Dear Editor
Temperature-sensitive (ts) mutations have provided fundamental insight into in our understanding of gene function in vivo. Although they are rare in higher organisms, temperature sensitive mutations in the pigmentation pathways include tyrosinase ts mutations found in oculocutaneous albinism (OMIM: 606952), in Siamese cats and the Himalayan mouse (Giebel et al., 1991), and in kit in zebrafish (Rawls and Johnson, 2001). Microphthalmia-associated transcription factor (Mitf) is the master melanocyte transcription factor that has critical functions in melanocyte development, melanocyte stem cell renewal and the tanning response, and is an important drug target(Hsiao and Fisher, 2014). Given the importance of MITF, it is critical to develop laboratory animals that enable conditional control of MITF activity in different temporal, cellular, developmental and disease contexts. Conditional or ts alleles in MITF have not been described in mammals. We (SLJ, JAL) have previously identified the first conditional MITF mutation (mitfavc7) in an animal in an ENU-based genetic screen in zebrafish, but the molecular mechanism underlying this mutation was unknown (Johnson et al., 2011). Here, we explain the molecular mechanism that enables the mitfavc7 mutation to confer conditional temperature dependent control of MITF activity in zebrafish, and describe an unusual intron mutation that leads to aberrant splicing of wild type and dominant negative splice variants at the restrictive temperatures.
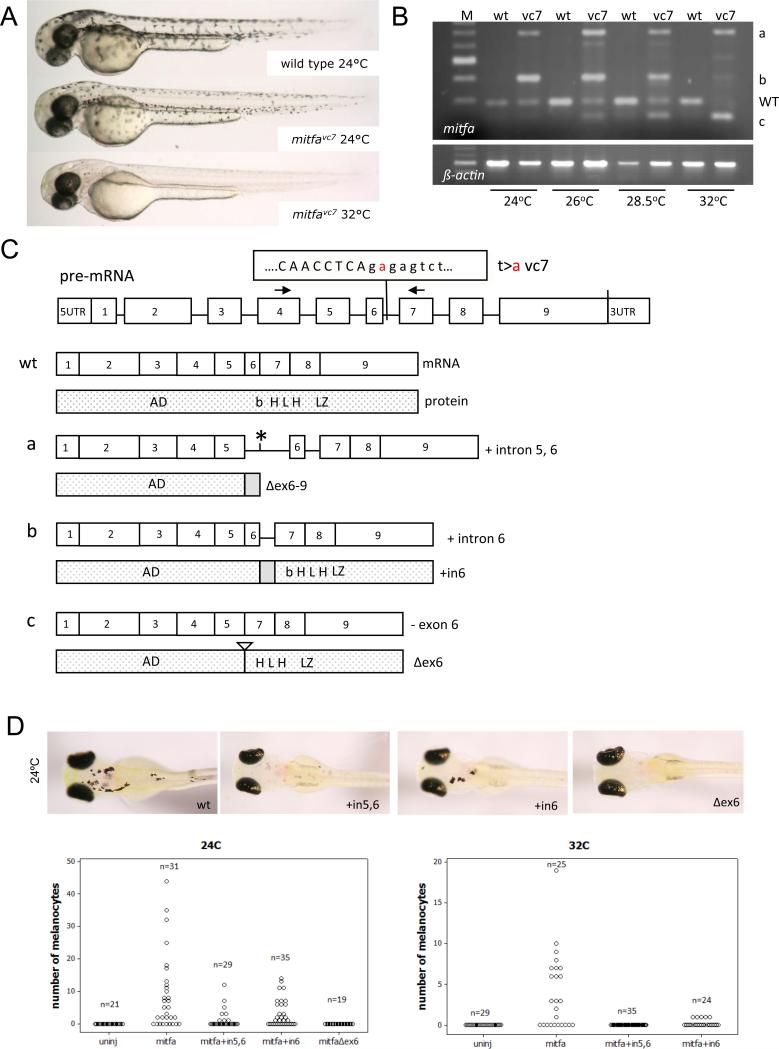
Most temperature-sensitive mutations affect exon sequences, but the mitfavc7 mutation is unusual because it is in an intron splice donor (t>a at position 39638 in mitfa genomic locus or position 2 in intron 6) (Johnson et al., 2011). We collected wild type and mitfavc7 mutant zebrafish embryos, and grew the animals at 24°C, 26°C, 28.5°C and 32°C. At higher temperatures zebrafish embryos grow more rapidly, and to compensate for this, for all experiments, zebrafish embryos were carefully stage-matched. At 32°C mitfavc7 zebrafish embryos lack all neural crest derived melanocytes due to loss of mitfa activity (Figure 1A). At 24°C, wild type and mitfavc7 zebrafish embryos develop similar numbers of clearly visible and pigmented melanocytes (Johnson et al., 2011), albeit with a delay in differentiation and/or cell size in the mitfavc7 mutant embryos at two days post-fertilization (Figure 1A). We examined the mitfa RNA encompassing exons 4-7, and found correct splicing occurs at low (permissive) temperatures along with aberrant splicing, while aberrant splice forms predominate with increasing temperature (Figure 1B; as described in the Supplementary Methods). Temperature-sensitive splicing at this locus had not previously been detected (Johnson et al., 2011), possibly due to PCR conditions. We cloned each of the splice forms (labeled a-c), and found that the aberrant splice forms include those that skip exons or retain introns (Figure 1C). Splice form a includes intron 5 and 6 (mitfa+in5,6), form b includes an in-frame intron 6 (mitfa+intron6), and splice form c leads to an in-frame deletion of exon6 (mitfaΔex6). At the highest temperature (32°C), almost no wild type m/t/a or mitfa+in6 was detectable, while there was strong expression of mitfaΔex6. These results demonstrate that aberrant of the mitfa RNA correlates with the temperature sensitivity of the mitfavc7 mutation.
Figure 1. The mitfavc7 intron 6 mutation causes defective temperature-dependent splicing of mitfa.
A. Images of mitfavc7 zebrafish embryos (2 days post fertilization) at 24°C (permissive temperature) and 32 °C (restrictive temperature). Pigmented melanocytes are clearly visible on the body of the zebrafish.
B. RT-PCR mitfa RNA expression at 24°C, 26°C, 28.5°C, and 32OC in wild type and mitfavc7 mutant embryos. Four types of mitfa transcripts (wt, a, b, c) were consistently observed in mitfavc7 embryo across all temperatures while only one transcript (wt) was observed in wild type embryo.
C. Schematic overview of mitfavc7 splice variants transcripts and their predicated MITF protein products.
D. Images and quantitation of zebrafish embryos following injection with transgenes expressing the mitfavc7 splice variants from the mitfa promoter at 24°C and 32°C. At 24OC, the number of melanocytes promoted by expression of the mitfa+in6 splice variant (2.97 (2.27 to 3.67) [mean (95% CI)]) was significantly less active compared to wild type mitfa (8.71 (6.66 to 10.76) [mean (95% CI)]) (p=0.007; ANOVA). In addition, the number of melanocytes promoted by expression of mitfa+in5,6 (1.138 (0.64 to 1.636) [mean (95% CI)]) was significantly less than mitfa (p=0.001; ANOVA) and compared to mitfa+in6 (p=0.045; ANOVA).
Aberrant splice variants can have partial or neomorphic function. To establish if the splice variants had activity, we cloned the splice variants under the control of the mitfa promoter, and microinjected them into zebrafish null mitfa (the mitfavc7 (nacre) mutation, a premature stop in exon 3, in which there are no neural crest derived melanocytes; Lister et al. 1999) (Figure 1C). Expression of wild type mitfa rescued the nacre mutation, and melanocytes were clearly visible at 5 dpf. In contrast, the mitfaΔex6 isoform was unable to stimulate melanocyte development in nacre mutants. The mitfa+in6 splice variant was functional in this assay, albeit with significantly reduced activity compared to wild type mitfa. In addition, the mitfa+in5,6 isoform demonstrated significantly reduced activity compared to mitfa and compared to mitfa+in6. At 32°C, mitfa+in6 was significantly reduced in its activity and mitfa+in5,6 had no function at all, demonstrating the temperature sensitivity of the splice products.
