Abstract
Sensory hair cells convert mechanical motion into chemical signals. Otoferlin, a six-C2 domain transmembrane protein linked to deafness in humans, is hypothesized to play a role in exocytosis at hair cell ribbon synapses. To date, however, otoferlin has been studied almost exclusively in mouse models, and no rescue experiments have been reported. Here we describe the phenotype associated with morpholino-induced otoferlin knockdown in zebrafish and report the results of rescue experiments conducted with full-length and truncated forms of otoferlin. We found that expression of otoferlin occurs early in development and is restricted to hair cells and the midbrain. Immunofluorescence microscopy revealed localization to both apical and basolateral regions of hair cells. Knockdown of otoferlin resulted in hearing and balance defects, as well as locomotion deficiencies. Further, otoferlin morphants had uninflated swim bladders. Rescue experiments conducted with mouse otoferlin restored hearing, balance, and inflation of the swim bladder. Remarkably, truncated forms of otoferlin retaining the C-terminal C2F domain also rescued the otoferlin knockdown phenotype, while the individual N-terminal C2A domain did not. We conclude that otoferlin plays an evolutionarily conserved role in vertebrate hearing and that truncated forms of otoferlin can rescue hearing and balance.
INTRODUCTION
Hair cells couple mechanical motion to neurotransmitter release at synapses (1). In contrast to conventional neural synapses, hair cell synapses release neurotransmitter continuously and, in a graded manner (2), possess synaptic ribbons (2–4), and lack synaptophysin (5), complexin (6–9), Munc13 (10), and the calcium sensors synaptotagmin I and II (11). In place of synaptotagmin, it is believed that otoferlin may confer calcium sensitivity to evoke neurotransmitter release (12, 13). Otoferlin is a six-C2 domain transmembrane protein expressed in inner, outer, and vestibular hair cells, as well as restricted regions of the brain (13–16). In humans, missense mutations in otoferlin have been linked to hearing loss (17, 18), and biochemical studies have determined that otoferlin binds calcium and lipids (12, 19), as well as membrane trafficking proteins (12, 20–23). Further, in vitro assays have demonstrated that otoferlin accelerates SNARE-mediated membrane fusion (12). Based upon this evidence, it is hypothesized that otoferlin functions as a calcium-sensitive regulator of neurotransmitter release in sensory hair cells.
However, the results of several studies have raised questions related to otoferlin's function. For instance, despite otoferlin expression in vestibular hair cells, knockout mice show no balance defects (24, 25) despite reduced exocytosis in vestibular type I hair cells (24, 26). This raises questions as to the importance of otoferlin in this system. Further, otoferlin did not rescue synchronous neurotransmitter release in synaptotagmin I knockout cultured neurons, indicating that otoferlin and synaptotagmin are not functionally redundant (27). It is also unclear as to whether otoferlin-related deafness can be rescued by introduction of a functional copy of the otoferlin gene, and no rescue experiments have been reported. Related to this, it is currently unknown as to which domains of the protein are critical for hearing, and whether truncated otoferlin protein can recapitulate the function of wild-type (WT) otoferlin. To date, almost all studies on otoferlin have used a mouse model (28), and while insightful, current mammalian models present obstacles to progress in understanding otoferlin, including the challenge of hair cell isolation and difficulties in transfection. To circumvent such difficulties, and to add to the general body of knowledge of otoferlin across species, we have turned to zebrafish for the study of otoferlin. In this study, we characterized otoferlin expression in zebrafish as well as the phenotype associated with knockdown (KD). We also conducted rescue experiments using full-length (FL) and truncated forms of otoferlin.
MATERIALS AND METHODS
Fish strains.
Tropical 5D strains of zebrafish (Danio rerio) were used for this study and reared according to Institutional Animal Care and Use Committee protocols at the Sinnhuber Aquatic Research Laboratory, Oregon State University. Adult fish were raised on a recirculating water system (28 ± 1°C) with a 14-h:10-h light-dark schedule. Spawning and embryo collection were conducted as described in reference 29.
Bioinformatic tools.
Protein sequences were obtained from Ensembl (Ensembl accession numbers ENSP00000272371 [human], ENSMUSP00000073803 [mouse], ENSDARP00000123935 and ENSDARP00000118166 [zebrafish], ENSRNOP00000046997 [rat], ENSOCUP00000013417 [rabbit], ENSCPOP00000000288 [guinea pig], and ENSXETP00000007211 [frog]). The NCBI blastp tool was used to detect percent identity in the peptide sequences across different species, with the human sequence as the query sequence. The blastp tool was further used to obtain percent identity across the different C2 domains of otoferlin, keeping the human C2 domains as the query sequences. ClustalW and PRALINE tools were used for sequence alignments. SMART and SWISS-MODEL tools were used for domain analysis. R software was used to create the dot plot.
qPCR.
Total RNA was extracted from wild-type (WT) embryos collected at different hours postfertilization (hpf) with RNAzol (Molecular Research Centre, OH), and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, CA). Gene-specific primers (see Table SA in the supplemental material) were designed from genomic sequences for otoferlin b and otoferlin a found in Ensembl (Ensembl accession numbers ENSDART00000149773 and ENSDART00000136255), and relative abundances were assessed by real-time quantitative reverse transcription-PCR (qPCR) performed using Power Sybr green PCR master mix (Applied Biosystems, CA). The data were normalized to otoferlin expression at 24 hpf for both isoforms. Also, the expression of myosin VIb (Ensembl accession number ENSDART00000088801), vglut3 (Ensembl accession number ENSDART00000080454), and sonic hedgehog a (shha; NCBI accession number NM_131063.3) was examined in the otoferlin single- and double-KD larvae with gene-specific primers (see Table SA). The data were normalized relative to expression of the control and beta-actin genes. Graphs were plotted with Prism software version 5.0.
Whole-mount immunohistochemistry.
WT and microinjected zebrafish embryos were collected at different times postfertilization and fixed in 4% paraformaldehyde overnight at 4°C. Mouse monoclonal antiotoferlin and 3A10 (anti-Mauthner neuron) primary antibodies (dilutions, 1:500, and 1:200; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) and rabbit polyclonal anti-myosin VI (dilution, 1:800; Proteus Biosciences, CA) were used. Alexa Fluor 488- and 555-conjugated goat anti-mouse secondary antibodies (dilution, 1:1,000; Molecular Probes, Invitrogen, Eugene, OR) and Alexa Fluor 594-conjugated goat anti-rabbit secondary antibodies (dilution, 1:500; Molecular Probes, Invitrogen) were used. Fixed embryos were washed with phosphate-buffered saline–Tween (PBST) and UltraPure distilled water (Invitrogen, CA). Collagenase (0.0001 g/ml of PBST; C9891; Sigma-Aldrich, MO) treatment was performed to permeabilize the embryos, followed by rinsing with phosphate-buffered saline. Permeabilized embryos were blocked with 10% normal goat serum (G6767; Sigma-Aldrich) for an hour and incubated with primary antibody overnight at 4°C. On the following day, samples were rinsed in phosphate-buffered saline and incubated with secondary antibody. Embryos were imaged with an inverted Zeiss Axiovert 200M epifluorescence microscope fitted with a Zeiss Axiocam HRm camera and a 5× objective.
Whole-mount ISH.
In situ hybridization (ISH) of otoferlin was performed with digoxigenin-labeled antisense RNA probes specific to zebrafish otoferlin a and otoferlin b on WT zebrafish embryos collected at different times postfertilization as described in reference 30. Furthermore, ISH was performed as described in reference 30 with digoxigenin-labeled antisense RNA probes specific to the mouse otoferlin (NCBI accession number NM_001100395.1) to detect expression of the hair cell-specific mouse otoferlin construct in larval zebrafish double morphants. To synthesize the probe, gene-specific primers (see Table SB in the supplemental material) with an RNA polymerase promoter were designed for amplifying the probe templates, and cDNA was prepared from RNA isolated from whole zebrafish at 48 hpf. Embryos were labeled with either fast red (31) or nitroblue tetrazolium (NBT) or 5-bromo-4-chloro-3-indolylphosphate (BCIP) stain. Stained embryos were imaged using an inverted Zeiss Axiovert 200M epifluorescence microscope and a Nikon SMZ 1500 stereomicroscope mounted with a Coolpix E500 digital camera for NBT or BCIP.
ISH of larval zebrafish paraffin sections.
WT 120-hpf larval zebrafish were fixed in 10% neutral buffer formalin overnight at 4°C. The fish were rinsed in phosphate-buffered saline and dehydrated in graded ethanol. Agar blocks were prepared using zebrafish metal molds (32), and prefixed zebrafish were arranged in the agar blocks. The agar blocks were sent to the Veterinary Diagnostic Lab, Oregon State University, Corvallis, OR, for paraffin embedding and sectioning. Five-micrometer sections were obtained, and these sections were used for ISH with digoxigenin-labeled antisense RNA probes specific to otoferlin a as described in references 33 and 30.
Plasmids and constructs.
