Abstract
Cyclin-dependent kinase 9 (CDK9) and CDK12 have each been demonstrated to phosphorylate the RNA polymerase II C-terminal domain (CTD) at serine 2 of the heptad repeat, both in vitro and in vivo. CDK9, as part of P-TEFb and the super elongation complex (SEC), is by far the best characterized of CDK9, CDK12, and CDK13. We employed both in vitro and in vivo assays to further investigate the molecular properties of CDK12 and its paralog CDK13. We isolated Flag-tagged CDK12 and CDK13 and found that they associate with numerous RNA processing factors. Although knockdown of CDK12, CDK13, or their cyclin partner CCNK did not affect the bulk CTD phosphorylation levels in HCT116 cells, transcriptome sequencing (RNA-seq) analysis revealed that CDK12 and CDK13 losses in HCT116 cells preferentially affect expression of DNA damage response and snoRNA genes, respectively. CDK12 and CDK13 depletion also leads to a loss of expression of RNA processing factors and to defects in RNA processing. These findings suggest that in addition to implementing CTD phosphorylation, CDK12 and CDK13 may affect RNA processing through direct physical interactions with RNA processing factors and by regulating their expression.
INTRODUCTION
The largest subunit of RNA polymerase II (Pol II), Rpb1, contains a C-terminal domain (CTD) consisting of 52 heptad repeats of the YSPTSPS consensus sequence in humans (1). The CTD is phosphorylated within these repeats, including at serines 2, 5, and 7 (Ser2, Ser5, and Ser7, respectively) (2). The CTD serves as a phosphorylation-regulated platform for the recruitment of transcription factors, RNA processing factors, and chromatin modifiers, which affect mRNA synthesis, cotranscriptional processing, and histone modifications during the transcription cycle (3).
The CTD undergoes a cycle of phosphorylation and dephosphorylation during the transcription cycle of initiation, elongation, and termination (4). During transcription initiation and early transcription, Ser5 of the CTD is phosphorylated by the cyclin-dependent kinase 7 (CDK7) subunit of the basal transcription factor TFIIH (2, 5). The positive transcription elongation factor, P-TEFb (comprised of CDK9 and cyclin T), regulates transcription elongation through phosphorylation of the CTD at Ser2 (6). P-TEFb also phosphorylates negative elongation factor (NELF) (7) and DRB sensitivity-inducing factor (DSIF) (8) during the transition to productive elongation.
In the budding yeast Saccharomyces cerevisiae, there are two complexes for CTD Ser2 phosphorylation: the Bur1/Bur2 complex and the Ctk1/Ctk2/Ctk3 (CTDK) complex. The Bur complex implements CTD phosphorylation early in the transcription cycle, while the Ctk complex implements CTD phosphorylation during the elongation phase of RNA Pol II (9). CDK9 was long considered to be the only CTD Ser2 kinase in metazoans, but recently the Drosophila dCdk12/dCyclin K complex was shown to be the major CTD Ser2 kinase implementing Ser2 phosphorylation during the elongation stage, analogous to the S. cerevisiae Ctk1/2 complex (10). In humans, there are two proteins (CDK12 and CDK13) that are homologous to dCdk12 (11). Two studies (11, 12) have demonstrated that human CDK12 can interact with CCNK and phosphorylate a recombinant CTD in in vitro CTD kinase assays. CDK12, CDK13, and CCNK are highly expressed in mouse embryonic stem cells and are required for stem cell self-renewal (13). Furthermore, in Caenorhabditis elegans, CDK12 was shown to be the predominant germ line Ser2 kinase, but its loss did not lead to bulk changes in CTD phosphorylation in somatic cells (14). CDK12 in mammalian cells has been shown to regulate DNA damage response genes and is required for genome stability (11, 15).
CDK12 and CDK13 each contain N-terminal arginine-serine (RS) dipeptide-rich regions, which are frequently found in splicing factors and regulators of RNA processing (16). Consistent with a role in RNA processing, CCNK, CDK12, and CDK13 are localized in nuclear speckles, which are subnuclear structures enriched for splicing factors (16, 17). CDK12 has been shown to regulate the splicing pattern of an E1a minigene in P19 cells (18), neurexin V in glial cells (19), and 3′-end processing of MYC in HeLa cells (20). CDK13 was demonstrated to interact with the splicing factor SRSF1 and to regulate the alternative splicing of HIV (21, 22). However, the relative roles of CDK12 and CDK13 in these processes are largely unknown.
In this study, we used biochemical and genome-wide approaches to better understand the cellular functions of CDK12 and CDK13. We found that CDK12 and CDK13 associate with a large number of splicing factors and that they positively regulate the expression of a large number of genes. Depletion of CDK12 or CDK13 could downregulate gene expression and result in defects in RNA processing, with little effect on global CTD phosphorylation. We propose that in mammalian cells, in addition to CTD phosphorylation, CDK12 and CDK13 may have additional roles in RNA processing through interacting directly with RNA processing factors and by regulating their expression.
(This work was performed to fulfill, in part, requirements for the Ph.D. thesis research of K. Liang as a student registered with the Open University.)
MATERIALS AND METHODS
Expression plasmids and cell lines.
Human CDK12 and CDK13 cDNAs were purchased from Open Biosystems. The cDNAs were PCR amplified and cloned into the pCDNA5/FRT-TO plasmid (Invitrogen) with a Flag tag at the N terminus. The expression plasmids were transfected into 293 Flp-in-TRex cells with the plasmid POG44 and selected with hygromycin to generate stable cell lines. The expression of Flag-tagged proteins was induced with 1 μg/ml tetracycline for 48 h. HEK-293T and HCT116 cells were purchased from ATCC. Flp-in-TRex (Invitrogen) and HCT116 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
Antibodies.