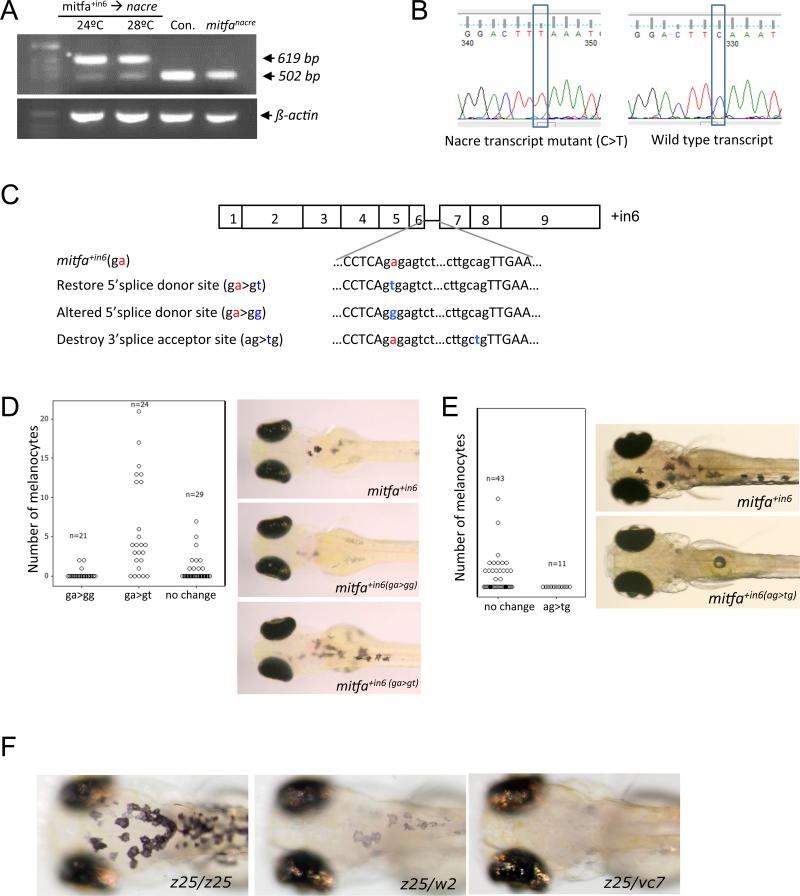
Given the activity of mitfa+in6, and the very weak activity of mitfa+in5,6, we hypothesized that some mitfa pre-RNA species can be correctly spliced in these transcripts to wild type mitfa and promote melanocyte development. To explore this idea, we examined the mitfa transcripts in the mitfa+in6 embryos at 24°C and 28°C and found transcripts that were the same size as wild type mitfa. However, the interpretation of this was complicated by the presence of mitfanacre transcripts that are also the same size as wild type mitfa (502 bp) (Figure 2A). Sequencing the 502 bp species in the mitfa+in6− expressing nacre mutant embryos revealed both the nacre and wild type mitfa transcripts indicating that mitfa+in6 splice variants could be correctly spliced to the wild type form in the nacre embryos (Figure 2B). While not tested, we anticipate a similar mechanism explains the few melanocytes that develop in the mitfa+in5,6-expressing nacre animals, rather than functional Mitfa activity of the protein product of the splice variant, because the encoded protein for the mitfaΔex6−9 lacks the DNA binding and dimerization domains (Figure 1C, D). Subsequent PCR analysis suggests that the reduction of the mitfanacre transcript in the mitfa+in6 injected embryos (Figure 2A) is due to PCR template competition in favour of the ectopically expressed mitfa+in6, rather than regulation of mitfanacre expression by Mitfa+In6 (data not snown).
Figure 2. mitfavc7 temperature-sensitivity is determined by reduction of wild type mitfa mRNA coupled with dominant negative activity of novel splice variants.
A. RT-PCR analysis of mitfa in nacre mutant zebrafish expressing the mitfa+in6 transgene, and in control wild type and uninjected nacre mutant zebrafish. The correctly spliced mitfa (502 bp) and the mitfa+in6 variant (619 bp) are indicated.
B. Sequencing traces of the correctly spliced mitfa PCR product identified in A. Cloned transcripts contained the nacre transcript with a mutation C>T and wild type mitfa correctly spliced from the mitfa+in6 transcript.
C. Illustration of the mitfa+in6 5′ splice donor site and the 3′ splice acceptor site, and engineered mutations.
D. E. Quantitation and images of nacre zebrafish embryos: D. expressing the mitfa+in6 transgene with the restored 5′ donor site (A>T), the altered 5′ donor site (A>G), E. and when the 3′ splice acceptor site is destroyed.
