Abstract
Chronic inflammation in obese adipose tissue is linked to endoplasmic reticulum (ER) stress and systemic insulin resistance. Targeted deletion of the murine fatty acid binding protein (FABP4/aP2) uncouples obesity from inflammation although the mechanism underlying this finding has remained enigmatic. Here, we show that inhibition or deletion of FABP4/aP2 in macrophages results in increased intracellular free fatty acids (FFAs) and elevated expression of uncoupling protein 2 (UCP2) without concomitant increases in UCP1 or UCP3. Silencing of UCP2 mRNA in FABP4/aP2-deficient macrophages negated the protective effect of FABP loss and increased ER stress in response to palmitate or lipopolysaccharide (LPS). Pharmacologic inhibition of FABP4/aP2 with the FABP inhibitor HTS01037 also upregulated UCP2 and reduced expression of BiP, CHOP, and XBP-1s. Expression of native FABP4/aP2 (but not the non-fatty acid binding mutant R126Q) into FABP4/aP2 null cells reduced UCP2 expression, suggesting that the FABP-FFA equilibrium controls UCP2 expression. FABP4/aP2-deficient macrophages are resistant to LPS-induced mitochondrial dysfunction and exhibit decreased mitochondrial protein carbonylation and UCP2-dependent reduction in intracellular reactive oxygen species. These data demonstrate that FABP4/aP2 directly regulates intracellular FFA levels and indirectly controls macrophage inflammation and ER stress by regulating the expression of UCP2.
INTRODUCTION
Obesity-linked metabolic disorders, including insulin resistance, fatty liver disease, and coronary arterial disease, share the common signature of chronic inflammation and endoplasmic reticulum (ER) stress (1, 2). Macrophage and T cell infiltration and activation in adipose tissue play a key role in affecting adipokine synthesis and secretion, thereby regulating systemic insulin resistance (3). Inflammatory cytokines increase oxidative stress and decrease the protein-folding efficiency of the ER, initiating a counterregulatory unfolded protein response (UPR) (4) involving pancreatic ER kinase (PERK), activating transcription factor-6 (ATF6), and inositol-requiring enzyme 1 (IRE1). Such concomitant activation leads to the downstream activation of response pathways and the induction of inflammatory signaling networks via c-Jun N-terminal kinase (JNK) and/or NF-κB (nuclear factor kappa B) (1).
Lipid metabolism in macrophages has been shown to play an important role in triggering inflammation and ER stress (5, 6) and has led to the identification of critical proteins that regulate the obesity-metabolic disease axis. For example, genetic ablation of the fatty acid binding protein (FABP4, also known as aP2) in macrophages alone is sufficient to protect mice from development of atherosclerosis and dyslipidemia (7). FABP4/aP2 is a cytoplasmic fatty acid (FA) carrier protein that mediates intracellular fatty acid trafficking, and a number of hypotheses have been proposed to explain why the loss of FABP4/aP2 results in metabolic improvement (6). Moreover, small molecules that target FABP4/aP2 have been developed as potential therapeutics (8). However, conflicting reports exist concerning the effectiveness of these inhibitors using cell-based and animal models (9). FABP4/aP2-deficient macrophages exhibit suppressed inflammatory signaling, attenuated activation of the NF-κB pathway, and decreased ER stress (6, 10). Consistent with a role for FABP4/aP2 as a key determinant in obesity-linked inflammation, genetic variation in the human FABP4/aP2 promoter that leads to decreased expression of the protein in adipose tissue is associated with lower serum triglyceride levels, reduced coronary disease, and type 2 diabetes (11). The biochemical processes underlying the effects of FABP4/aP2 deficiency on macrophage lipid metabolism and ER stress and inflammatory pathways are not understood but may be linked to the accumulation of intracellular unsaturated fatty acids, particularly palmitoleic acid (6, 10).
The investigation herein describes the novel finding that UCP2 (uncoupling protein 2) is upregulated selectively in macrophages from FABP4/aP2 null mice and that increased expression of UCP2 plays an important and essential role in alleviating ER stress and decreasing inflammation (12, 13). Unlike its structural homolog UCP1 that is highly expressed in brown fat, UCP2 is more broadly expressed in various tissues and cells, functions as a sensor of mitochondrial oxidative stress, and is generally considered to be cytoprotective (14). Moreover, unsaturated fatty acids increase UCP2 expression in macrophage cells, suggesting that the FABP-fatty acid equilibrium is central to mediating metabolic homeostasis.
MATERIALS AND METHODS
Cell lines.
FABP4/aP2 knockout and wild-type macrophage cells were maintained in RPMI 1640 medium (Invitrogen) with 5% fetal bovine serum (FBS). Raw 264.7 macrophages as well as UCP2 knockdown Raw 264.7 macrophages were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% FBS. Peritoneal macrophages were isolated from C57BL/6J animals. A total of 1 × 106 to 2 × 106 cells were plated and incubated overnight (15).
Intracellular fatty acid analysis.