The monoclonal antibody E7, recognizing alpha tubulin, was purchased from the Developmental Studies Hybridoma Bank. The M2 Flag monoclonal antibody was purchased from Sigma. The cyclin K (CCNK) antibody (A301-939A) was purchased from Bethyl Laboratories. Pol II unphosphorylated (8WG16), Ser2P (H5), and Ser5P (H14) CTD antibodies were purchased from Covance. Pol II Ser2P (3E10), Ser5P (3E8), and Ser7P (4E12) antibodies were purchased from Millipore. His-tagged human CDK12 (amino acids [aa] 1207 to 1481) and CDK13 (aa 1137 to 1463) C-terminal regions were used to generate their respective antisera in rabbits.
RNA interference (RNAi) and reverse transcription-quantitative PCR (RT-qPCR).
TRC lentiviral human CDK12 (RHS4533; accession no. NM_015083), CDK13 (RHS4533; accession no. NM_003718), and CCNK (RHS4533; accession no. NM_001099402) short hairpin RNA (shRNA) sets were purchased from Open Biosystems. Lentiviral particle preparation and infection were performed as previously described (23). Total RNA was extracted with TRIzol reagent (Invitrogen), and residual DNA was digested with RNase-free DNase I (NEB). RNA levels were measured with Power SYBR master mix (Invitrogen).
Primers for RT-qPCR had the following sequences (5′ to 3′): for CDK9, AGC TCG CCA AGA TCG GCC AAG and ATC TCC CGC AAG GCT GTA ATG GG; for CDK12, TGG ACT TGC TCG GCT CTA TAA CTC and CCC AAG AAT ACA TCC ACA GCT CCA; for CDK13, ACG TGT CCC CTA GTC CCT ACA and GCC TAG ATG AAT ACG GGC TTC TG; for CCNK, CTC CCA AAG AAG AGA ACA AAG CA and AGG CAA CGG TGG ATG AGT G; for BRCA1, TTC ACC CTC TGC TCT GGG TA and TGG TCA CAC TTT GTG GAG ACA; for APEX1, GAG TAG GGC AAC GCG GTA AA and TTC TTT GCG GCC GTC TTA CT; and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), AGC CAC ATC GCT CAG ACA C and GCC CAA TAC GAC CAA ATC C.
Flag purification, MudPIT analysis, and size-exclusion chromatography.
Nuclear extract preparation and Flag affinity purifications were performed with M2 anti-Flag–agarose (Sigma) in the presence of Benzonase (Sigma). Flag peptide eluates were precipitated with trichloroacetic acid (TCA). After washing with acetone, the protein mixtures were digested with endoproteinase Lys-C and trypsin (Roche) and analyzed by multidimensional protein identification technology (MudPIT) as previously described (24). The eluates from the Flag purifications of CDK12 and CDK13 were individually subjected to a Superose 6 HR size-exclusion chromatography column (GE Healthcare) containing size-exclusion buffer (40 mM HEPES [pH 7.5], 350 mM NaCl, 10% glycerol, and 0.1% Tween 20). The fractions were collected and analyzed by silver staining and Western blotting.
Pol II CTD kinase assays.
Kinase activities of CDK12 and CDK13 complexes were measured in the presence of 100 ng His-tagged Pol II CTD (Calbiochem) or His-tagged CTD (Abcam) and 2 μCi [γ-32P]ATP in 20 μl kinase buffer (20 mM HEPES [pH 7.9], 8 mM MgCl2, 0.5% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol [DTT]) (with or without 0.1 μM, 1 μM, or 10 μM flavopiridol). After incubation at 37°C for 1 h or 10 h, reactions were stopped by adding 2× SDS loading buffer, and reaction mixtures were subjected to SDS-PAGE. Phosphorylated proteins were visualized by autoradiography or immunoblotted with phosphorylation-specific antibodies.
Transcriptome sequencing (RNA-seq) analysis.
Two independent shRNAs were used to generate lentiviral particles. Total RNA was prepared from lentivirus-transduced HCT116 cells. Total RNA was depleted of rRNA by use of Ribo-zero (Illumina), and libraries were made with a Tru-Seq mRNA kit (Illumina) and subjected to Hi-Seq (Illumina) analysis. Sequencing data were acquired through the default Illumina pipeline by using Casava v1.8. Reads from two biological replicates for each sample were aligned to the human genome UCSC hg19 and to gene annotations from Ensembl 67 by using TopHat v2.0.10 (25). Cuffdiff v1.3.0 was used to perform differential expression analysis, with a false discovery rate (FDR) cutoff of <0.05 (26). Certain types of RNA (protein-encoding RNA, long interspersed noncoding RNA [lincRNA], microRNA [miRNA], snoRNA, snRNA, and noncoding RNA) were selected for further analysis. In addition, R package edgeR 3.0.8 was used to perform differential expression analysis, using a P value of <0.01 to determine significance (27). snRNA and snoRNA analysis was based on the edgeR output.
Microarray data accession number.
RNA-seq data have been deposited in the Gene Expression Omnibus under accession number GSE58107.
RESULTS
Flag purification of CDK12 and CDK13 complexes.