The cDNA encoding mouse otoferlin was a gift from Christine Petit (Institut Pasteur, Paris, France). The p5E-pmyo6b vector used in cloning was a gift from Teresa Nicolson (Oregon Health and Science University, OR). The p5E-pmyo6b vector contains a hair cell-specific promoter for myosin VIb. Full-length (FL) and truncated constructs of otoferlin were cloned downstream of the promoter at SacII and NotI sites. Clones were screened by colony PCR and verified by sequencing.
Microinjections.
A pair of morpholinos (MOs) targeting exon/intron boundaries of otoferlin a and otoferlin b and a standard negative control was obtained from GeneTools, Philomath, OR (see Table SD in the supplemental material). Approximately 2 nl of 0.6 mM otoferlin a MO, 0.7 mM otoferlin b MO, and both MOs diluted with RNase-free ultrapure distilled water and 3% phenol red were pressure injected into WT embryos at the one-cell stage. For verification of splicing pattern and efficacy of knockdown, total RNA was extracted and cDNA was synthesized from injected and WT zebrafish embryos collected at different times postfertilization as described below. Gene-specific primers (see Table SE in the supplemental material) were used for PCR with KOD Hot Start DNA polymerase (Novagen, USA), and the products were separated on a 1.25% agarose gel.
For the rescue, approximately 600 pg of the vector construct with either mouse FL otoferlin or truncated mouse otoferlin (construct with mouse putative C2DEF domains with the transmembrane, C2F domain with the transmembrane, C2EF domains with the transmembrane, and C2A) were coinjected with the morpholinos. Capped RNA was synthesized with the mMessage mMachine transcription kit (Ambion, TX) and the PCR template. The PCR template was amplified from pcDNA3 vector with the coding region for mouse full-length otoferlin using primers containing a T7 RNA polymerase promoter site. The amplified PCR template was purified with QIAquick PCR purification kit (Qiagen, CA). Approximately 250 pg of synthesized mRNA was coinjected with the morpholinos. Larvae were screened for rescue of the balance phenotype and acoustic startle response, including rescue of uninflated swim bladder, for further analysis.
Staining with vital dyes.
FM1-43FX dye (Life Technologies, NY) labeling of neuromast hair cells was performed on live zebrafish at 120 hpf. Zebrafish larvae were immersed in 3 μM FM1-43FX dye in embryo medium and rinsed off. The fish were washed several times with embryo medium and anesthetized with 0.2 mg/ml of tricaine solution for confocal imaging.
YO-PRO-1 (Life Technologies, NY) staining of the neuromast hair cells was performed on live morpholino-injected fish at 120 hpf. Zebrafish larvae were incubated for an hour at 28°C in 2 μM YO-PRO-1 dye in embryo medium. The fish were washed three times with the embryo medium and anesthetized with 0.2 mg/ml of tricaine solution for imaging with the inverted Zeiss Axiovert 200M epifluorescence microscope fitted with a Zeiss Axiocam HRm camera and 5×, 10×, and 20× objectives.
Confocal image acquisition and processing.
Whole-mount immunohistochemistry preparations were mounted with 1% agar on a 35-mm glass-bottomed petri dish and imaged with a confocal laser scanning microscope fitted with a 40× oil immersion objective (Zeiss LSM 510 Meta) with Alexa Fluor 555 filter sets. For live zebrafish stained with vital dye FM1-43, fish were immobilized with 1% agarose containing 0.2 mg/ml of tricaine on a glass-bottomed petri dish and imaged with a 63× water immersion objective (Zeiss LSM 510 Meta) with appropriate filter sets. Stacks of confocal images were taken and reconstructed with ImageJ software.
Larval behavior tests.
Injected zebrafish larvae were tested in a 96-well plate with a ZebraBox (Viewpoint Life Sciences, Lyon, France). Locomotor activity was measured using the Viewpoint tracker by subjecting the larvae to alternate phases of light and dark. Behavioral differences between the different injected groups were determined by comparing the distance moved during the dark period. Briefly, 96-hpf zebrafish larvae were loaded in a 96-well plate at least 3 h prior to the experiment to give them sufficient time to acclimatize. Larvae were subjected to alternate phases of light followed by dark and finally light during which the Viewpoint tracker recorded fish movement from the individual wells. Raw data files obtained from the Viewpoint tracker were processed using a python script and JMP software to average the total distance traveled during the dark phase for each group. Graphs were plotted and statistically analyzed with GraphPad Prism software version 5.0.
Acoustic startle response.
Injected zebrafish larvae from different groups were subjected to a startle stimulus assay at 120 hpf. Larvae were individually placed on a 100-mm petri dish filled with embryo medium and startled with a push solenoid that generated a sudden tap when activated. Movement was recorded with a digital video camera (Sony; HDR CX22) for 30 s after startling. The distances moved from the point of origin by different groups of larvae were compared by analyzing the video outputs from the camera with Noldus EthoVision XT (version 8.5) tracking software. Graphs were plotted and statistically analyzed with GraphPad Prism software version 5.0.
RESULTS
Zebrafish have two copies of otoferlin.
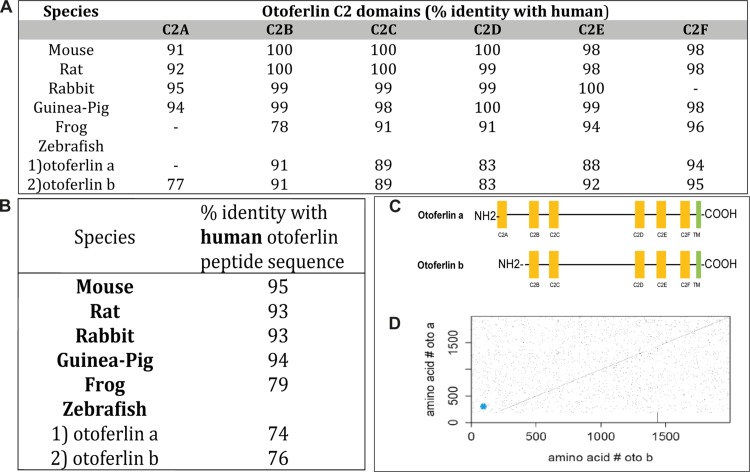
In contrast to that of mammals, the zebrafish genome contains two copies of otoferlin, located on chromosomes 17 and 20. The transcript (∼8 kb) encoded by chromosome 17 is referred to here as otoferlin b, with otoferlin a referring to the transcript encoded by chromosome 20 (∼7 kb). A comparative study between the human otoferlin amino acid sequence with sequences from other species indicates that otoferlin is highly conserved (Fig. 1A and B). Overall, the zebrafish otoferlin isoforms show 74% (otoferlin b) and 76% (otoferlin a) identity with human otoferlin (Fig. 1B). Even higher identity was found when the comparison was restricted to sequences predicted to form C2 domains (Fig. 1A). Sequence identities between zebrafish otoferlin a and human otoferlin are 77% in the C2A domain, 91% in the C2B domain, 89% in C2C, 83% in C2D, 92% in C2E, and 95% in C2F. Zebrafish otoferlin b is 91% similar in C2B, 89% in C2C, 83% in C2D, 88% in C2E, and 94% in the C2F domain with human otoferlin. Zebrafish otoferlin a appears to be a closer representation of the human otoferlin because of the presence of all six C2 domains, unlike otoferlin b, which lacks the C2A domain (Fig. 1A and C). The identity between amino acid sequences of the two zebrafish otoferlin isoforms is ∼80%. From the diagonal in the center of the dot plot (Fig. 1D), it can be discerned that the zebrafish otoferlin peptide sequences are identical in most regions except in the first ∼200 amino acids. On further comparison by sequence alignments (data not shown), it was confirmed that the peptide sequence of otoferlin b was ∼180 amino acids shorter on the N-terminal side than that of otoferlin a. In summary, the sequences of the C2 domains are more conserved than those of the non-C2 domain regions, and the C2A domain is the least conserved of the C2 domains.
FIG 1.
Sequence identity of otoferlin across different species. (A) Percent identity of predicted otoferlin C2 domains of various species compared to human otoferlin. (B) Comparison of overall sequence identity of otoferlin with human otoferlin. (C) Zebrafish otoferlin isoforms with putative C2 domains. Otoferlin b lacks the C2A domain. (D) Dot plot showing the identity of amino acid sequences between the two zebrafish otoferlin proteins: otoferlin a and otoferlin b. The diagonal across the plot indicates highly identical sequnces. Breaks along the diagonal indicate regions which are nonidentical. The blue dot designates the absence of the C2A domain (amino acids 3 to 97).
Otoferlin expression and localization in zebrafish.
