALER switches controlled by HeLa-specific miR21 and indicated HEK293-specific microRNA. The bar chart shows the fraction of engineered HEK293 and HeLa in EYFP+ or mKate2+ cell population. Each bar shows mean ± SD from three independent replicates. FACS data is shown in the scatter plot. The upper row shows distribution of EYFP+ cells on iRFP-TagBFP scatter plots, and the lower row shows distribution of mKate2+ cells on iRFP-TagBFP scatter plots. Circles indicate engineered HEK293 or HeLa cell population.
A microRNA expression database40 indicates that the expression level of miR18a, miR191 and miR19ab (i.e. miRNA19a plus miRNA19b) are high in HEK293 cells but low in HeLa cells (Supplementary Fig. 10a). A fluorescent reporter assay in the mixed cell population suggested that miR18a, but not miR191 or miR19ab, displays strong RNAi knockdown in engineered HEK293 and HeLa cells (Supplementary Fig. 10b and 10c) and miRNA sensors with no mutual inhibition did not efficiently classify cells (Supplementary Fig. 10d). We then replaced the FF4 target sites downstream of TALER14 with target sites for either miR18a, miR191, or miR19ab. As shown in Fig. 5b and Supplementary Fig. 11a, the TALER14-TALER9 sensory switch with target sites for HEK293-specific miR18a and HeLa-specific miR21 provided the best cell classification among all three pairs tested, suggesting that TALER switches can be efficiently regulated by endogenous microRNA inputs. To compare the behavior of the mutual-inhibiting TALER switch to an open-loop counterpart which represents a simplified version of our previous classifier circuit18, we replaced TALER14 with TALER10 in the TALER switch where TALER9 represses the promoter of TALER10 but not vice versa (Supplementary Fig. 11b). The results demonstrated that although both the closed-loop and open-loop switch displayed similar accuracy for identification of engineered HeLa cells, the closed-loop circuit showed an improved accuracy for identification of engineered HEK293 cells with ~2.5-fold less misclassified HeLa:TagBFP cells over the open-loop control (Supplementary Fig. 11c). In addition, the mKate2 level of the closed-loop switch in engineered HeLa cells is 7 fold higher over the open-loop counterpart (Supplementary Fig. 11c and 11d).
Discussion
Engineering complex synthetic gene circuits is often hindered by a lack of orthogonal and reversible transcriptional repressors. Here, we introduce a framework for quickly engineering reversible and orthogonal TALE repressors and their cognate promoters in mammalian cells that is based on steric hindrance of key transcriptional initiation elements. Given the modularity of TALE DNA binding domains bearing different variable di-residues (RVDs) combinations41, the TALER library can be readily extended by generating a large number of TALERs with different repression activities in a short time with high throughput TALE assembly methods32, 33, 42. It may also be possible to engineer TALE repressors to target endogenous genes using a similar steric-hindrance strategy when transcription initiation sites of the target genes are well understood.
Synthetic biology aims to apply engineering principles for modular construction of synthetic gene circuits based on well-defined devices. However, quantitative description of synthetic components and prediction of assembled circuits in mammalian cells remains a significant challenge. In this work, based on a multi-fluorescent Dox-inducible system, we measured the input and output levels of TALER devices and established color models that allow us convert between input and output fluorescent levels. We showed that experimentally derived input-output transfer curves of individual TALERs can be used to quantitatively predict the behavior of modularly assembled TALER cascades and switches. For further improvements, binning analysis is likely to help eliminate the copy number variations introduced by transient transfection12. It will also be necessary to consider TALER dynamics for predictions of a variety of other circuit motifs.
It has been demonstrated that coordinated transcriptional and microRNA-mediated posttranscriptional regulation often occurs in feedback and feed-forward motifs, which plays an important role in controlling gene expression in natural systems43. In addition, coupled transcriptional and microRNA-mediated motifs may display different systematic properties than their solely transcriptional counterparts12. However, studying the underlying design principles of core regulatory motifs is often hampered by undesired cross regulation between the core motif and surrounding nodes in the natural genetic network. Our library of orthogonal and well-characterized TALERs serves as a valuable tool for constructing isolated synthetic circuits that couple transcriptional and microRNA-mediated post-transcriptional regulation to help elucidate the design principles of core regulatory motifs. For instance, we demonstrated that the closed-loop TALER switch displays a superior state transition in response to an shRNA input as compared to the open-loop counterpart. It will be interesting to compare behavior of a switch with solely transcriptional regulation to that of a switch with coupled transcriptional and microRNA-mediated post-transcriptional regulation. Furthermore, it has been shown that three minimal core motifs including a motif with mutual inhibition can generate spontaneous polarity on cell membrane14. Similarly, evaluating the effect of variant topologies on the performance of a TALER switch by adding or eliminating appropriate positive and negative feedback would enable an improved understanding of the design principles for robust TALER switches.