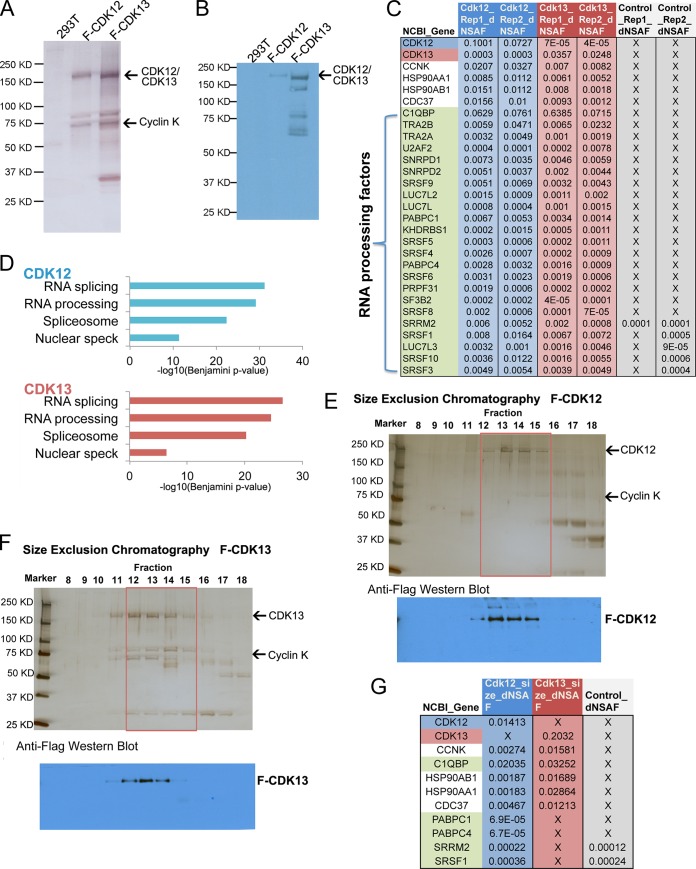
In order to further investigate CDK12 and CDK13 activities and functions, we generated inducible stable cell lines expressing Flag epitope-tagged CDK12 or CDK13 by using site-specific integration. Flag-CDK12 and Flag-CDK13 complexes were analyzed by silver staining and mass spectrometry (Fig. 1A to C). Significant amounts of CCNK were reproducibly detected in the MudPIT analysis of both the CDK12 and CDK13 complexes. In addition, both the splicing factor SRSF1 and its inhibitory subunit, C1QBP (28), were enriched in both the CDK12 and CDK13 purifications compared to HEK293T cells. This is consistent with previous studies (21, 22) that demonstrated that CDK13 could bind and phosphorylate SRSF1 and C1QBP. Moreover, we found numerous other RNA processing proteins that were significantly enriched in CDK12 and CDK13 purifications compared to HEK293T cells (Fig. 1C). Flag-CDK12 and Flag-CDK13 purifications showed significant enrichment for 113 and 89 proteins, respectively. Gene ontology analysis of the identified proteins by use of DAVID (29) showed that RNA processing factors, the spliceosome, and nuclear speckle components were significantly enriched in both CDK12 and CDK13 purifications (Fig. 1D). These results are consistent with the presence of arginine- and serine-rich (RS) domains within CDK12 and the localization of CDK12 and CDK13 to nuclear speckles, in which splicing factors are highly enriched (16, 17, 30). To more rigorously purify CDK12 and CDK13 complexes, the Flag eluates were applied to a Superose 6 size-exclusion column. Silver staining and anti-Flag Western blotting (Fig. 1E and F) of the fractionated complexes demonstrated that both complexes eluted in fractions 12 to 15, similarly to endogenous CDK12 and CDK13 (data not shown). The peak fractions were pooled and subjected to MudPIT analysis (Fig. 1G). Significant enrichments of C1QBP, CCNK, HSP90AB1, HSP90AA1, and HSP90 cochaperone CDC37 peptides were identified. HSP90 and CDC37 were previously shown to form a kinase-specific chaperone complex with CDK9 that is essential for the assembly of the CDK9/cyclin T1 complex (31). These chaperones may serve a similar role in CDK12 and CDK13 complex formation with CCNK.
Fig 1.
Purification of CDK12 and CDK13 complexes. (A and B) Flag-purified CDK12 and CDK13 complexes were analyzed by silver staining (A) and Western blotting with the M2 Flag monoclonal antibody (B). Arrows indicate the positions of Flag-CDK12, Flag-CDK13, and cyclin K. (C) MudPIT analysis of CDK12 and CDK13 complexes. Flag-CDK12 and Flag-CDK13 purifications identified numerous RNA processing factors. (D) Gene ontology analysis showed that the CDK12- and CDK13-associated proteins are significantly enriched for factors involved in RNA processing, splicing, the spliceosome, and nuclear splicing speckles (nuclear speck). (E and F) Superose 6 size-exclusion chromatography of Flag-CDK12 and Flag-CDK13 purifications. Silver staining and anti-Flag Western blotting demonstrated that both CDK12 and CDK13 complexes peaked in fractions 13 and 14 (∼1 to 2 MDa). (G) MudPIT analysis of fractions 12 to 15 (red boxes in panels E and F) was performed, and the top interacting proteins are shown.
We found that the splicing regulator C1QBP was highly enriched in CDK12 and CDK13 fractions 12 to 15. SRSF1, polyadenylate-binding protein 1, polyadenylate-binding protein 4, and serine/arginine repetitive matrix 2 were enriched in CDK12 fractions 12 to 15 but not in the corresponding CDK13 fractions. These RNA processing proteins may interact more specifically with CDK12 complexes, or they may dissociate during size-exclusion chromatography. Together, these data support previous reports of CDK12 and/or CDK13 functioning in RNA processing (18, 21).
Characterization of CDK12- and CDK13-dependent Pol II CTD phosphorylation.