Monolayers of cells were washed with phosphate-buffered saline (PBS) and harvested into 2 ml of 100 mM sodium acetate (pH 3.9). Lipids were extracted into hexane-isopropanol-H2O (3:2:2) and centrifuged at 3,000 rpm for 10 min to achieve phase separation. The aqueous phase was dried under nitrogen, and lipids were solubilized in 1 ml of chloroform. Samples were loaded onto an equilibrated HF Bond Elut NH2 column (Agilent Technology) and washed with chloroform-isopropanol (2:1) to remove neutral lipids, and fatty acids were eluted with 2% acetic acid in diethyl ether. The fatty acid eluate was dried and resolubilized in isopropanol for measurement of fatty acid abundance (NEFA kit; Wako) or submitted to the Metabolomics Resources Core of the Mayo Clinic (Rochester, MN) for fatty acid composition analysis. The fatty acid composition analysis was carried out by liquid chromatography-mass spectrometry (LC-MS) with C17:0 spiked in each sample as an internal standard.
Isolation of stromal vascular cells.
Epididymal fat pads were dissected from wild-type C57BL/6J and FABP4/aP2 knockout (KO) mice (n = 6) maintained on a high-fat diet for 12 weeks (16). Briefly, fat pads were minced and digested with type I collagenase in Krebs-Ringers-HEPES (KRH) buffer supplemented with 10 mg/ml bovine serum albumin (BSA). After incubation at 37°C for 1 h, the mixture was filtered with a cell strainer (100-μm-pore-size nylon; Falcon) to remove undigested tissues. The stromal vascular fraction (SVF) was collected by centrifugation at 500 × g for 10 min. The stromal vascular fraction was washed, and TRIzol reagent was used for RNA isolation. All experimental procedures using animals were reviewed and approved by the University of Minnesota Institutional Animal Care and Use Committee.
shRNA knockdown of UCP2 in macrophages.
Raw 264.7 and FABP4/aP2 KO macrophages were transduced with a short hairpin RNA (shRNA) lentivirus as described previously (17). Green fluorescent protein (GFP) scrambled and Ucp2 targeting sequences were obtained from Open Biosystems. The following were used: Ucp2 (GenBank accession number NM_011671) targeting sequence (UCP2 knockdown [UCP2 kd]), 5′-CCGGTCTCCCAATGTTGCCCGTAATCTCGAGATTACGGGCAACATTGGGAGATTTTTG-3′; alternative UCP2 targeting sequence (UCP2-2 kd), 5′-CCGGCCCAGCCTACAGATGTGGTAACTCGAGTTACCACATCTGTAGGCTGGGTTTTTG-3′; scrambled sequence, 5′-AACGTACGCGGAATACTTCGA-3′.
Expression analysis by qPCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse transcribed to cDNA using iScript according to the manufacturer's protocol (Bio-Rad). Real-time quantitative PCR (qPCR) amplification was performed on a Bio-Rad CFX 96 real-time system using SYBR green Supermix (Bio-Rad). Transcription factor II E (TFIIE) was used as an internal control to normalize expression unless specified otherwise. Primer sequences are provided in Table 1.
TABLE 1.
Real-time qPCR primer sequences
| Target | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| UCP2 | ACTGTGCCCTTACCATGCTCC | ATTGGTAGGCAGCCATTAGGG |
| PPARγ | GCCATTGAGTGCCGAGTC | TGTGGATCCGGCAGTTAAG |
| LXRα | TCAAGGGAGCACGCTATGTC | TTCCTCTTCTTGCCGCTTC |
| CD36 | TGGAGCTGTTATTGGTGCAG | TGGGTTTTGCACATCAAAGA |
| Arginase | AACACGGCAGTGGCTTTAACC | GGTTTTCATCTGGCGCATTC |
| SCD1 | CCTACGACAAGAACATTCAATCCC | CAGGAACTCAGAAGCCCAAAGC |
| iNOS | AGCGAGTTGTGGATTGTCC | TCTCTGCCTATCCGTCTCG |
| Catalase | CCAGCGACCAGATGAAGCAG | CCACTCTCTCAGGAATCCGC |
| Gpx4 | GCTGTGCGCGCTCCAT | CCATGTGCCCGTCGATGT |
| SOD1 | GCCAATGATGGAATGCTCTCC | CAATCTGACTGCTGGAAAGGAC |
| SOD2 | CCGAGGAGAAGTACCACGAG | GCTTGATAGCCTCCAGCAAC |
| Prd3 | TTA AACATGGTTAGTTGCTAGTACAAGGA | TTGAGACATGATCTAAGAATAGCCTCCTA |
| ALDH2 | TTTATCCAGCCCACCGTGTTC | CAAGCCCATACTTAGAATCATTGG |
| XBP-1s | CTGAGTCCGAATCAGGTGCAG | GTCCATGGGAAGATGTTCTGG |
| XBP-1u | TGGCCGGGTCTGCTGAGTCCG | GTCCATGGGAAGATGTTCTGG |
| F4/80 | TTTGGCTATGGGCTTCCAGTC | TCAGCAACCTCGTGTCCTTGAG |
| TFIIE | CAAGGCTTTAGGGGACCAGATAC | CATCCATTGACTCCACAGTGACAC |
RT-PCR analysis.