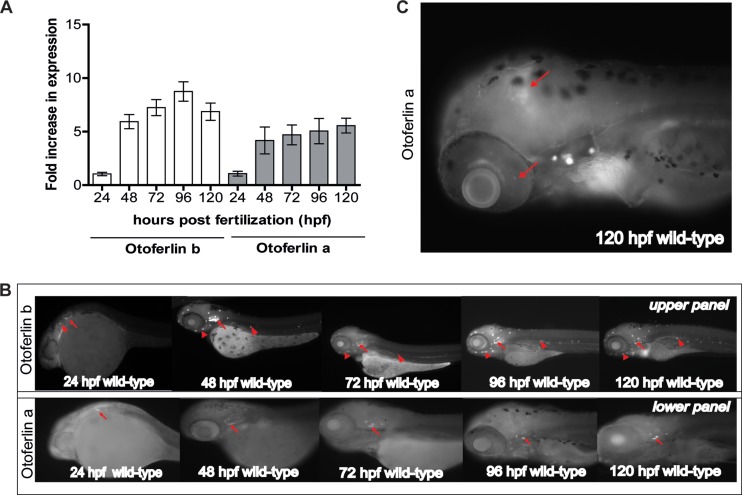
The amino acid sequences of otoferlins are similar across species, including zebrafish, suggesting a conserved function. Given the ease with which the organism can be genetically manipulated, we chose zebrafish as our model for use in the study of otoferlin. To ascertain the developmental expression profile of otoferlin, we conducted qPCR on wild-type (WT) zebrafish samples collected every 24 h during the first 120 h postfertilization (hpf). We found that 24-hpf zebrafish larvae express both copies of otoferlin (Fig. 2A), with an increase in relative abundance of both transcripts at 48 hpf (Fig. 2A). Expression plateaus after 72 hpf. The rise in otoferlin expression during the first 72 h of embryonic development coincides with the deposition of neuromasts and formation of the anterior lateral line (aLL) and posterior lateral line (pLL). It also coincided with formation of the semicircular canals and otic vesicle occurring during this stage of development (34). We next sought to characterize the spatial pattern of otoferlin expression in zebrafish larvae using whole-mount in situ hybridization (Fig. 2B). Transcripts of otoferlin b were detected in the otic placodes of 24-h-old larvae (Fig. 2B). The otic placode eventually forms sensory patches and develops as the zebrafish inner ear (35). There was also weak expression of otoferlin b in primordial cells that form neuromasts of the zebrafish lateral line organ system (Fig. 2B). The expression of otoferlin b becomes pronounced in the neuromasts of the pLL, aLL, and the inner ear region as the zebrafish larvae continues to develop through 120 hpf (Fig. 2B). Transcripts of otoferlin a were detected at around 24 hpf in the otic placode (Fig. 2B) and were restricted to the sensory patches of the inner ear as the larvae continued to develop through 120 hpf (Fig. 2B). We observed a relatively weak and diffuse expression in the zebrafish brain region and in the retina starting at around 48 hpf that became prominent at 120 hpf (Fig. 2C). Analyses of sectioned 120-hpf larval zebrafish confirmed expression of otoferlin transcripts in the midbrain and the retinal ganglion cell layer (see Fig. S1 in the supplemental material). However, no expression of the otoferlin a transcript was detected in the hair cells of the aLL and pLL neuromasts (Fig. 2B).
FIG 2.
mRNA expression of otoferlin in developing zebrafish. (A) Fold expression of otoferlin a and b transcripts at 24 to 120 hpf. Expression is normalized to that at 24 hpf. Error bars indicate 95% confidence intervals of the sample means (n = 5). (B) Whole-mount in situ hybridization images showing expression of otoferlin in 24- to 120-hpf wild-type zebrafish larvae. The upper panel shows expression of otoferlin b. Arrows indicate sensory patches in the inner ear. Arrowheads indicate anterior and posterior lateral line neuromasts. The lower panel shows expression of otoferlin a. Arrows indicate sensory patches in the inner ear. (C) Expression of otoferlin a in the brain region and retina in a 120-hpf zebrafish larva. Arrows indicate the brain and retina.
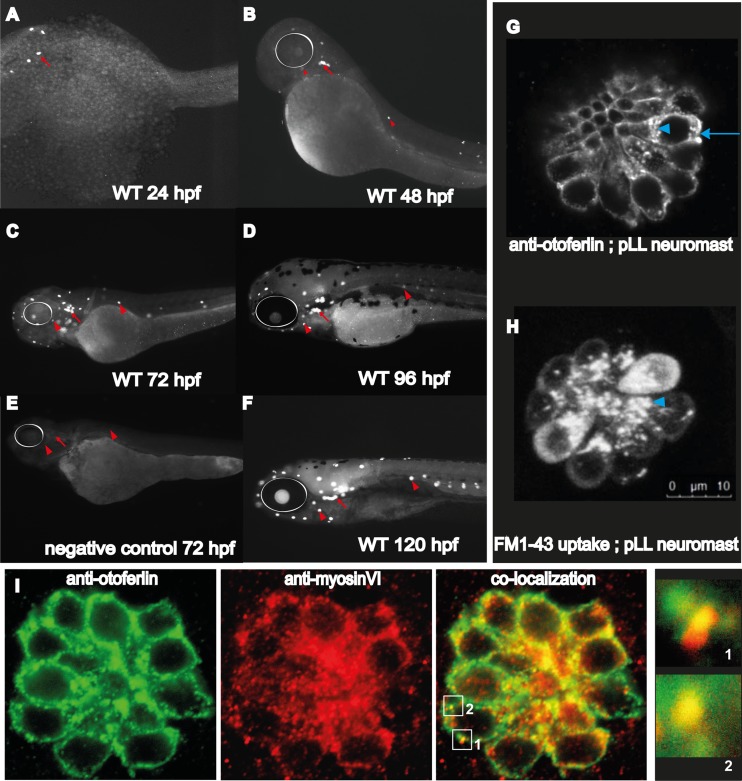
Whole-mount immunohistochemistry on WT zebrafish larvae with the antiotoferlin HCS-1 antibody was consistent with the mRNA expression profile in the inner ear and lateral line (Fig. 2B) during the first 120 h of zebrafish development (36). Strong immunoreactivity in the nascent hair cells of the zebrafish otic region at 24 hpf was detected (Fig. 3A), and immunoreactivity was detectable in the hair cells of both the pLL and aLL at 48 hpf (Fig. 3B), becoming more pronounced between 72 and 120 hpf (Fig. 3C, D, and F). Negative controls at 72 hpf (Fig. 3E) with no primary antibody confirmed that the labeling observed was due only to binding of the secondary antibody to the HCS-1 antibody. Overall, the onset and increase of otoferlin expression correlate with the formation and development of the zebrafish inner ear and the pLL and aLL system (34).
FIG 3.
Otoferlin protein expression in developing zebrafish. (A to F) Whole-mount immunohistochemistry on wild-type larval zebrafish at indicated developmental time points (24 to 120 hpf). (E) Negative control with no primary antibody in 72-hpf wild-type larvae. White circles indicate the position of the eye, arrows indicate sensory patches of the ear, and arrowheads indicate anterior and lateral line neuromasts. (G) Confocal image showing subcellular distribution of otoferlin in a 120-hpf hair cell neuromast cluster. The arrowhead indicates the supranuclear region of the hair cell; the arrow indicates the basolateral compartment of the hair cell (scale bar, 10 μm). (H) Confocal image showing uptake of FM1-43 dye uptake in the apical end (arrowhead) of hair cells within a neuromast of the posterior lateral line. Dye incubation time, 3 min. (I) Dual immunofluorescence with wild-type 120-hpf zebrafish showing colocalization of otoferlin in the hair cells of the neuromast with another hair cell marker, myosin VI. Boxes 1 and 2 indicate regions of colocalization. Insets of regions 1 and 2 show otoferlin colocalizing with myosin VI.
To examine the subcellular localization of otoferlin in zebrafish hair cells, confocal images of 120-hpf larvae were collected. Pronounced immunolabeling in both the supranuclear and basolateral compartments, including a punctate distribution throughout the cytoplasm, was observed in hair cells of the neuromast (Fig. 3G). This subcellular distribution is similar to observations made on mouse hair cells (20, 23, 37). Probing 120-h wild-type zebrafish with the FM1-43 dye (Fig. 3H) showed uptake in the apical end of the hair cells of a posterior lateral line neuromast cluster, indicating active vesicle recycling (38). This overlap in otoferlin distribution and FM1-43 dye uptake in the apical hair cell compartment raises the possibility of another role for otoferlin in the apical region in addition to the established function in synaptic transmission at the basolateral region (13, 37). To validate expression of otoferlin in the zebrafish hair cells, dual immunofluorescence was performed on wild-type 120-hpf larval zebrafish with antiotoferlin and anti-myosin VI antibodies. Myosin VI is a marker for hair cells, and fluorescence images confirm that otoferlin colocalizes with myosin VI in larval zebrafish hair cells (Fig. 3I) (39, 40).
Knockdown of otoferlin in the zebrafish hair cells.