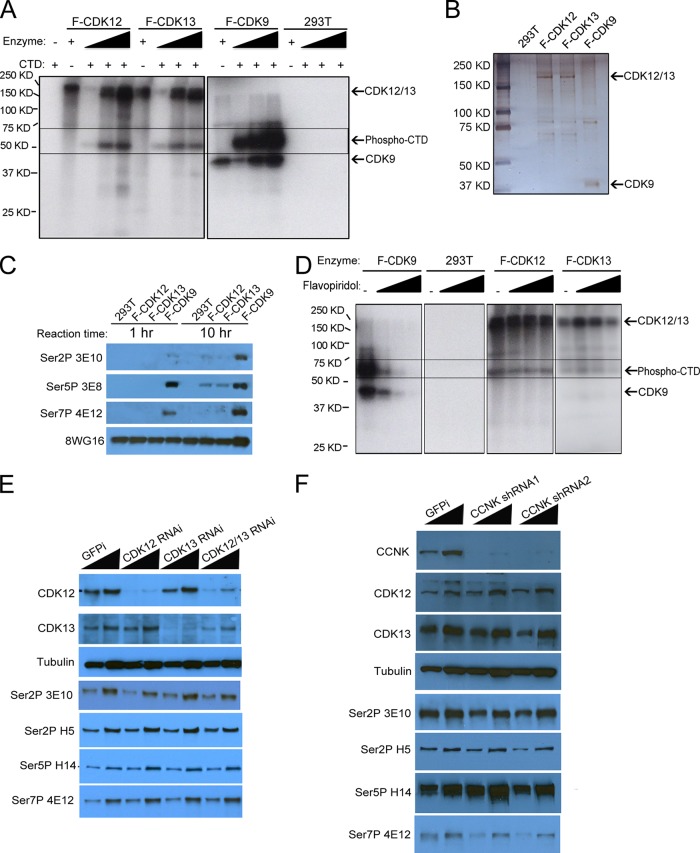
To characterize CDK12 and CDK13 as CTD kinases, we performed in vitro CTD kinase assays (Fig. 2A and B) with the Flag-purified CDK12 and CDK13 complexes, along with Flag-purified CDK9 complexes as a positive control. CDK12, CDK13, and CDK9 complexes were normalized by silver staining so that similar amounts of CDKs were used in the kinase assays (Fig. 2B). The purified CDK12 and CDK13 complexes could phosphorylate CTD in vitro; however, the kinase activities were much weaker than those of CDK9 complexes. CDK12, CDK13, and CDK9 were each autophosphorylated in the kinase assay. In order to determine the sites of phosphorylation of the Pol II CTD implemented by CDK12 and CDK13, we performed Western blotting of Pol II CTD kinase assay products by using the following phosphorylation site-specific monoclonal antibodies: 3E10 (specific for Ser2P), 3E8 (recognizes Ser5P and, to a lesser extent, Ser2P), and 4E12 (specific for Ser7P) (32). The CTD phosphorylated by CDK9 was recognized by all three antibodies (Fig. 2C). However, none of the phospho-CTD monoclonal antibodies recognized the CTD phosphorylated by CDK12 or CDK13, although the 8WG16 signal remained strong. We considered that the discrepancy between the hot kinase and Western blot kinase assays may have been due to the difference in sensitivity of these two methods. Therefore, we extended the reaction time from 1 h to 10 h. As shown in Fig. 2C, we detected low levels of Ser2P and Ser5 phosphorylation by CDK12 and CDK13. These results are consistent with recent studies that observed low levels of CTD phosphorylation implemented by CDK12 (33).
FIG 2.
Characterization of CDK12 and CDK13 activities on Pol II CTD phosphorylation. (A) Pol II CTD kinase activities of CDK12, CDK13, and CDK9 complexes. In vitro CTD kinase assays were performed with purified CDK complexes, recombinant His-tagged CTD, and [γ-32P]ATP. The reaction products were subjected to SDS-PAGE and autoradiography to assess phosphorylated Pol II levels. Triangles indicate increasing amounts of the indicated enzyme used in the assay mixtures. Arrows indicates the positions of CDK9, CDK12, CDK13, and the His-tagged CTD. (B) Silver staining of CDK9, CDK12, and CDK13 complexes used in the CTD kinase assays. (C) Western blotting of phosphorylated recombinant CTD by use of phosphorylation-specific antibodies. A reaction time of 1 h gave no detectable signal for the CDK12- or CDK13-phosphorylated CTD with the 3E10 (Ser2P), 3E8 (Ser5P), and 4E12 (Ser7P) monoclonal antibodies, while the CDK9-phosphorylated CTD was recognized by all of these antibodies. The 8WG16 monoclonal antibody preferentially recognizes unphosphorylated CTD repeats. After extension of the reaction time to 10 h, modest Ser2P and Ser5P signals could be detected with CDK12 and CDK13 complexes. (D) CDK12 and CDK13 complexes are not as sensitive as CDK9 to flavopiridol inhibition. Flavopiridol (100 nM, 1 μM, and 10 μM) was incubated with the indicated CDK, recombinant Pol II CTD, and [γ-32P]ATP. Phosphorylated CTD levels were measured by SDS-PAGE and autoradiography. (E and F) CDK12, CDK13, and CCNK depletion by shRNA knockdown in HCT116 cells did not affect bulk Pol II CTD phosphorylation levels as assayed with the indicated antibodies. The nontargeting shRNA (GFPi) was used as a negative control. Cell lysates were made at 3 days posttransduction and were analyzed by Western blotting. N20 is a polyclonal antibody recognizing total Pol II levels. Tubulin served as a loading control.
Flavopiridol is a widely used CDK9 inhibitor and has been shown to inhibit CTD phosphorylation and gene transcription in vivo (34, 35). To find if flavopiridol could inhibit CDK12 and CDK13, we tested different concentrations of flavopiridol (100 nM, 1 μM, and 10 μM) in CTD kinase assays (Fig. 2D). CDK9 kinase activity could be blocked completely by 1 μM and 10 μM flavopiridol. However, CDK12 and CDK13 complexes were less sensitive to flavopiridol, as the kinase activities exhibited only modest decreases in activity with increased concentrations of flavopiridol. Therefore, despite CDK12 and CDK13 being the CDKs most highly related to CDK9 (36), their differential sensitivity to flavopiridol indicates differences in the catalytic sites between CDK9 and CDK12/13.