Sophisticated control of living cells demands synthetic gene circuits that are capable of sensing multiple endogenous molecular signals. RNA interference provides a versatile interface that allows synthetic gene circuits to modularly sense and integrate endogenous molecular inputs in mammalian cells18, 44-48. In this work, we demonstrated that endogenous microRNAs can be used to control the state of TALER sensory switches and that the sensitivity of TALER switches to shRNAs is tunable by adjusting the ratio of the two primary components. We have also shown that two cell-type specific microRNAs can tightly regulate the TALER switch outputs, resulting in precise cell-type classification for a mixed cell population. Accordingly, our results can facilitate construction of TALER switches for sensing cell-type specific microRNAs expressed at different levels by selecting appropriate TALERs from our TALER library. In addition, TALER switches can be used as building blocks together with combinatorial logics21 for the construction of more sophisticated logic circuits, for example sensing a multi-input cell-type specific microRNA profile by using the logic design framework as described previously18, as well as for engineering reversible memory devices to track cellular events and signal processing5, 7, 8. In the future, our TALER switches could be used in a variety of biomedical applications, provided that hurdles such as efficient in vivo delivery to cells and long-term functional stability are addressed.
Online methods
Reagents and enzymes
Restriction endonuclease, polynucleotide kinase (PNK), T4 DNA ligase, and Quick DNA ligase were purchased from New England Biolabs. Accuprime pfx supermix DNA polymerase (Life technologies) and Phusion High-Fidelity DNA polymerase (New England Biolabs) were used in PCR amplification. Oligonucleotides were synthesized by Integrated DNA Technologies and Genewiz. Doxycycline was purchased from Clontech. Gateway BP reaction (Life technologies) and LR reaction (Life technologies) were performed by following manufacturer's protocol.
Plasmid DNA constructs
All plasmid sequences were submitted to GenBank, with accession number KM486811 ~ KM486934 and KM519784 ~ KM519788.
TALER RVD sequences are listed in Supplementary Table 1. When required, equal molar amounts of oligonucleotides were annealed in 1× PNK buffer by heating to 95 °C and gradually cooling down (− 1 °C per min) to 37 °C, and then 1 μM of annealed product was phosphorylated by 0.5 unit/μL PNK in presence of 0.5 mM ATP (New England Biolabs).
The vectors for making stable cell line in Supplementary Fig. 2d were made as described35. Briefly, plasmid DNAs were pooled 7 fmol each and digested with I-SceI. The digested DNA mixture was added to a Gibson reaction buffer with integrative carrier vector and adapter vector.
Cell culture and transfection
HEK293 (293-H) cell line was purchased from Invitrogen. HeLa (CCL.2) cell line was originally obtained from ATCC. HEK293 and HeLa cells were cultured in high-glucose DMEM complete media (Dulbecco's modified Eagle's medium (DMEM), 4.5 g/L glucose, 0.045 units/mL of penicillin and 0.045 g/mL streptomycin and 10% FBS (Invitrogen)) at 37 °C, 100% humidity and 5% CO2.
In transfection experiments with individual cell lines, ~ 6 × 104 HEK293 cells in 0.5 mL of high-glucose DMEM complete media were seeded into each well of 24-well plastic plates (Falcon) and grown for ~ 24 h. In transfection experiments with cell line mixtures, ~ 1.5 × 104 HEK293:iRFP_shRNA-FF4 cells were mixed with ~ 4.5 × 104 HeLa:TagBFP cells in 0.5 mL of DMEM complete media, then the mixture was seeded into each well of 24-well plastic plates and grown for ~ 24 h. Shortly before transfection, the medium was replaced with fresh DMEM complete media. Attractene transfection reagent (Qiagen) was used in experiments for Fig. 1 and Supplementary Fig. 2 as described in the manual. Lipofectamine LTX (Life technologies) was used in the rest of experiments in this study by following manufacturer's protocol. Briefly, a sample mixture was prepared by mixing the required amounts of plasmid DNAs with 1.5 μL Attractene and incubated at room temperature for 20 min before adding to cells. Alternatively, a sample mixture was prepared by mixing the required amounts of plasmid DNAs with Plus reagent and incubated at room temperature for 5 min, then the mixture was added with 1.2 μL Lipofectamine LTX and incubated for 25 min before adding to cells. pDT7004 (pUBI-linker-NOS) that contains a maize ubiquitin promoter (UBI) followed by a NOS terminator with no protein-coding sequences between UBI and NOS was used to ensure equal amount of plasmid DNA18. The amount of plasmid DNA and the final concentration of Doxycycline added to each well are listed in the Supplementary Table 2. Cells were cultured for 2 days before flow cytometry analysis as described18.