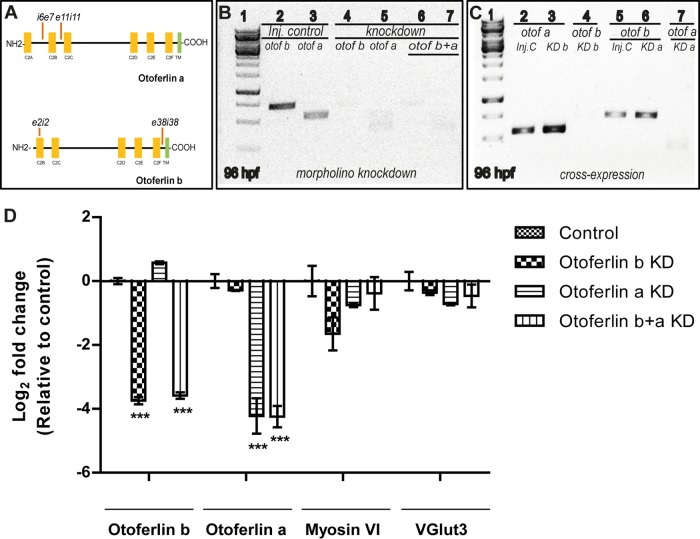
To determine the function of otoferlin in vivo, we used antisense splice-blocking morpholinos that targeted both isoforms, including their splice variants. Two splice-blocking MOs for each otoferlin gene targeting exon-intron boundaries (otoferlin b MOs, e2i2 and e38i38; otoferlin a MOs, i6e7 and e11i11) (Fig. 4A) were designed and microinjected at the one-cell stage. Comparable phenotypes were observed with each pair of morpholinos, and the e38i38 MO targeting otoferlin b and the i6e7 MO targeting otoferlin a were used for subsequent experiments in this study. We evaluated mRNA expression to assess knockdown of the otoferlin a and b transcripts in the single and double morphants. Analysis by qPCR showed that the expression of otoferlin a was significantly reduced in the otoferlin a and otoferlin b+a KD groups (P < 0.001) (Fig. 4D). Similarly, otoferlin b was significantly reduced in the otoferlin b and otoferlin b+a KD groups (P < 0.001) (Fig. 4D). qPCR further supported the conclusion that both the single and double knockdowns were effective at 96 hpf (Fig. 4B; lanes 4, 5, 6, and 7) compared with age-matched microinjected controls (Fig. 4C, lanes 1 and 2) and lasted up to 120 hpf (see Fig. S2A in the supplemental material). As the otoferlin b and a sequences are ∼80% identical (Fig. 1C), it was necessary to validate the specificity of MOs that were designed to target each isoform separately. Analysis of qPCR data indicates that knockdown of otoferlin b does not significantly affect the expression of otoferlin a and vice versa (Fig. 4D). qPCR further supported the conclusion that the MOs specifically block the targeted otoferlin without affecting expression of the other isoform at up to 120 hpf (Fig. 4C; see also Fig. S2B).
FIG 4.
Morpholino knockdown of otoferlin in zebrafish larvae. (A) Diagram of four splice-blocking morpholinos used in this study targeting otoferlin a and otoferlin b (e, exon; I, intron). (B) qPCR gel image of otoferlin KD zebrafish larvae at 96 hpf. Lane 1, molecular weight marker; lane 2, negative control tested for otoferlin b; lane 3, negative control injected tested for otoferlin a; lane 4, otoferlin b KD larva tested for otoferlin b; lane 5, otoferlin a KD tested for otoferlin a; lane 6, otoferlin b+a double-KD larva tested for otoferlin b; lane 7, otoferlin b+a double-KD larva tested for otoferlin a. (C) Cross expression studies with 96-hpf zebrafish larvae by qPCR. Lane 1, molecular weight marker; lane 2, otoferlin a in control; lane 3, otoferlin a in otoferlin b KD larva; lane 4, otoferlin b in otoferlin b KD larva; lane 5, otoferlin b in control; lane 6, otoferlin b in otoferlin a KD larva; lane 7, otoferlin a in otoferlin a KD larva. Inj. C, injected control; KD, knockdown; otof a, otoferlin a; otof b, otoferlin b; otof b+a, otoferlin b+a. (D) qPCR bar graph showing relative levels of expression of otoferlin a, otoferlin b, myosin VI, and VGlut3 genes in 96-hpf larval zebrafish across control, otoferlin a KD, otoferlin b KD, and otoferlin b+a KD groups where each gene was normalized with respect to the corresponding control. For the myosin VI and Vglut3 genes, no statistical deviation in expression was observed among the different KD groups. Expression of otoferlin a was significantly reduced (***, P ≤ 0.001) in the otoferlin a and otoferlin b+a KD groups but not in the otoferlin b KD group. Otoferlin b expression was significantly reduced in the otoferlin b and otoferlin b+a KD groups (P < 0.001) but not in the otoferlin a KD group. The statistical significance was calculated through Bonferroni multiple comparisons in Prism software. Error bars indicate 95% confidence intervals of the sample means (n = 3).
Since otoferlin interacts with the hair cell marker myosin VI (21, 40), and a reduction in otoferlin decreases the immunofluorescence of synaptic vesicle marker VGlut3 (37), qPCR was conducted on otoferlin single and double morphants to measure the relative expression changes of both myosin VI and VGlut3. Analysis indicates that otoferlin knockdown does not significantly affect the expression of myosin VI or VGlut3 in larval zebrafish at 96 hpf (Fig. 4D).
Immunohistochemistry of 120-hpf injected control (Fig. 5A) and morphant larvae (Fig. 5B to D) was also conducted. Embryos injected with the otoferlin b morpholino showed staining in the hair cells of the inner ear (Fig. 5B). However, there was no detectable signal in the hair cells of the neuromasts of the pLL and aLL (Fig. 5B). In contrast to otoferlin b, the otoferlin a morphants show staining in the hair cells of the inner ear, with strong immunolabeling in the hair cells of the neuromasts of the pLL and aLL (Fig. 5C). Visual inspection of the otic region reveals that, as in the control zebrafish larvae (Fig. 5E), otoferlin a is distributed in the hair cells of the sensory patches of the anterior macula. It is also present in the hair cells of the posterior, anterior, and medial cristae (Fig. 5F). However, otoferlin b is distributed in hair cells of the sensory patches of both the anterior and posterior maculae but absent from the cristae (Fig. 5G). This suggests that the otoferlin isoforms might play distinctive roles in the otic region. Finally, otoferlin b and a double morphants showed no antiotoferlin signal in any of the sensory patches of the inner ear and neuromasts of the lateral line (Fig. 5D). Confocal microscopy images of immunolabeled 120-hpf injected controls and double morphants were also collected (see Fig. S3A and B in the supplemental material). In contrast to findings for control siblings, a compressed z-stack image of the head region of double morphants showed an absence of antiotoferlin in the otic region. This indicates a complete knockdown of both isoforms of the zebrafish otoferlin in the otoferlin b+a double morphants to the limits of detection.
FIG 5.
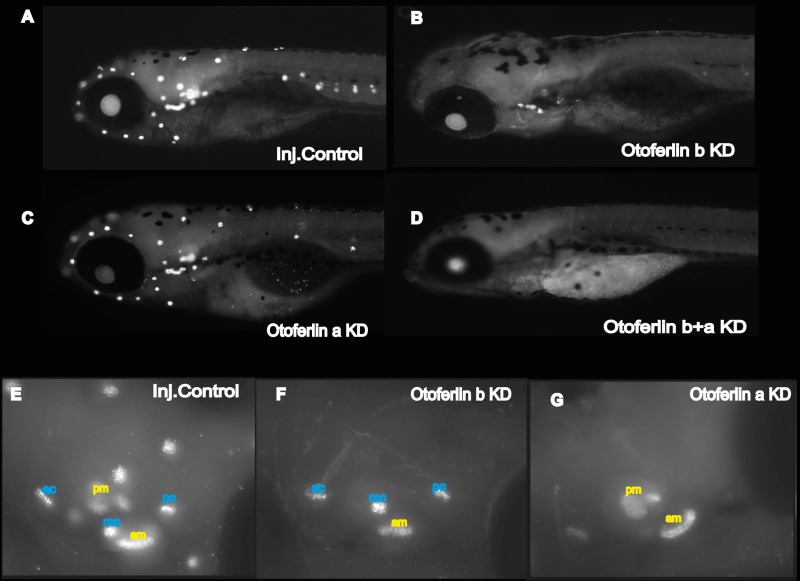
Whole-mount immunohistochemistry with the HCS-1 antiotoferlin antibody. (A to D) Images of showing otoferlin expression in 120-hpf larvae as follows: control injected (A), otoferlin b KD (B), otoferlin a KD (C), and otoferlin b+a KD (D). (E to F) Fluorescent maximum-intensity projection image of 120-hpf larval otic region of injected control (E), otoferlin b KD (F), and otoferlin a KD (G) larvae showing distinct distribution of otoferlin in the sensory patches of cristae and maculae. ac, anterior crista; mc, medial crista; pc, posterior crista; pm, posterior macula; am, anterior macula.
Phenotypic analysis of the otoferlin single- and double-knockdown larvae.