F. Images of mitfa mutant zebrafish embryos at 32°C. mitfaz25 is a hypomorphic allele that results in fewer and pale melanocytes as mitfaz25/w2, while no body melanocytes are present in mitfavc7/z25.
Next, we altered the splice acceptor and donor sites to test if the mitfa+in6 splice variants can have activity independently of splicing to the wild type form (Figure 2C-E). Restoration of the splice donor site (ga>gt) in the mitfa+in6 cDNA was sufficient to restore Mitf activity in the transgene, while change to another nucleotide (ga>gg) resulted in further reduced activity (Figure 2D). In contrast, when we maintained the mitfa+in6 mutation, but destroyed the 3’ splice acceptor site (ag>tg) it was non-functional in the nacre mutants (Figure 2E). Taken together, these experiments indicate that the mitfa+in6 can be spliced to the wild type mitfa form, and that the activity of mitfa+in6 is due to the minor accumulation of the wild type species. The particular mutation (t>a) appears to be crucial to the unique temperature sensitive splicing.
MITF binds to DNA as a homodimer, and the aberrant splice variants could interfere with wild type MITF activity. Deletion of exon 6 is predicted to truncate the basic region (Figure 1C). The dominant negative alleles in mouse cluster in the basic region, which is necessary for DNA binding, while retaining dimerization capability (reviewed in Steingrimsson et al. 2004). Heterozygous mitfavc7 mutants appear similar to wild type zebrafish at 5 dpf (Johnson et al., 2011), suggesting that the dominant negative activity of the vc7 mutation is not sufficient to produce a robust phenotype by interfering with wild type protein. However, when combined with the weakly active mitfaz25 mutation (a substitution, 1219F, in the first helix of the HLH domain)(Johnson et al., 2011), the mitfaz25/vc7 mutant embryos failed to develop melanocytes at 32°C, while the mitfaz25/w2 mutant embryos were able to develop a few and weakly pigmented melanocytes (Figure 2F). These results indicate that the mitfavc7 splice variants have some weak dominant interfering activity that may contribute to complete loss of mitfa activity at 32°C.
Temperature sensitive mutations are classic genetic tools that enable functional and temporal control of gene action. Our zebrafish mitfavc7 temperature sensitive mutant is the only conditional MITF mutation in vertebrates, and has already provided insight into the function of MITF in melanocyte stem cells (Johnson et al., 2011), melanocyte development and differentiation (Johnson et al., 2011; Taylor et al., 2011), and in melanoma (Lister et al., 2014). Unusually, rather than increasing temperature affecting the protein function directly as is the case with most temperature sensitive mutations, mitfavc7 is an intron mutation that leads to aberrant splice forms. Few examples of temperature sensitive splicing due to intron mutations are found in the literature. 1n human disease, temperature-dependent splicing in ß-globin pre-mRNA of thalassemic patients is caused by a mutation in intron 2 (Gemignani et al., 2002), and temperature- sensitive aberrant splicing of Type 111 Procollagen transcripts is caused by mutations in splice-donor sites within introns in patients with Ehler-Danlos Syndrome Type IV (Lee et al., 1991; and references therein). In Arabidopsis, an exon mutation close to a 5’-splice site confers temperature-sensitivity of RNA splicing in the floral homeotic gene APETALA3 (called the ap3-1 mutant) that controls stamen and petal development(Sablowski and Meyerowitz, 1998). Conditional mutants have been cleverly engineered by the addition of temperature sensitive DEGRON (Dohmen et al., 1994) and self-excising intein (excising protein) sequences (Tan et al., 2009; Zeidler et al., 2004). We have tested if the intron 6 can confer temperature sensitivity to GFP in zebrafish but have thus far been unsuccessful: additional exonic sequences may be required to enable temperature-sensitive splicing to be engineered into other genes.