Construction of stable cell lines
Information for plasmid DNA constructs used in Supplementary Fig. 2d is provided in the Supplementary Information. The assembly of circuits was performed as described35. Briefly, an integration vector was co-transfected with the recombinase expressing vector into HEK293 cells. Stable clones were selected with 1 μg/mL of Puromycin (InvivoGen) and 10 μg/mL Blasticidin (InvivoGen). Cells were sorted on mKate2 negative population using flow cytometry. Subsequently, cells were cultured in the presence of doxycycline (1 μg/mL). Then cells were sorted on EYFP positive, and mKate2 negative using BD FACSAria III cell sorter.
FACS measurement
Cells are trypsinized 48 h post-transfection and are then centrifuged at 250 g for 10 min at 4 °C. The supernatant is removed and the cells are resuspended in 1× PBS that does not contain calcium or magnesium. Fortessa flow analyzers (BD Biosciences) were used for fluorescence activated cell sorting (FACS) analysis at different experimental sites with the following set of settings. EBFP2 and TagBFP were measured using a 405 nm laser and a 450/50 filter with a photomultiplier tube (PMT) 260 V. EYFP was measured with a 488 nm laser and a 530/30 filter using a PMT 260 V. mKate2 was measured with a 561 nm laser and a 670/30 filter using a PMT 400 V. iRFP was measured using a 640 nm laser and a 780/60 filter with a PMT 450 V. DsRed was measured using a 561 nm laser and a 586/15 filter with a PMT 400 V. For each sample, ~ 1 × 104 to ~ 1 × 105 cell events were collected. In parallel, Rainbow Calibration Particles (Spherotech Inc.) were measured in order to standardize the data between different instruments and settings (see Data analysis below).
Data analysis
We used Rainbow Calibration Particles to convert relative fluorescent units to standardized units as described with minor modifications18. As a rule we did not resolve all the peaks but a few brightest ones. We plotted peak intensity in units of absolute fluorescence (MECY for mKate2 channel, MEFL for EYFP channel, MEAPCY7 for iRFP channel and MEBFP for TagBFP or EBFP2) versus measured peak intensity for a given set of machine settings. We connected adjacent dots on the plot with straight lines, creating a linear calibration curve that translates machine units into absolute fluorescence intensity. For example, a linear relationship of MEFL versus EYFP was established in logarithmic scale using mean EYFP fluorescence of particle peaks and their absolute MEFL units. Similar linear relationships were established for MEBFP versus TagBFP or EBFP2, MECY versus mKate2 and MEAPCY7 versus iRFP. Scatter plots were drawn using beads-calibrated data in Matlab (MathWorks) with a modified Flow Cytometry Data Reader and Visualization tool.
The normalized fluorescence (NFL) was calculated using the following formula in order to compensate for differences in transfection efficiencies between different samples, where a constitutively expressed fluorescence reporter was used as a transfection internal control.
where, mean(FL) is the mean value of MECY, MEFL or MEBFP in the measured cell population, and mean(Control) is the mean value of the constitutively expressed fluorescent reporter in the measured cell population. Then we calculated percentage (p) of repression and repression fold as the following:
To establish a color model that maps the relationship between different fluorescence used as either input or output in TALER devices, we measured intensity of fluorescent reporters without or with a 2A-tag using a Dox-inducible system (Supplementary Fig. 3). A linear regression in logarithmic scale was established using normalized fluorescence for EYFP versus EBFP2 and EYFP versus mKate2, respectively (Supplementary Fig. 3), which allows unit conversion between different fluorescent reporters.
A similar Dox-inducible system was used to measure TALER transfer function curves (Fig. 2 and Supplementary Fig. 5) with a variety of Dox concentrations as indicated in Supplementary Table 2. With an assumption that TALER input and output reached equilibrium when measured, beads-calibrated and normalized fluorescent intensities were used for modeling the Hill function of TALERs as follows.