Compared to age-matched injected controls (Fig. 6A), otoferlin single-knockdown larvae show no noticeable defects in gross morphology at 120 hpf (Fig. 6B and C). However, after 96 hpf the double morphants failed to develop an inflated swim bladder (Fig. 6D), in contrast to single-knockdown larvae and injected controls. The phenotypic defects were visible around 72 hpf in the double morphants, when they failed to maintain an upright position, in contrast to injected control siblings (Fig. 6E and F). This suggests a balance defect (41) and indicates a critical contribution of otoferlin to balance and vestibular function. Moreover, about 80% of the 96- and 120-h double morphants swam on their sides or back, landed head-on, and often floated vertically head-up. On touching with a hair, double-KD larvae exhibited a circling and looping motion (data not shown) but swam back to the source of the stimulus rather than escaping. Beyond 120 hpf, double morphants gradually developed a curved spine, in contrast to injected controls (Fig. 6G and H), that enhanced the circling movement. These phenotypic abnormalities are comparable to the “circler mutants” (39, 41, 42) that were determined to be mutations in sensory hair cell-related genes. Since we observed a defective escape response with the double morphants, we examined the morphological differences in the patterning of the lateral line with hair cell marker YO-PRO-1. We did not observe differences in staining between the control and otoferlin b+a double morphants (Fig. 7A). Further, bright-field images of the otic region indicate that, like those in the controls, the semicircular canal folds and otolith formed normally in 120-hpf otoferlin double morphants (Fig. 7B). A similar swim bladder phenotype was observed in the double morphants that had been injected with a second set of morpholinos, e11i11 for otoferlin a and e2i2 for otoferlin b (see Fig. S4A in the supplemental material).
FIG 6.
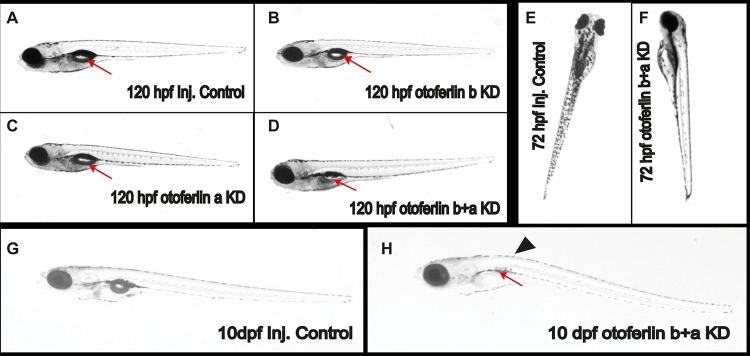
Observable phenotypes associated with the otoferlin KD 120-hpf larval zebrafish. (A) Control injected larva; (B) otoferlin b KD larva; (C) otoferlin a KD larva; (D) otoferlin b+a double KD larva (arrows indicate the swim bladder). The otoferlin double-KD larva failed to develop an inflated swim bladder. (E) Control larva at 72 hpf maintains an upright posture. (F) An otoferlin b+a double KD larva at 72 hpf failed to maintain an upright posture. (G) Ten-day-old injected control larva. (H) A 10-day-old otoferlin b+a double-KD zebrafish failed to inflate its swim bladder (arrow) and developed a curved spine (arrowhead).
FIG 7.
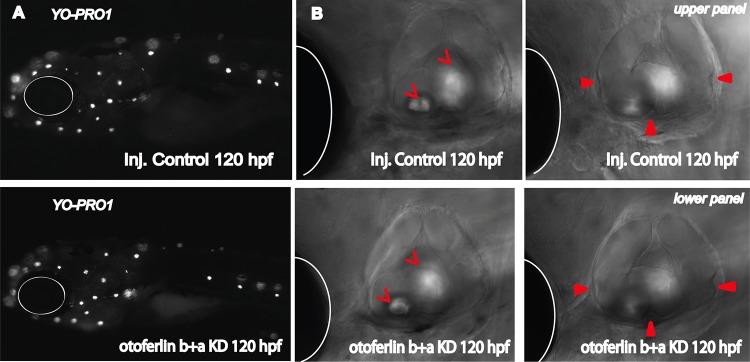
Otoferlin KD effect on structure and formation of lateral line, otic vesicle, and semicircular canals. (A) (Upper panel) YO-PRO-1 uptake in injected control larval zebrafish at 120 hpf; (lower panel) YO-PRO-1 uptake in otoferlin b+a double KD larval zebrafish at 120 hpf. (B) Bright-field images showing otic region in 120-h-old injected control (upper panel) and otoferlin b+a double-KD (lower panel) larvae. Open arrowheads point to the otoliths, filled arrowheads point to the semicircular canals, and white circles and hemicircles indicate the position of the eye.
Given that the double morphants show an uninflated swim bladder phenotype, we tested whether otoferlin knockdown affected swim bladder development using qPCR analysis of shha in double morphants at 72 hpf. shha is a swim bladder developmental marker (43), and results indicate that relative to that of the control, the expression level of the shha gene does not change in the double morphants (see Fig. S4B in the supplemental material).
Since both isoforms of otoferlin are expressed in hair cells of the otic region and lateral line, both copies may play a role in hearing and balance. To test whether otoferlin knockdown impairs hearing, acoustic startle reflex assays were conducted on 120-hpf single and double morphants. Acoustic startle assays are widely used to evaluate hearing-induced escape response in larval zebrafish (41). As shown in Fig. 8C, the distance traveled by control larvae (n = 23) was 63.94 mm, while otoferlin b KD larvae (n = 18) traveled 66.03 mm and otoferlin a KD larvae (n = 17) traveled 66.79 mm. When the single-KD larvae were compared to control larvae using Dunn's multiple comparison with a standard 5% significance level, the test showed no significant difference. In contrast, double-KD larvae (n = 16) traveled only 22.92 mm after startling. This value is significantly lower (Dunn's multiple comparison, P < 0.001) than the distance moved by the single-KD larvae and age-matched controls. A summary comparing the startle between otoferlin single and double morphants as well as the control group is included in Fig. S5 in the supplemental material. These results suggest that there is redundancy of function between the two isoforms and that the startle escape is significantly attenuated when the levels of expression of both the isoforms are reduced. To ensure that the larval zebrafish movement is not random but coincides with the startle stimuli, the average startle velocity as a function of time was plotted. The movement of the larval fish coincided with the startle stimuli for both the control and the double morphants (Fig. 8D). Furthermore, the control fish showed a marked increase in velocity after startling, followed by a decrease, while the double morphants show some slight movement after startling which quickly declined relative to that of the control (Fig. 8D). These data are consistent with the defective escape response observed in the double morphants compared to that of age-matched controls.
FIG 8.
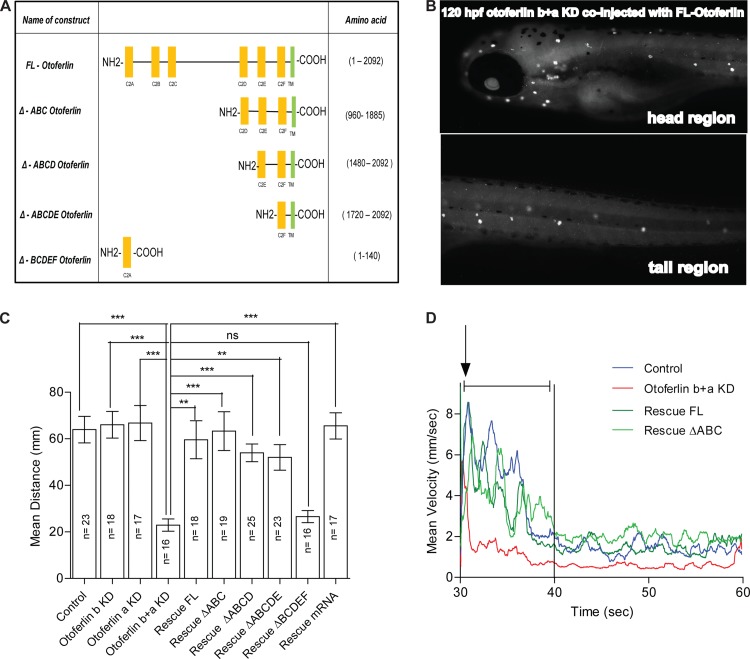
Rescue of zebrafish otoferlin knockdown with mouse otoferlin. (A) Schematic of the truncated mouse otoferlin constructs used in this study. Amino acid numbers are indicated. (B) Whole-mount immunohistochemistry on 120-hpf larval zebrafish double morphants coinjected with the mouse full-length otoferlin construct under the control of the hair cell-specific promoter. Otoferlin expression only in the hair cells at 120 hpf is shown. (C) Otoferlin deficiency causes defects in startle escape response in 120-hpf larval zebrafish. A plot of distance after startling is shown for the following groups: control (n = 23; mean distance = 63.94 mm), otoferlin b KD (n = 18; mean distance = 66.03 mm), otoferlin a KD (n = 17; mean distance = 66.79 mm), otoferlin b+a KD (n = 16; mean distance = 22.92 mm), rescue FL (n = 18; mean distance = 59.58 mm), rescue ΔABC (n = 19; mean distance = 63.32 mm), rescue ΔABCD (n = 25; mean distance = 53.94 mm), rescue ΔABCDE (n = 23; mean distance = 52.00 mm), rescue ΔBCDEF (n = 16; mean distance = 26.57 mm), and rescue mRNA (n = 17); mean distance = 65.55 mm (Dunn's multiple comparison with the standard 5% significance between otoferlin a+b KD and other groups; ***, P ≤ 0.001; **, P ≤ 0.01). Error bars indicate 95% confidence intervals of the sample means. ns, not significant. (D) Mean velocity traces of different groups: control (n = 23), otoferlin b+a KD (n = 16), rescue FL (n = 18), and rescue ΔABC (n = 19). The time point of startle is denoted with an arrow, and the first 10 s after startling is denoted with a capped line.