Since previous reports on Drosophila, C. elegans, and mammalian cells indicated that CDK12/CDK13 homologs are required for the Ser2P CTD modification in vivo, we performed Western blotting with HCT116 cells after CDK12 and CDK13 RNAi. Under these conditions, despite obtaining significant reductions in CDK12 and CDK13 protein levels, we did not observe large decreases in the signals from the phosphorylation-specific antibodies (Fig. 2E), although a previous study of HCT116 cells found modest reductions in bulk Ser2 phosphorylation (11). Therefore, we used shRNAs to knock down CCNK, the cyclin partner for both CDK12 and CDK13, in HCT116 cells. As shown in Fig. 2F, more than 90% of the CCNK was depleted by use of two independent shRNAs. However, we still did not observe large decreases in CTD phosphorylation, suggesting possible differences from what was previously observed for Drosophila CDK12 (10). However, we cannot rule out the possibility that these enzymes regulate CTD phosphorylation in these cells, since it is possible that the levels of knockdown achieved in our studies were not sufficient to result in bulk decreases in CTD phosphorylation.
CDK9, CDK12, and CDK13 have positive effects on gene expression.
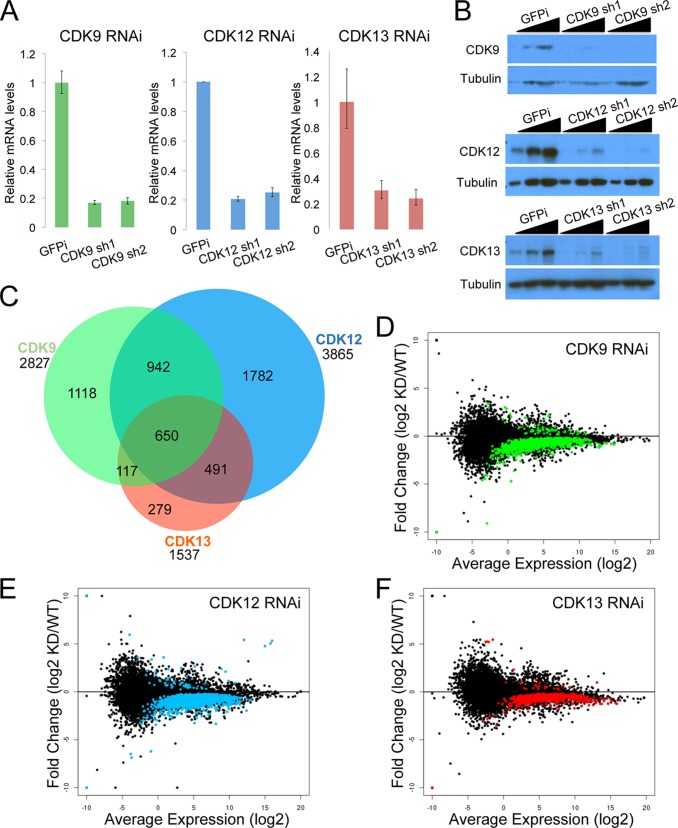
Although we did not observe changes in CTD phosphorylation in CDK12 and CDK13 knockdown cells, we wished to determine if there were any effects from the reduced levels of these proteins. Therefore, we performed RNA-seq analysis in the presence and absence of CDK9, CDK12, and CDK13. We established stable knockdown HCT116 cell lines with nontargeting (GFPi), CDK9, CDK12, and CDK13 shRNAs. Each CDK was targeted by two different shRNAs. Knockdown efficiencies measured by RT-qPCR indicated that the RNAi achieved about 80% suppression of the targets (Fig. 3A). Western blotting confirmed a strong reduction of CDK protein levels with these shRNAs (Fig. 3B). All of these shRNAs were designed to be specific to their targets, and no cross-reaction with other CDKs was observed (data not shown).
FIG 3.
CDK9, CDK12, and CDK13 have positive effects on gene expression in vivo. (A) RT-qPCR results showing the efficiencies of CDK9, CDK12, and CDK13 knockdowns by two individual shRNAs in HCT116 cells. (B) Knockdown efficiency was further confirmed by Western blotting with the indicated antibodies. (C) Venn diagram of the differentially expressed transcripts after RNAi of CDK9, CDK12, and CDK13. The majority of transcripts (74%) affected by CDK13 knockdown were affected by CDK12 knockdown. (D to F) Log2 ratio (M) versus mean average (A) plots showing that most differentially regulated transcripts (indicated in color) were downregulated by CDK9 (D), CDK12 (E), or CDK13 (F) depletion. The y axis of each plot shows the log2 fold change in transcript expression of the knockdown over the wild type. The x axis of each plot shows the log2 average expression level.
RNA-seq was performed on cells, using two nontargeting replicates and two different shRNAs for each CDK knockdown. Differentially expressed genes were called with Cuffdiff, using an FDR cutoff of <0.05. In total, 2,827 genes were differentially expressed in the CDK9 knockdown cells (2,766 downregulated), 3,865 genes in the CDK12 knockdown cells (3,804 downregulated), and 1,537 genes in the CDK13 (1,510 downregulated) knockdown cells (Fig. 3C). Interestingly, 650 of the genes affected by CDK9 knockdown were also changed by CDK12 and CDK13 knockdown, corresponding to ∼23.0%, ∼16.8%, and ∼42.3% of CDK9, CDK12, and CDK13 genes, respectively. Furthermore, CDK12 and CDK13 knockdown cells had 1,141 genes with altered expression in common, corresponding to ∼29.5% and ∼74.2% of CDK12 and CDK13 genes. A total of 1,118 genes with changed expression in CDK9 knockdown cells were not differentially expressed in either the CDK12 or CDK13 knockdown sample. A total of 39.5% of the CDK9-responsive genes were distinct, while 46.1% of CDK12-responsive genes (1,782 genes) and 18.2% of CDK13-responsive genes (279 genes) were unique to their respective knockdown conditions. Taken together, these observations indicate that different CDK proteins control distinct subsets of genes in vivo, with CDK12 and CDK13 sharing more overlap in function than either protein with CDK9.