Where B is the fluorescent intensity of the output, A is the fluorescent intensity of the input which represents the concentration of TALER, β2 represents the maximum production rate from TALER promoter, β1 represents the leakage production rate from TALER promoter, k is the input concentration that gives 50% repression, n is the Hill coefficient, and γ is the degradation rate.
For cascade fold prediction, the input and output of each TALER were first standardized to MEBFP by linear interpolation. Then, the output from the first TALER was used as the input of the second TALER. The cascade circuit was simulated with the input range set to that of the experimentally measured input of the first TALER and the maximum fold change was calculated from the output of the second TALER.
In nullcline analysis, transfer curves for a pair of TALERs were plotted together with one transfer curves inverted around the y=x diagonal line, effectively flipping its input and output. With the standardized unit used for both axes, line intersections of two transfer curves were the predicted equilibrium states of TALER switches. To evaluate the prediction accuracy of TALER switches using the nullcline analysis, a linear regression was established using the ratio of EYFP to mKate2 at the intersection points in nullcline analysis shown in Supplementary Fig. 5g and the ratio of mean value of EYFP to mean value of mKate2 in the measured cell population shown in Fig. 3b.
Modeling of TALER sensory switches
We modeled the TALER sensory switch by combining two cross-repressing single TALER models, of which one is stronger than the other. The shRNA regulatory effect is modeled into varying TALER degradation rate, which is dependent on shRNA dosage.
Where B is the concentration of the weaker TALER, A represents the concentration of stronger TALER, which is regulated by shRNA, β2 represents the maximum production rate from TALER promoter, β1 represents the leakage production rate from TALER promoter, k is the input concentration that gives 50% repression, n is the Hill coefficient, and γ is the degradation rate. shRNA silencing is also modeled using Hill functions, in which shRNA represents the concentration of the shRNA, θ is the maximum degradation rate by shRNA relative to the degradation rate without shRNA, and m is the Hill coefficient for shRNA regulation.
We considered two conditions when there is no shRNA and when there is maximum amount of shRNA. In the first condition, we set the shRNA = 0, and assumed that the stronger TALER can fully repress the weaker TALER. We can obtain
We considered two conditions when there is no shRNA and when there is maximum amount of shRNA. In the first condition, we set the shRNA = 0, and assumed that the stronger TALER can fully repress the weaker TALER. We can obtain In the maximum shRNA repression condition, we set the shRNA >> k2, and we can derive
The fold change for A is
Compared to open loop circuit, whose fold change is 1 + θ, the increase in fold change is
We experimentally measured the transfer function of shRNA-FF5 knockdown on a TALER and obtained the fitting parameters. We then simulated an shRNA sensor made of TALER9 and TALER14, which is regulated by shRNA-FF5, with β2 and β1 in the Hill functions scaled to compensate for the difference.
To model the cell classification circuit based on TALER switch, we combined two transfer functions using the same method described in nullcline analysis. We scaled the parameters β2 and β1 in the hill function to simulate the effect of shRNA/miRNA regulation. We defined the correct classification to be high TALERb output and low TALERa output for the case where the miRNA regulating TALERa is present and the miRNA regulating TALERb is absent.
Construction of stable cell lines used in cell mixture experiments
For production of lentiviral particles, ~ 2 × 105 HEK293-FT cells in 1 mL of DMEM complete media were plated into 12-well and grown for ~ 24 h. Then cells were co-transfected with the expression vector (KM519787 or KM519788), the packaging plasmid pCMV-dR8.2 (Addgene) and the envelope plasmid pCMV-VSV-G (Addgene) using Lipofectamine LTX with Plus reagent. Media containing viral particles produced from transfected HEK293-FT cells were harvested ~ 48 h post-transfection and centrifuged to remove cell debris. 2 mL of the supernatant and 10 μg/mL of polybrene (Millipore) were added to ~ 2 × 105 HEK293 (KM519788) or HeLa (KM519787) cells in 24-well plate seeded 24 h prior infection. After 72 h, Blasticidin (InvivoGen) was added into media to a final concentration of 5 μg/mL and the cells were grown for another 6 days. After Blasticidin concentration was raised to 10 μg/mL for 2 days, FACS analysis confirmed that ~ 95% of HeLa:TagBFP cells were TagBFP positive (Supplementary Fig. 10a) and ~ 40% of HEK293:iRFP_shRNA-FF4 cells were iRFP positive. To enrich iRFP positive cells in HEK293:iRFP_shRNA-FF4 stable cell line, cells were trypsinized and centrifuged at 300 g for 5 min. Then, cells were resuspended in 1× PBS with 10% FBS (Invitrogen) and 1% sodium pyruvate (Invitrogen). HEK293:iRFP_shRNA-FF4 cells were sorted on a BD AriaII equipped with a Red laser tuned to 640 nm for the excitation wavelength with a 780/60 bandpass filter in APC-Cy7 channel. The top of ~ 10% iRFP positive HEK293:iRFP_shRNA-FF4 cells were collected in DMEM complete media, and plated into 6-well and cultured at 37 °C, 100% humidity and 5% CO2. We used FACS analysis to measure iRFP positive percentage in the enriched HEK293:iRFP_shRNA-FF4 cells (Supplementary Fig. 9a). HEK293:iRFP_shRNA-FF4 and HeLa:TagBFP cells are distinguished by using iRFP/TagBFP scatter plot (Supplementary Fig. 9a).