Studies on mouse models have established a link between otovestibular defects and locomotion using a dark-light test (44, 45). Upon finding that zebrafish otoferlin morphants exhibit balance defects and an abnormal startle escape response, dark-light locomotory tests were conducted on control and KD larval fish. Otoferlin single (n = 72) and double (n = 72) morphants at 96 hpf, as well as age-matched controls (n = 72), were used for the dark-light assay. Compared to controls, single-KD larvae traveled less during the dark phase (mean distance traveled: control, 62.73 mm; otoferlin b KD larvae, 45.67 mm; and otoferlin a KD larvae, 46.68 mm; Dunn's multiple-comparison test, P < 0.05) (see Fig. S6 in the supplemental material), and double-KD fish travel even less (distance for otoferlin double-KD fish, 35.61 mm; Dunn's multiple-comparison test, P < 0.001) (see Fig. S6). Since the zebrafish single and double morphants shows a defective dark-light response, this might indicate a direct or indirect role for otoferlin on neuronal wiring in the larval zebrafish. However, comparison of immunofluorescently labeled Mauthner cells that mediate the escape response (46–48) did not reveal any gross defects in the double morphants (n = 3) compared to age-matched controls (n = 2) (see Fig. S5E and F).
Rescue of zebrafish otoferlin knockdown with mouse otoferlin.
From comparison it is evident that there is a high sequence similarity between otoferlins of different species, suggesting functional conservation (Fig. 1A and B). To test for functional conservation, we coinjected double morphants with a p5E-pmyo6b vector encoding mouse otoferlin and a hair cell-specific myosin VIb promoter and tested for rescue of the knockdown phenotype. Several constructs, including the full-length mouse otoferlin (FL otoferlin) as well as truncated forms of the protein, were used in this study to identify the domain(s) critical for otoferlin function (Fig. 8A). Expression of mouse otoferlin was validated by performing whole-mount immunohistochemistry on zebrafish double morphants coinjected with the FL otoferlin construct. Figure 8B shows that the expression of the FL otoferlin construct is restricted only in the hair cells of the otic region and the lateral line, thereby confirming that the myosin VIb promoter recapitulates endogenous otoferlin expression. We also performed whole-mount in situ hybridization experiments on zebrafish double morphants coinjected with the mouse FL otoferlin construct. Figure S5A to C in the supplemental material show the presence of otoferlin transcripts in the otic region, pLL, and aLL. Figure 8C and D and Fig. S6 in the supplemental material show that the FL otoferlin was able to rescue the startle escape response (mean distance moved = 59.58 mm; n = 18; no significant difference with control group in Dunn's multiple-comparison test with the standard 5% significance level) and dark-light behavior in the zebrafish double morphants (mean distance traveled during dark phase = 59.89 mm; n = 72; no significant difference with control group in Dunn's multiple-comparison test with the standard 5% significance level). Mouse FL otoferlin also rescues the swim bladder defects observed in 120-hpf zebrafish double morphants (Fig. 9A). Furthermore, FM1-43 uptake in the rescue larvae was indistinguishable from that in WT larvae (data not shown). These results suggest that the mouse otoferlin was sufficient to correct for the disorders associated with depletion of both otoferlin isoforms found in larval zebrafish.
FIG 9.
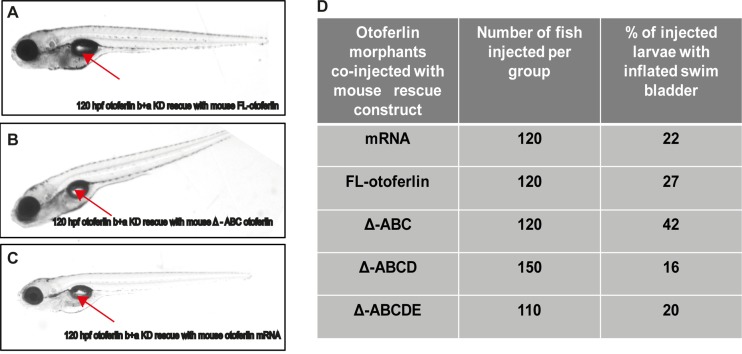
Rescue of otoferlin KD swim bladder phenotype with mouse otoferlin constructs. (A) Rescue of swim bladder defect in 120-hpf otoferlin b+a KD larva with FL otoferlin. (B) Rescue of swim bladder defect in 120 hpf otoferlin b+a KD larva with the ΔABC construct. (C) Rescue of swim bladder defect in 120-hpf otoferlin b+a double-KD zebrafish with otoferlin mRNA. The arrow indicates the inflated swim bladder. (D) Table showing percentage of fish rescued at 120 hpf coinjected with mouse otoferlin constructs including otoferlin mRNA.
Remarkably, we were able to rescue the double morphants with a truncated form of mouse otoferlin lacking the first three putative C2 domains (ΔABC). Zebrafish double morphants coinjected with the ΔABC construct were found to have inflated swim bladders, as observed at 120 hpf (Fig. 9B). The startle reflex abnormalities (Fig. 8C and D) were also rescued (mean distance moved = 63.32 mm; n = 19; no significant difference with control group in Dunn's multiple-comparison test with the standard 5% significance level). In addition, dark-light behavior was also rescued (mean distance traveled during dark phase = 59.81 mm; n = 48; no significant difference with control group in Dunn's multiple-comparison test with the standard 5% significance level) (see Fig. S6 in the supplemental material). This suggests that the first three C2 domains are not required to correct for the balance and hearing deficits that were observed with the zebrafish double morphants.
Several additional truncated constructs (Fig. 8A) either lacking the first 4 or 5 C2 domains (ΔABCD or ΔABCDE, respectively) or consisting of only the N-terminal C2A domain (ΔBCDEF) were also tested for recovery of the uninflated swim bladder (see Fig. S7A and B in the supplemental material) and acoustic startle response (Fig. 8C). The ΔABCD (n = 25, mean distance = 53.94 mm) and ΔABCDE (n = 23, mean distance = 52.00 mm) groups displayed no significant difference relative to the control group in Dunn's multiple-comparison test with the standard 5% significance level (Fig. 8C). Coinjection of the double morphants with the ΔBCDEF construct did not rescue the acoustic startle responses, however (n = 16; mean distance = 26.57 mm; no significant difference with otoferlin b+a KD group in Dunn's multiple-comparison test with the standard 5% significance level) (Fig. 8C).
Finally, as an alternative method of rescue, zebrafish double morphants were coinjected with mouse otoferlin mRNA encoding the full-length protein. The mRNA injection completely rescued the swim bladder defect (Fig. 9C), including recovery of the startle escape response (mean distance moved = 65.55 mm; n = 17; no significant difference with control group in Dunn's multiple-comparison test with the standard 5% significance level) (Fig. 8C), supporting the conclusion that mouse otoferlin can rescue morpholino knockdown in zebrafish. A table showing the percentage of larvae with the inflated swim bladder rescue phenotype for the double morphants coinjected with the different otoferlin constructs is included in Fig. 9D.
DISCUSSION
Based upon sequence, expression patterns, knockout phenotype, and rescue studies, we conclude that otoferlin plays a conserved role in hair cells. Despite divergence of the zebrafish and mammalian genomes approximately 420 million years ago (49), the amino acid sequences of otoferlin are highly similar, and mouse otoferlin successfully compensates for loss of endogenous otoferlin expression in zebrafish. This suggests both a conserved function of the protein in hair cells and a conserved set of binding partners for neurotransmitter release. Indeed, the loss in hearing we observed in otoferlin knockdown zebrafish matches mouse knockout models. We also note that while zebrafish have two otoferlin genes and mammals have one, multiple splice isoforms of the gene have been reported for mice and humans (23, 50).
Analysis of sequence identity of the C2 domains between species showed greater conservation among the C-terminal C2D, C2E, and C2F domains than for the N-terminal C2A domain. We speculate that these C-terminal domains may play a functionally conserved or redundant role in otoferlin function. Indeed, functional redundancy among the C2 domains has been noted in reconstituted membrane fusion assays (12). Experiments we report indicate that shortened forms of otoferlin lacking the N-terminal domains rescue the knockdown phenotype, in agreement with the idea of functional redundancy. However, it is possible that the N-terminal C2 domains play a role that was not detected in our assays or a function found in mammals but not in zebrafish. Indeed, our results seem to conflict with reports of missense mutations in the C2B and C2C domains that have been linked to hearing loss in mammals (51, 52) We speculate that these missense mutations may reduce the structural stability of the protein, resulting in lower levels of protein expression. Knockdown of both zebrafish otoferlin genes was required for an observable phenotype in our studies, despite differing in the first N-terminal 183 amino acids, corresponding to the C2A domain, again suggesting that some domains may be dispensable. Interestingly, biophysical studies on the C2A domain have determined that this is the only domain in otoferlin that does not bind calcium (12, 53). However, studies on the otoferlin orthologues myoferlin and dysferlin have found that the C2A domains of these proteins do bind calcium, supporting the idea that the calcium binding activity of the C2A domain may have diverged among the ferlins (12, 19, 54, 55).