For each CDK knockdown, most of the differentially expressed genes were downregulated, with a very small subset of genes being upregulated (Fig. 3D to F). These data indicate that CDK9, CDK12, and CDK13 have positive effects on gene expression, but with differences in the gene sets that are differentially expressed.
CDK12 and CDK13 losses preferentially affect DNA damage response and snRNA gene expression, respectively.
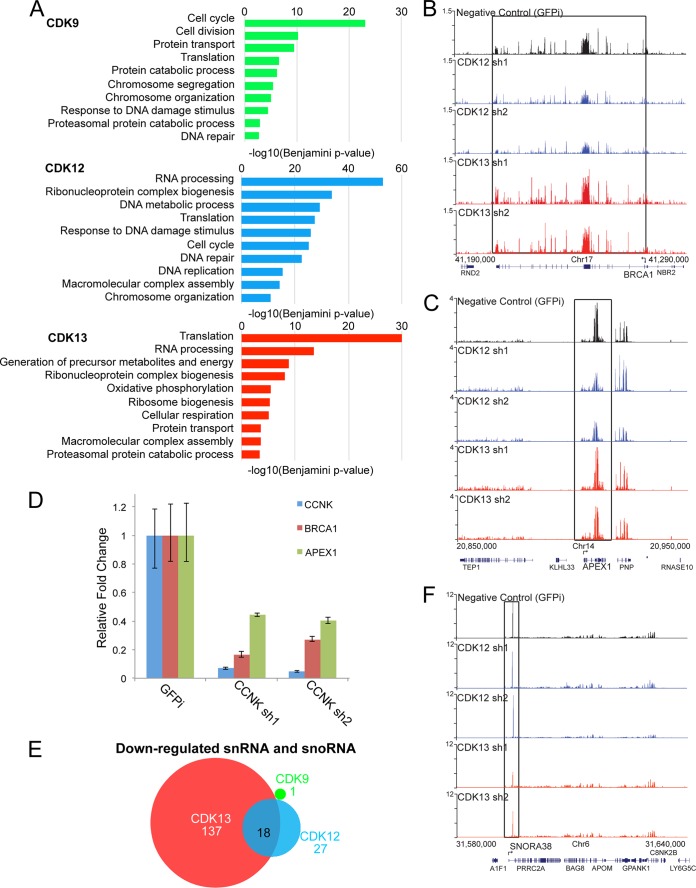
To determine which classes of genes are regulated by each CDK, we performed gene ontology analysis on the differentially expressed genes (Fig. 4A). The set of differentially expressed genes upon CDK9 knockdown were enriched for a broad range of cellular processes, including the cell cycle, cell division, protein transport, translation, DNA repair, and chromosome organization. CDK12 knockdown also affected genes involved in DNA repair, chromosome organization, and the cell cycle. DNA damage response genes were not significantly enriched under the CDK13 knockdown condition. For example, genome browser tracks for the DNA damage genes BRCA1 (Fig. 4B) and APEX1 (Fig. 4C) show that the mRNA levels of BRCA1 and APEX1 were specifically reduced by depletion of CDK12 but not CDK13. To verify the RNA-seq analysis, we used quantitative RT-PCR to determine the BRCA1 and APEX1 mRNA levels after CCNK knockdown (Fig. 4D). Two independent shRNAs were able to reduce CCNK mRNA more than 90%. Both BRCA1 and APEX1 levels were reduced in the CCNK knockdown cells as assayed by quantitative RT-PCR (Fig. 4D).
Fig 4.
CDK12 and CDK13 losses affect expression of different sets of genes. (A) Gene ontology analysis of CDK-regulated genes. CDK12 loss preferentially affected genes involved in RNA processing and DNA damage, while CDK13 loss preferentially affected genes involved in translation. (B and C) Genome browser track examples for DNA damage repair genes BRCA1 (B) and APEX1 (C), whose expression was affected by CDK12 but not CDK13 knockdown. (D) Depletion of CCNK also resulted in reduced expression of BRCA1 and APEX1 in HCT116 cells. (E) CDK13 loss affected snRNA and snoRNA gene expression more than CDK9 or CDK12 loss did. The Venn diagram shows the numbers of downregulated snRNA and snoRNA genes after knockdown of CDK9, CDK12, and CDK13. (F) Levels of snoRNA38 were downregulated by depletion of CDK13 but not CDK12.
Both CDK12 and CDK13 knockdowns affected the expression of genes involved in RNA processing. However, the CDK13-regulated gene set also showed enrichment for genes functioning in energy metabolism in the mitochondrion that were not affected by CDK9 or CDK12 knockdown, further demonstrating some nonoverlapping requirements for the CDKs. We noticed that expression of 137 snRNA and snoRNA genes was affected in the CDK13 knockdown cells, compared to 27 genes in the CDK12 knockdown cells and 1 gene in the CDK9 knockdown cells (Fig. 4E). For example, levels of snoRNA38 (Fig. 4F) were downregulated after depletion of CDK13 but not CDK12. Thus, CDK13 appears to be required for the proper expression of diverse classes of small noncoding RNA genes, each of which has its RNA processed by mechanisms distinct from that for protein-encoding genes.
Altered processing of SRSF1 by CDK12 knockdown.