TALE binding sequence analysis
We performed a blast analysis using TALER DNA binding sequences against promoter sequences around transcription start site (−200 to +100) of human and mouse genome obtained from EPD (eukaryotic promoter database) database. The number of match for each TALER DNA binding sequence in the blast results was counted.
RNA library preparation and sequencing
HEK293 cells were transfected in triplicate using the same procedure described in the cell culture and transfection method. The transfection mixture of TALER device is the same as that used in Fig. 1 and is described in Supplementary Table 2. The transfection mixture of control circuit is the same as that of the TALER device except that EYFP-2A-Hygro is used instead of EYFP-2A-TALER. Three days after transfection, cells were harvested. Total RNA was extracted using RNeasy Plus Mini Kit (Qiagen) and was eluted in to 50 μL TE buffer and 1 μL of extracted RNA was processed for cDNA synthesis. From each sample a cDNA library was prepared following the SMART-seq2 protocol as described49, only replacing the reverse transcriptase enzyme with 0.1 μL of Maxima H Minus enzyme (200 U/μL, Thermo Scientific), and scaling down the PCR reaction to a volume of 25 μL. The tagmentation reaction and final PCR amplification were done using the Nextera XT DNA Sample preparation kit (Illumina), with the following modifications: all reaction volumes were scaled down by a factor of 4, and the libraries were pooled after the PCR amplification step by taking 2.5 μL of each sample. The pooled libraries were cleaned and size-selected using two rounds of 0.7 volume of AMPure XP SPRI bead cleanup (Beckman Coulter Genomics). Samples were loaded on a High-Sensitivity DNA chip (Agilent) to check the quality of the library, while quantification was done with Qubit High-Sensitivity DNA kit (Invitrogen). The pooled libraries were diluted to a final concentration of 4 nM and 12 pM and were sequenced using the Illumina MiSeq Personal Sequencer (Life Technologies) with 50 bp paired end reads.
RNA library data analysis
Bowtie2 index was created based on the human UCSC genome and known Gene transcriptome19, and paired-end reads were aligned directly to this index using Bowtie with command line options -q --phred33-quals -n 2 -e 99999999 -l 25 -I 1 -X 1000 -a -m 200 –p 4 --chunkmbs 512. Next, RSEM v1.27 was run with default parameters on the alignments created by Bowtie to estimate expression levels. The gene expression level estimates (tau) calculated by the RSEM program were multiplied by 1,000,000 to obtain transcript per million (TPM) estimates for each gene. Genes were filtered if the number of samples having TPM higher than 5 is less than 2. Based on this criterion, 7296 genes were filtered out. A gene is considered detected if its TPM is higher than 2, and there are 9043 genes detected on average for each sample. Then, samples were normalized using normalization scaling factors calculated in DESeq package50. To find differentially expressed genes (DE) in samples transfected with each TALER device compared to samples transfected with control circuit, two-sample t-test (Matlab V2013b) was used on log transformed data (log(TPM+1)), and correction for multiple hypothesis testing was performed (Benjamini Hochberg procedure, FDR<20%). Then, for each TALER device, genomic sequences including 1000 bp upstream and downstream of identified DE genes were used in searching for potential TALER binding sites. Hamming distances were calculated for each position along genomic sequences of DE genes in a sliding window of the size of a TALER binding site, and minimal Hamming distances were found for each DE genes for each TALER.