Unexpectedly, otoferlin knockdown zebrafish displayed severe deficits in balance and uninflated swim bladders, suggesting a critical role for otoferlin in balance. While otoferlin is expressed in vertebrate vestibular hair cells, neither otoferlin knockout mice nor human patients with otoferlin mutations suffer from balance deficits (24). Our results clearly demonstrate a critical role for otoferlin in zebrafish balance and vestibular hair cell function. That calcium-dependent neurotransmitter release is attenuated but not completely abrogated in knockout mouse vestibular hair cells (24) may indicate that compensatory or redundant calcium sensors exist in mammalian hair cells that are not active in zebrafish cells. Thus, zebrafish may serve as a model for future characterization of otoferlin's contribution to vestibular hair cells.
Despite the loss of morpholino knockdown efficiency and expression of both otoferlin genes around 120 hpf, most zebrafish did not show any signs of swim bladder inflation or recovery of balance even at 10 dpf, and many developed a curved spine. This suggests that otoferlin may play a developmental role and that the lack of otoferlin during a certain developmental window may have permanent effects on zebrafish physiology. In support of this, a recent study reported abnormally small ventral cochlear nuclei in otoferlin knockout mice (56). Future studies should focus on the developmental effects linked to otoferlin loss of function.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Medical Research Foundation (MRF) award to C.P.J., as well as Oregon State University, and NIH grant P30 000210 (R.L.T.).
We thank Teresa Nicolson for providing us with the p5E-pmyo6b vector, Christine Petit for providing us with mouse otoferlin cDNA, the Veterinary Diagnostic Lab, Oregon State University, OR, for help with the paraffin sections, Carrie Barton for help with fish maintenance and embryo collection, Mike T. Simonich for assembling the startle apparatus, Kai Tao and Anne-Marie Girard for help with confocal microscopy, Siba R. Das for advice on the Noldus software, Sean M. Bugel for advice on molecular biology, Pamela Noyes for advice on GraphPad software, Leah Wehmas for advice on agar block preparation, Jacob Thomas Huegel for help with colocalization images, Justin Sanders for advice on in situ hybridization on zebrafish paraffin sections, and Ananda S. Roy for help with the R program.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01439-14.
REFERENCES
- 1.LeMasurier M, Gillespie PG. 2005. Hair-cell mechanotransduction and cochlear amplification. Neuron 48:403–415. doi: 10.1016/j.neuron.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Nouvian R, Beutner D, Parsons TD, Moser T. 2006. Structure and function of the hair cell ribbon synapse. J Membr Biol 209:153–165. doi: 10.1007/s00232-005-0854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs PA. 2005. Time and intensity coding at the hair cell's ribbon synapse. J Physiol 566:7–12. doi: 10.1113/jphysiol.2004.082214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khimich D, Nouvian R, Pujol R, Tom Dieck S, Egner A, Gundelfinger ED, Moser T. 2005. Hair cell synaptic ribbons are essential for synchronous auditory signalling. Nature 434:889–894. doi: 10.1038/nature03418. [DOI] [PubMed] [Google Scholar]
- 5.He S, Yang J. 2011. Maturation of neurotransmission in the developing rat cochlea: immunohistochemical evidence from differential expression of synaptophysin and synaptobrevin 2. Eur J Histochem 55(1):e2. doi: 10.4081/ejh.2011.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMahon HT, Missler M, Li C, Sudhof TC. 1995. Complexins—cytosolic proteins that regulate snap receptor function. Cell 83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 7.Reim K, Mansour M, Varoqueaux F, McMahon HT, Sudhof TC, Brose N, Rosenmund C. 2001. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell 104:71–81. doi: 10.1016/S0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 8.Reim K, Wegmeyer H, Brandstatter JH, Xue MS, Rosenmund C, Dresbach T, Hofmann K, Brose N. 2005. Structurally and functionally unique complexins at retinal ribbon synapses. J Cell Biol 169:669–680. doi: 10.1083/jcb.200502115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strenzke N, Chanda S, Kopp-Scheinpflug C, Khimich D, Reim K, Bulankina AV, Neef A, Wolf F, Brose N, Xu-Friedman MA, Moser T. 2009. Complexin-I is required for high-fidelity transmission at the endbulb of held auditory synapse. J Neurosci 29:7991–8004. doi: 10.1523/JNEUROSCI.0632-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uthaiah RC, Hudspeth AJ. 2010. Molecular anatomy of the hair cell's ribbon synapse. J Neurosci 30:12387–12399. doi: 10.1523/JNEUROSCI.1014-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beurg M, Michalski N, Safieddine S, Bouleau Y, Schneggenburger R, Chapman ER, Petit C, Dulon D. 2010. Control of exocytosis by synaptotagmins and otoferlin in auditory hair cells. J Neurosci 30:13281–13290. doi: 10.1523/JNEUROSCI.2528-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson CP, Chapman ER. 2010. Otoferlin is a calcium sensor that directly regulates SNARE-mediated membrane fusion. J Cell Biol 191:187–197. doi: 10.1083/jcb.201002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux I, Safieddine S, Nouvian R, Grati M, Simmler MC, Bahloul A, Perfettini I, Le Gall M, Rostaing P, Hamard G, Triller A, Avan P, Moser T, Petit C. 2006. Otoferlin, defective in a human deafness form, is essential for exocytosis at the auditory ribbon synapse. Cell 127:277–289. doi: 10.1016/j.cell.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 14.Schug N, Braig C, Zimmermann U, Engel J, Winter H, Ruth P, Blin N, Pfister M, Kalbacher H, Knipper M. 2006. Differential expression of otoferlin in brain, vestibular system, immature and mature cochlea of the rat. Eur J Neurosci 24:3372–3380. doi: 10.1111/j.1460-9568.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 15.Valiyaveettil M, Alamneh Y, Miller SA, Hammamieh R, Wang Y, Arun P, Wei Y, Oguntayo S, Nambiar MP. 2012. Preliminary studies on differential expression of auditory functional genes in the brain after repeated blast exposures. J Rehabil Res Dev 49:1153–1162. doi: 10.1682/JRRD.2011.09.0182. [DOI] [PubMed] [Google Scholar]
- 16.Żak M, Bress A, Brandt N, Franz C, Ruth P, Pfister M, Knipper M, Blin N. 2012. Ergic2, a brain specific interacting partner of Otoferlin. Cell Physiol Biochem 29:941–948. doi: 10.1159/000188338. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez-Ballesteros M, Reynoso R, Olarte M, Villamar M, Morera C, Santarelli R, Arslan E, Meda C, Curet C, Volter C, Sainz-Quevedo M, Castorina P, Ambrosetti U, Berrettini S, Frei K, Tedin S, Smith J, Cruz Tapia M, Cavalle L, Gelvez N, Primignani P, Gomez-Rosas E, Martin M, Moreno-Pelayo MA, Tamayo M, Moreno-Barral J, Moreno F, del Castillo I. 2008. A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat 29:823–831. doi: 10.1002/humu.20708. [DOI] [PubMed] [Google Scholar]
- 18.Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C. 1999. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet 21:363–369. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
- 19.Marty NJ, Holman CL, Abdullah N, Johnson CP. 2013. The C2 domains of otoferlin, dysferlin, and myoferlin alter the packing of lipid bilayers. Biochemistry 52:5585–5592. doi: 10.1021/bi400432f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncker SV, Franz C, Kuhn S, Schulte U, Campanelli D, Brandt N, Hirt B, Fakler B, Blin N, Ruth P, Engel J, Marcotti W, Zimmermann U, Knipper M. 2013. Otoferlin couples to clathrin-mediated endocytosis in mature cochlear inner hair cells. J Neurosci 33:9508–9519. doi: 10.1523/JNEUROSCI.5689-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidrych P, Zimmermann U, Kuhn S, Franz C, Engel J, Duncker SV, Hirt B, Pusch CM, Ruth P, Pfister M, Marcotti W, Blin N, Knipper M. 2009. Otoferlin interacts with myosin VI: implications for maintenance of the basolateral synaptic structure of the inner hair cell. Hum Mol Genet 18:2779–2790. doi: 10.1093/hmg/ddp213. [DOI] [PubMed] [Google Scholar]