To conclude, our work explains the temperature sensitivity of the mitfavc7 mutation to be due to an unusual intron 6 mutation that leads to reduced levels of wild type mitfa RNA. The production of interfering variants may also ensure that no melanocytes develop at 32°C. We suggest that the mitfavc7 mutation compromises base-pairing with the small nuclear RNAs of the spliceosome, and that this interaction becomes destabilized at the restrictive temperatures. Alternative splicing is an integral feature of MITF pre-RNA processing, and gives rise to multiple M1TF spice variants that are both melanocyte specific and relevant in melanocyte development and melanoma (Bharti et al., 2010; Cronin et al., 2009; Debbache et al., 2012; Simmons et al., 2014). Given the importance of MITF in melanocytes, the mitfavc7 allele enables careful examination of MITF activity at multiple stages of melanocyte development, stem cells and melanoma, and is a unique means to explore the function of aberrant pre-RNA splicing in zebrafish.
Supplementary Material
Acknowledgements
We are grateful to Professor Ian Jackson for many helpful discussions; Dr. Karthika Paranthaman and Wei Qing for zebrafish husbandry. This work was funded by the NIH (to SLJ, grant number RO1GM056988), the MRC (to ZZ, EEP), and by Concern Foundation for Cancer Research (to JAL).
References
- Bharti K, Debbache J, Wang X, Arnheiter H. The basic-helix-loop-helix-leucine zipper gene Mitf: analysis of alternative promoter choice and splicing. Methods Mol Biol. 2010;647:237–250. doi: 10.1007/978-1-60761-738-9_14. [DOI] [PubMed] [Google Scholar]
- Cronin JC, Wunderlich J, Loftus SK, Prickett TD, Wei X, Ridd K, Vemula S, Burrell AS, Agrawal NS, Lin JC, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22:435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbache J, Zaidi MR, Davis S, Guo T, Bismuth K, Wang X, Skuntz S, Maric D, Pickel J, Meltzer P, et al. In vivo role of alternative splicing and serine phosphorylation of the microphthalmia-associated transcription factor. Genetics. 2012;191:133–144. doi: 10.1534/genetics.111.135996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Gemignani F, Sazani P, Morcos P, Kole R. Temperature-dependent splicing of beta-globin pre-mRNA. Nucleic Acids Res. 2002;30:4592–4598. doi: 10.1093/nar/gkf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel LB, Tripathi RK, King RA, Spritz RA. A tyrosinase gene missense mutation in temperature-sensitive type I oculocutaneous albinism. A human homologue to the Siamese cat and the Himalayan mouse. J Clin Invest. 1991;87:1119–1122. doi: 10.1172/JCI115075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao JJ, Fisher DE. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch Biochem Biophys. 2014 doi: 10.1016/j.abb.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Nguyen AN, Lister JA. mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev Biol. 2011;350:405–413. doi: 10.1016/j.ydbio.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Vitale E, Superti-Furga A, Steinmann B, Ramirez F. G to T transversion at position +5 of a splice donor site causes skipping of the preceding exon in the type III procollagen transcripts of a patient with Ehlers-Danlos syndrome type IV. J Biol Chem. 1991;266:5256–5259. [PubMed] [Google Scholar]
- Lister JA, Capper A, Zeng Z, Mathers ME, Richardson J, Paranthaman K, Jackson IJ, Patton EE. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J Invest Dermatol. 2014;134:133–140. doi: 10.1038/jid.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Johnson SL. Requirements for the kit receptor tyrosine kinase during regeneration of zebrafish fin melanocytes. Development. 2001;128:1943–1949. doi: 10.1242/dev.128.11.1943. [DOI] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM. Temperature-sensitive splicing in the floral homeotic mutant apetala3-1. Plant Cell. 1998;10:1453–1463. doi: 10.1105/tpc.10.9.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons JL, Pierce CJ, Boyle GM. Evidence for an alternatively spliced MITF exon 2 variant. J Invest Dermatol. 2014;134:1166–1168. doi: 10.1038/jid.2013.426. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the micro-phthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Tan G, Chen M, Foote C, Tan C. Temperature-sensitive mutations made easy: generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges. Genetics. 2009;183:13–22. doi: 10.1534/genetics.109.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KL, Lister JA, Zeng Z, Ishizaki H, Anderson C, Kelsh RN, Jackson IJ, Patton EE. Differentiated melanocyte cell division occurs in vivo and is promoted by mutations in Mitf. Development. 2011;138:3579–3589. doi: 10.1242/dev.064014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler MP, Tan C, Bellaiche Y, Cherry S, Hader S, Gayko U, Perrimon N. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat Biotechnol. 2004;22:871–876. doi: 10.1038/nbt979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.