Variance analysis of TALER devices and cascade circuits
For the systems of individual TALERs and TALER cascades (Supplementary Fig. 4a and 4b), all cell events were firstly sorted on EYFP intensity (MEFL) and then EYFP+ events were divided into 51 bins at equal logarithmic MEFL scale. In each bin, mean value of EBFP2 in MEBFP and coefficient of variance (CV) of mKate2 in MECY were calculated. The moving average of mean (EBFP2) and CV (mKate2) values calculated in Matlab with default span are plotted in Supplementary Fig. 4.
Supplementary Material
Acknowledgments
We thank members of Xie lab and Weiss lab for helpful discussions. We thank Huiya Huang and Tingting Wang for technical support. We thank Cong Le and Feng Zhang for providing TALE1 and TALE2 as gifts. We thank Jacob Beal and Michael Q. Zhang for insightful discussions. The research is supported by Beijing Natural Science Foundation (5152013, Z.X.), National Key Basic Research Program of China (2014CB745200, Z.X.), Junior “1000 Plan” Program (Z.X.), THU-NTHU Joint Grant (2012THZ0, Z.X.), and US National Institute of Health Grants (5R01CA155320-04, P50GM098792, and 1R01CA173712-01, R.W.).
Footnotes
Author Contributions
Z.X., R.W. and Y.L. conceived of the ideas implemented in this work. Y.L. and Z.X. performed TALER promoter optimization experiments. Y.L. constructed the TALER library and circuits for stable cell lines, performed orthogonality analysis, and initial experiments for transfer function curves, TALER switches, and stable cell lines. Y.J. performed experiments for transfer function curves, color model, TALER switches and cell classification. H.C. performed initial experiments for TALER switches and cell classification. W.L. performed experiments for stable cell lines and cell classification. Z. L. performed initial tests for TALE transfer function curves. Z.X., Y.L., W.L. and R.W. performed data analysis. Z.X. and R.W. supervised the project. Z.X., Y.L. and R.W. wrote the paper.
Competing Financial Interests
A pending patent application recently submitted by the authors is related to certain aspects of the work described in this manuscript.
Supplementary Information
Supplementary information includes nine figures and four tables.
References
- 1.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 3.Prindle A, et al. A sensing array of radically coupled genetic ‘biopixels’. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 5.Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 6.Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci U S A. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galloway KE, Franco E, Smolke CD. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341:1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, et al. Sequential establishment of stripe patterns in an expanding cell population. Science. 2011;334:238–241. doi: 10.1126/science.1209042. [DOI] [PubMed] [Google Scholar]
- 11.Bacchus W, et al. Synthetic two-way communication between mammalian cells. Nat Biotechnol. 2012;30:991–996. doi: 10.1038/nbt.2351. [DOI] [PubMed] [Google Scholar]
- 12.Bleris L, et al. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol Syst Biol. 2011;7:519. doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei P, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–388. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chau AH, Walter JM, Gerardin J, Tang C, Lim WA. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell. 2012;151:320–332. doi: 10.1016/j.cell.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–623. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 17.Lienert F, et al. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013;41:9967–9975. doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 19.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 20.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 21.Gaber R, et al. Designable DNA-binding domains enable construction of logic circuits in mammalian cells. Nat Chem Biol. 2014;10:203–208. doi: 10.1038/nchembio.1433. [DOI] [PubMed] [Google Scholar]
- 22.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 23.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–5187. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Scientific reports. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 31.Rinaudo K, et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller J, Oehler S, Muller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 35.Guye P, Li Y, Wroblewska L, Duportet X, Weiss R. Rapid, modular and reliable construction of complex mammalian gene circuits. Nucleic Acids Res. 2013;41:e156. doi: 10.1093/nar/gkt605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Zhu X, Hood L, Ao P. From Phage lambda to human cancer: endogenous molecular-cellular network hypothesis. Quantitative Biology. 2013;1:32–49. [Google Scholar]
- 38.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 39.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 42.Liang J, Chao R, Abil Z, Bao Z, Zhao H. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth Biol. 2014;3:67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- 43.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol Syst Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Z, Liu SJ, Bleris L, Benenson Y. Logic integration of mRNA signals by an RNAi-based molecular computer. Nucleic Acids Res. 2010;38:2692–2701. doi: 10.1093/nar/gkq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nat Nanotechnol. 2010;5:666–670. doi: 10.1038/nnano.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2011;39:2981–2994. doi: 10.1093/nar/gkq954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashida S, Inoue T, Saito H. Three-dimensionally designed protein-responsive RNA devices for cell signaling regulation. Nucleic Acids Res. 2012;40:9369–9378. doi: 10.1093/nar/gks668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 50.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.