- 22.Ramakrishnan NA, Drescher MJ, Drescher DG. 2009. Direct interaction of otoferlin with syntaxin 1A, SNAP-25, and the L-type voltage-gated calcium channel Cav13 J Biol Chem 284:1364–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakrishnan NA, Drescher MJ, Morley BJ, Kelley PM, Drescher DG. 2014. Calcium regulates molecular interactions of otoferlin with SNARE proteins required for hair cell exocytosis. J Biol Chem 289:8750–8766. doi: 10.1074/jbc.M113.480533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dulon D, Safieddine S, Jones SM, Petit C. 2009. Otoferlin is critical for a highly sensitive and linear calcium-dependent exocytosis at vestibular hair cell ribbon synapses. J Neurosci 29:10474–10487. doi: 10.1523/JNEUROSCI.1009-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwander M, Sczaniecka A, Grillet N, Bailey JS, Avenarius M, Najmabadi H, Steffy BM, Federe GC, Lagler EA, Banan R, Hice R, Grabowski-Boase L, Keithley EM, Ryan AF, Housley GD, Wiltshire T, Smith RJH, Tarantino LM, Muller U. 2007. A forward genetics screen in mice identifies recessive deafness traits and reveals that pejvakin is essential for outer hair cell function. J Neurosci 27:2163–2175. doi: 10.1523/JNEUROSCI.4975-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent PF, Bouleau Y, Safieddine S, Petit C, Dulon D. 2014. Exocytotic machineries of vestibular type I and cochlear ribbon synapses display similar intrinsic otoferlin-dependent Ca2+ sensitivity but a different coupling to Ca2+ channels. J Neurosci 34:10853–10869. doi: 10.1523/JNEUROSCI.0947-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reisinger E, Bresee C, Neef J, Nair R, Reuter K, Bulankina A, Nouvian R, Koch M, Buckers J, Kastrup L, Roux I, Petit C, Hell SW, Brose N, Rhee JS, Kugler S, Brigande JV, Moser T. 2011. Probing the functional equivalence of otoferlin and synaptotagmin 1 in exocytosis. J Neurosci 31:4886–4895. doi: 10.1523/JNEUROSCI.5122-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leibovici M, Safieddine S, Petit C. 2008. Mouse models for human hereditary deafness. Curr Top Dev Biol 84:385–429. doi: 10.1016/S0070-2153(08)00608-X. [DOI] [PubMed] [Google Scholar]
- 29.Westerfield M. 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 4th ed. University of Oregon Press, Eugene, OR. [Google Scholar]
- 30.Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 31.Novak AE, Ribera AB. 2003. Immunocytochemistry as a tool for zebrafish developmental neurobiology. Methods Cell Sci 25:79–83. doi: 10.1023/B:MICS.0000006894.43940.b1. [DOI] [PubMed] [Google Scholar]
- 32.Sabaliauskas NA, Foutz CA, Mest JR, Budgeon LR, Sidor AT, Gershenson JA, Joshi SB, Cheng KC. 2006. High-throughput zebrafish histology. Methods 39:246–254. doi: 10.1016/j.ymeth.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Javelle M, Marco CF, Timmermans M. 2011. In situ hybridization for the precise localization of transcripts in plants. J Vis Exp 2011(57):e3328. doi: 10.3791/3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 35.Waterman RE, Bell DH. 1984. Epithelial fusion during early semicircular canal formation in the embryonic zebrafish, Brachydanio rerio. Anat Rec 210:101–114. doi: 10.1002/ar.1092100113. [DOI] [PubMed] [Google Scholar]
- 36.Goodyear RJ, Legan PK, Christiansen JR, Xia B, Korchagina J, Gale JE, Warchol ME, Corwin JT, Richardson GP. 2010. Identification of the hair cell soma-1 antigen, HCS-1, as otoferlin. J Assoc Res Otolaryngol 11:573–586. doi: 10.1007/s10162-010-0231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pangrsic T, Lasarow L, Reuter K, Takago H, Schwander M, Riedel D, Frank T, Tarantino LM, Bailey JS, Strenzke N, Brose N, Muller U, Reisinger E, Moser T. 2010. Hearing requires otoferlin-dependent efficient replenishment of synaptic vesicles in hair cells. Nat Neurosci 13:869–876. doi: 10.1038/nn.2578. [DOI] [PubMed] [Google Scholar]
- 38.Griesinger CB, Richards CD, Ashmore JF. 2002. Fm1-43 reveals membrane recycling in adult inner hair cells of the mammalian cochlea. J Neurosci 22:3939–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kappler JA, Starr CJ, Chan DK, Kollmar R, Hudspeth AJ. 2004. A nonsense mutation in the gene encoding a zebrafish myosin VI isoform causes defects in hair-cell mechanotransduction. Proc Natl Acad Sci U S A 101:13056–13061. doi: 10.1073/pnas.0405224101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roux I, Hosie S, Johnson SL, Bahloul A, Cayet N, Nouaille S, Kros CJ, Petit C, Safieddine S. 2009. Myosin VI is required for the proper maturation and function of inner hair cell ribbon synapses. Hum Mol Genet 18:4615–4628. doi: 10.1093/hmg/ddp429. [DOI] [PubMed] [Google Scholar]
- 41.Nicolson T, Rusch A, Friedrich RW, Granato M, Ruppersberg JP, Nusslein-Volhard C. 1998. Genetic analysis of vertebrate sensory hair cell mechanosensation: the zebrafish circler mutants. Neuron 20:271–283. doi: 10.1016/S0896-6273(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 42.Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Sollner C, Duncan RN, Boehland A, Nicolson T. 2008. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci 28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, Gong Z. 2009. Development of zebrafish swimbladder: the requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol 331:222–236. doi: 10.1016/j.ydbio.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 44.Khan Z, Carey J, Park HJ, Lehar M, Lasker D, Jinnah HA. 2004. Abnormal motor behavior and vestibular dysfunction in the stargazer mouse mutant. Neuroscience 127:785–796. doi: 10.1016/j.neuroscience.2004.05.052. [DOI] [PubMed] [Google Scholar]
- 45.Lindema S, Gernet M, Bennay M, Koch M, Loscher W. 2008. Comparative analysis of anxiety-like behaviors and sensorimotor functions in two rat mutants, ci2 and ci3, with lateralized rotational behavior. Physiol Behav 93:417–426. doi: 10.1016/j.physbeh.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 46.Burgess HA, Granato M. 2007. Sensorimotor gating in larval zebrafish. J Neurosci 27:4984–4994. doi: 10.1523/JNEUROSCI.0615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faber DS, Fetcho JR, Korn H. 1989. Neuronal networks underlying the escape response in goldfish. General implications for motor control. Ann N Y Acad Sci 563:11–33. [DOI] [PubMed] [Google Scholar]
- 48.Zottoli SJ, Faber DS. 2000. The Mauthner cell: what has it taught us? Neuroscientist 6:26–38. doi: 10.1177/107385840000600111. [DOI] [Google Scholar]
- 49.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assuncao JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Threadgold G, Harden G, Ware D, Mortimore B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Urun Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberlander M, Rudolph-Geiger S, Teucke M, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nusslein-Volhard C, Hubbard TJ, Roest Crollius H, Rogers J, Stemple DL, Begum S, Lloyd C, Lanz C, Raddatz G, Schuster SC. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasunaga S, Grati M, Chardenoux S, Smith TN, Friedman TB, Lalwani AK, Wilcox ER, Petit C. 2000. OTOF encodes multiple long and short isoforms: genetic evidence that the long ones underlie recessive deafness DFNB9. Am J Hum Genet 67:591–600. doi: 10.1086/303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Longo-Guess C, Gagnon LH, Bergstrom DE, Johnson KR. 2007. A missense mutation in the conserved C2B domain of otoferlin causes deafness in a new mouse model of DFNB9. Hear Res 234:21–28. doi: 10.1016/j.heares.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirghomizadeh F, Pfister M, Apaydin F, Petit C, Kupka S, Pusch CM, Zenner HP, Blin N. 2002. Substitutions in the conserved C2C domain of otoferlin cause DFNB9, a form of nonsyndromic autosomal recessive deafness. Neurobiol Dis 10:157–164. doi: 10.1006/nbdi.2002.0488. [DOI] [PubMed] [Google Scholar]
- 53.Helfmann S, Neumann P, Tittmann K, Moser T, Ficner R, Reisinger E. 2011. The crystal structure of the C(2)A domain of otoferlin reveals an unconventional top loop region. J Mol Biol 406:479–490. doi: 10.1016/j.jmb.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 54.Abdullah N, Padmanarayana M, Marty NJ, Johnson CP. 2014. Quantitation of the calcium and membrane binding properties of the C2 domains of dysferlin. Biophys J 106:382–389. doi: 10.1016/j.bpj.2013.11.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Therrien C, Di Fulvio S, Pickles S, Sinnreich M. 2009. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry 48:2377–2384. doi: 10.1021/bi802242r. [DOI] [PubMed] [Google Scholar]
- 56.Wright S, Hwang Y, Oertel D. 24 September 2014. Synaptic transmission between end bulbs of held and bushy cells in the cochlear nucleus of mice with a mutation in otoferlin. J Neurophysiol doi: 10.1152/jn.00522.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.