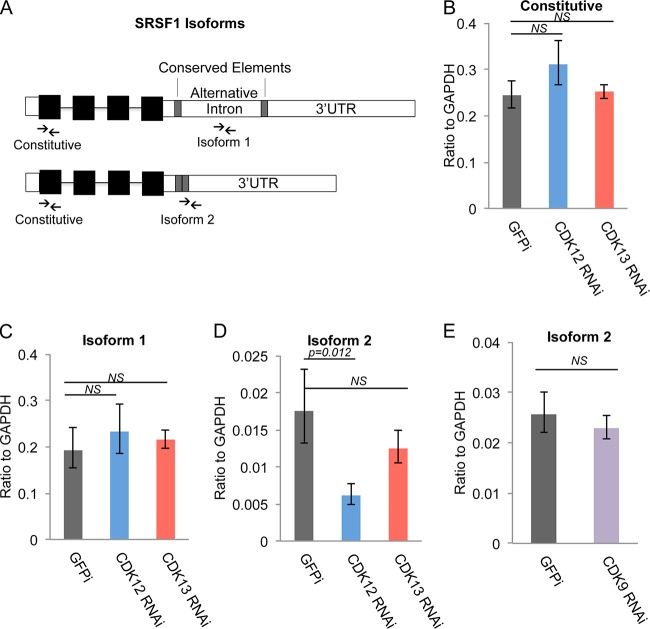
Since CDK12 and CDK13 have RS domains and associate with RNA processing and splicing factors, we tested the effects of CDK12 and CDK13 knockdowns on the processing of the SRSF1 gene, which is known to be alternatively spliced in HCT116 cells (37). SRSF1, which is upregulated in breast and lung cancers, is an oncogenic splicing factor which is targeted by MYC and mediates MYC-induced transformation (38, 39). Akaike and colleagues reported that skipping of an alternative intron in the SRSF1 3′ untranslated region (3′ UTR) increases transcript stability in HCT116 cells (37). To determine if CDK12 and CDK13 affected SRSF1 alternative splicing, we measured the levels of SRSF1 isoform 1 and isoform 2, which lacks the alternative intron, by using RT-qPCR analysis of CDK12 and CDK13 knockdown cells (Fig. 5A). Neither CDK12 nor CDK13 depletion affected the global levels of constitutive SRSF1 exons and isoform 1, but CDK12 knockdown led to reduced levels of minor isoform 2, which lacks the alternative intron (Fig. 5B to D). As a control, CDK9 knockdown did not change the relative expression of SRSF1 isoform 2 (Fig. 5E).
FIG 5.
Altered SRSF1 alternative splicing by CDK12 knockdown. (A) SRSF1 has two different isoforms in HCT116 cells. Compared to isoform 1, the isoform 2 transcripts lack an intron in the 3′ UTR. Arrows indicate the primers for RT-qPCR analysis of constitutive and isoform-specific SRSF1 transcripts. (B to D) Relative expression levels of SRSF1 and its specific isoforms after CDK12 and CDK13 depletion for 3 days. The CDK12 knockdown led to a significant decrease in expression of isoform 2, which lacks the 3′-UTR intron. (E) Relative expression levels of SRSF1 isoform 2 after CDK9 knockdown for 3 days. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
Our in vitro kinase results and in vivo depletion of CDK12 and CDK13 were not able to provide support for a role for CDK12 and CDK13 as major Ser2P CTD kinases in HCT116 cells. Interestingly, while the manuscript was under preparation, it was reported that CDK12's ability to phosphorylate a peptide containing only canonical repeats was stimulated by preexisting Ser7P but was still not as active as CDK9, suggesting that there may be some functional differences between the mammalian and Drosophila CDK12 homologs (33). Although our knockdowns of CDK12, CDK13, and CCNK were not sufficient to decrease Ser2-CTD phosphorylation, the levels of knockdown were sufficient for observation of numerous defects in gene expression, including DNA damage response genes and genes for RNA processing factors. The association of CDK12 and CDK13 with a large number of RNA processing factors raises the possibility that the RNA processing factors are themselves substrates of these kinases and that some of the gene expression defects are consequences of RNA processing defects.
Attempts at determining genome-wide binding profiles of CDK12 and CDK13 were unsuccessful using antibodies to CDK12 and CDK13 or using Flag antibodies with the Flag-CDK12 and -CDK13 HEK293T stable cell lines. These failures may have been due either to an inability of the antibodies to work for chromatin immunoprecipitation or to these factors interacting too transiently and indirectly with chromatin to be cross-linked efficiently. Consistent with the latter explanation was an immunofluorescence analysis demonstrating that CDK12 and CDK13 are localized to nuclear speckles, which are believed to be sites of storage and exchange of RNA processing factors (16, 40).
Analysis of gene expression after RNAi-mediated knockdown of CDK9, CDK12, and CDK13 showed that most of the differentially expressed genes were downregulated, indicating that all three factors have positive effects on gene expression. Gene ontology analysis showed that CDK12 and CDK13 have distinct gene specificities even though they show extensive sequence similarity. We confirmed that CDK12 regulates DNA damage response genes (11) and found that CDK13 is more specific for snRNA and snoRNA gene regulation. snoRNAs are small noncoding RNAs involved in posttranscriptional modification of rRNA. Most snoRNAs are located in introns of genes encoding proteins involved in translation (41). snoRNAs are transcribed under the control of the promoter of the protein-encoding gene and are cotranscriptionally processed to form the mature RNA (42). We found that CDK13 regulates the expression of snoRNA genes but not the corresponding protein-encoding genes, suggesting that defects in their RNA processing, not transcription, are the primary cause of the reduced levels of snoRNAs.
Altered RNA processing was also observed for the oncogenic splicing factor SRSF1, which plays an important role in breast and lung cancer development (38). We found that CDK12 could stabilize SRSF1 mRNA transcripts through skipping of an alternative intron in the 3′ UTR. It is conceivable that CDK12 affects RNA splicing through CTD phosphorylation, since CTD phosphorylation affects the recruitment of RNA processing factors (3, 20). However, in our RNAi experiments, gene expression and RNA processing defects were observed more readily than were changes in CTD phosphorylation. A precedent for a factor modifying the CTD and broadly regulating RNA processing is the arginine methyltransferase CARM1. CARM1 methylates a single conserved residue in the CTD, interacts with and methylates numerous proteins involved in RNA processing, and affects the expression of genes involved in RNA processing (43, 44).
The CDK12 gebe is one of the most frequently mutated genes in ovarian cancer and also plays an important role in the development of breast cancer (16). CDK13 was reported to be necessary for megakaryocyte development, and its expression was increased in some patients with refractory anemia (16). A previous study by Blazek and colleagues (11) showed that cells depleted of CCNK or CDK12 are sensitive to various DNA damaging agents, including camptothecin, mitomycin C, and etoposide. CDK12 regulates DNA damage response genes, which may be required for maintenance of genome stability through detection and repair of DNA lesions. Therefore, mutation of CDK12 may result in genome instability and contribute to oncogenesis. The misregulation of RNA processing has also been implicated in oncogenesis (45), and elucidating the roles of CDK12 and CDK13 functions in RNA processing may give insight into the treatment of cancers.
ACKNOWLEDGMENTS
We thank the Molecular Biology Core Facility at the Stowers Institute for creating and sequencing libraries for next-generation sequencing and the Tissue Culture Facility for large-scale cell culture and establishment of stable cell lines. We thank Alex Garruss for the initial analysis of the RNA-seq data. We thank Laura Shilatifard and Lisa Lassise for editorial assistance.
This study was supported by grants from the National Institutes of Health (grants 5R01CA089455 and 5R01GM069905).
REFERENCES
- 1.Corden JL. 1990. Tails of RNA polymerase II. Trends Biochem Sci 15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 2.Phatnani HP, Greenleaf AL. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 3.Hsin JP, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev 26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komarnitsky P, Cho EJ, Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall NF, Peng J, Xie Z, Price DH. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51. doi: 10.1016/S0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 8.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, Buratowski S, Handa H. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood A, Shilatifard A. 2006. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle 5:1066–1068. doi: 10.4161/cc.5.10.2769. [DOI] [PubMed] [Google Scholar]
- 10.Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM. 2011. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng SW, Kuzyk MA, Moradian A, Ichu TA, Chang VC, Tien JF, Vollett SE, Griffith M, Marra MA, Morin GB. 2012. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol Cell Biol 32:4691–4704. doi: 10.1128/MCB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai Q, Lei T, Zhao C, Zhong J, Tang YZ, Chen B, Yang J, Li C, Wang S, Song X, Li L, Li Q. 2012. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J Biol Chem 287:25344–25352. doi: 10.1074/jbc.M111.321760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowman EA, Bowman CR, Ahn JH, Kelly WG. 2013. Phosphorylation of RNA polymerase II is independent of P-TEFb in the C. elegans germline. Development 140:3703–3713. doi: 10.1242/dev.095778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. 2014. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem 289:9247–9253. doi: 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohoutek J, Blazek D. 2012. Cyclin K goes with Cdk12 and Cdk13. Cell Div 7:12. doi: 10.1186/1747-1028-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko TK, Kelly E, Pines J. 2001. CrkRS: a novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J Cell Sci 114:2591–2603. [DOI] [PubMed] [Google Scholar]
- 18.Chen HH, Wang YC, Fann MJ. 2006. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol Cell Biol 26:2736–2745. doi: 10.1128/MCB.26.7.2736-2745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues F, Thuma L, Klambt C. 2012. The regulation of glial-specific splicing of neurexin IV requires HOW and Cdk12 activity. Development 139:1765–1776. doi: 10.1242/dev.074070. [DOI] [PubMed] [Google Scholar]
- 20.Davidson L, Muniz L, West S. 2014. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev 28:342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berro R, Pedati C, Kehn-Hall K, Wu W, Klase Z, Even Y, Geneviere AM, Ammosova T, Nekhai S, Kashanchi F. 2008. CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J Virol 82:7155–7166. doi: 10.1128/JVI.02543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Even Y, Durieux S, Escande ML, Lozano JC, Peaucellier G, Weil D, Geneviere AM. 2006. CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo. J Cell Biochem 99:890–904. doi: 10.1002/jcb.20986. [DOI] [PubMed] [Google Scholar]
- 23.Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. 2012. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol 32:2608–2617. doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petersen-Mahrt SK, Estmer C, Ohrmalm C, Matthews DA, Russell WC, Akusjarvi G. 1999. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J 18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Bingham PM. 1991. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 31.O'Keeffe B, Fong Y, Chen D, Zhou S, Zhou Q. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J Biol Chem 275:279–287. doi: 10.1074/jbc.275.1.279. [DOI] [PubMed] [Google Scholar]
- 32.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 33.Bosken CA, Farnung L, Hintermair C, Merzel Schachter M, Vogel-Bachmayr K, Blazek D, Anand K, Fisher RP, Eick D, Geyer M. 2014. The structure and substrate specificity of human Cdk12/cyclin K. Nat Commun 5:3505. doi: 10.1038/ncomms4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao SH, Price DH. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 36.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ. 2009. Cyclin-dependent kinases: a family portrait. Nat Cell Biol 11:1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akaike Y, Kurokawa K, Kajita K, Kuwano Y, Masuda K, Nishida K, Kang SW, Tanahashi T, Rokutan K. 2011. Skipping of an alternative intron in the srsf1 3′ untranslated region increases transcript stability. J Med Invest 58:180–187. doi: 10.2152/jmi.58.180. [DOI] [PubMed] [Google Scholar]
- 38.Das S, Anczukow O, Akerman M, Krainer AR. 2012. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep 1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR. 2012. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol 19:220–228. doi: 10.1038/nsmb.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tollervey D, Kiss T. 1997. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol 9:337–342. doi: 10.1016/S0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 42.Kiss T. 2004. Biogenesis of small nuclear RNPs. J Cell Sci 117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 43.Cheng D, Cote J, Shaaban S, Bedford MT. 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Sims RJ III, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ III, Eick D, Reinberg D. 2011. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim MY, Hur J, Jeong S. 2009. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep 42:125–130. doi: 10.5483/BMBRep.2009.42.3.125. [DOI] [PubMed] [Google Scholar]