Reverse transcription-PCR (RT-PCR) was used to identify UCP1, UCP2, and UCP3 expression in macrophages. The following primers were used: UCP1 forward, 5′-GCCAGGCTTCCAGTACCATTA-3′; UCP1 reverse, 5′-TGGTACGCTTGGGTACTGTCC-3′; UCP2 forward, 5′-CCAGAGCACTGTCGAAGCCT-3′; UCP2 reverse, 5′-GCAGCCATTAGGGCTCTTTTG-3′; UCP3 forward, 5′-AGAACCCAGGGGCTCAGAG-3′; UCP3 reverse, 5′-AAAACGGAGATTCCCGCAGTA-3′.
Cellular respiratory assay.
A macrophage respiratory assay was performed on an XF24 extracellular flux analyzer (Seahorse Biosciences) (18). Macrophages were plated on V7 microplates at a density of 300,000 cells per plate and incubated overnight, and then cells were treated with either vehicle or lipopolysaccharide (LPS; 100 ng/ml) for 4 h. During the assay, cells were exposed to compounds in the following order: 2 μM oligomycin, 0.4 μM FCCP [carbonyl cyanide-p-(trifluoromethoxy)phenylhydrazone], and 4 μM antimycin A.
H2O2 assay.
Hydrogen peroxide (H2O2) quantification was determined using an Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen) according to the manufacturer's protocol with modification. Briefly, cells were scraped into phosphate buffer (pH 7.4) and inactivated at 95°C for 10 min. After spin-down of cell debris, 50 μl of supernatant was loaded with 50 μl of working solution. Following a 30-min incubation, fluorescence was measured using a microplate reader with excitation at 540 nm and emission at 590 nm.
Membrane potential measurement.
Mitochondrial proton motive force was measured by tetramethylrhodamine methyl ester (TMRM) staining (Invitrogen). Briefly, cells were washed with PBS and incubated in 1 ml of KRH buffer (pH 7.4) with a 20 nM final concentration of TMRM for 30 min. Then cells were washed with PBS and harvested into 300 μl of KRH buffer. Samples (150 μl each) were loaded into 96-well plates, and fluorescence was measured using a microplate reader with excitation at 531 nm and emission at 572 nm.
Mitochondrial isolation.
Cells were scraped into isolation buffer (20 mM Tris [pH 7.4], 220 mM mannitol, 70 mM sucrose, 1 mM EDTA, 0.1 mM EGTA) supplemented with protease inhibitors and lysed with 20 strokes of a Dounce homogenizer. Homogenates were centrifuged at 700 × g for 10 min to remove nuclei and unbroken cells. Mitochondria were pelleted by centrifugation at 12,000 × g for 15 min.
Immunoblotting.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors. Equal amounts of protein were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane. After a blocking step, membranes were incubated with primary antibody overnight at 4°C. Membranes were washed and incubated with secondary antibody conjugated to Li-Cor IRDye for 1 h and visualized using Odyssey infrared imaging (Li-Cor Biosciences). The antibodies used were anti-UCP2 (C-20; Santa Cruz Biotechnology), anti-4-hydroxynonenal (anti-4-HNE) (Millipore), anti-DDIT3 (anti-CHOP) (Abcam), anti-GPR78 (Bip) (H-129; Santa Cruz Biotechnology), anti-cyclooxygenase-2 (anti-Cox2) and anti-inducible nitric oxide synthase (anti-iNOS) (BD Transduction Laboratories), anti-β-actin (Sigma-Aldrich), and anti-ATP synthase-α subunit (MitoSciences).
Cytokine TNF-α measurement.
Secreted tumor necrosis factor alpha (TNF-α) in medium (8 h) was measured with a mouse TNF enzyme-linked immunosorbent assay (ELISA) set from BD Biosciences according to the manufacturer's instructions.
Fatty acid oxidation assay.
Fatty acid oxidation was carried out as described by Wiczer and Bernlohr (19). Briefly, cells were incubated for 1 h at 37°C in Krebs-Ringers-HEPES buffer (pH 7.4) containing 5.4 mM glucose and 400 μM [14C]palmitic acid bound to 100 μM fatty acid-free BSA. Cells were scraped from the plates and transferred with medium into 20-ml glass reaction vials containing a center reaction tube filled with 400 μl of 1 M sodium hydroxide. Perchloric acid (70%) was added to the medium (final concentration of 7%), and samples were incubated for 1 h with shaking at 80 rpm. After incubation, the content of the center tube was transferred into 10 ml of liquid scintillation fluid, and the 14CO2 level was determined by liquid scintillation counting.
Statistical analysis.
All data in the paper are expressed as means ± standard deviations (SDs). Statistical significance was determined using an unpaired, two-tailed Student t test.
RESULTS
Loss or inhibition of FABP4/aP2 in macrophages leads to increased UCP2 expression.
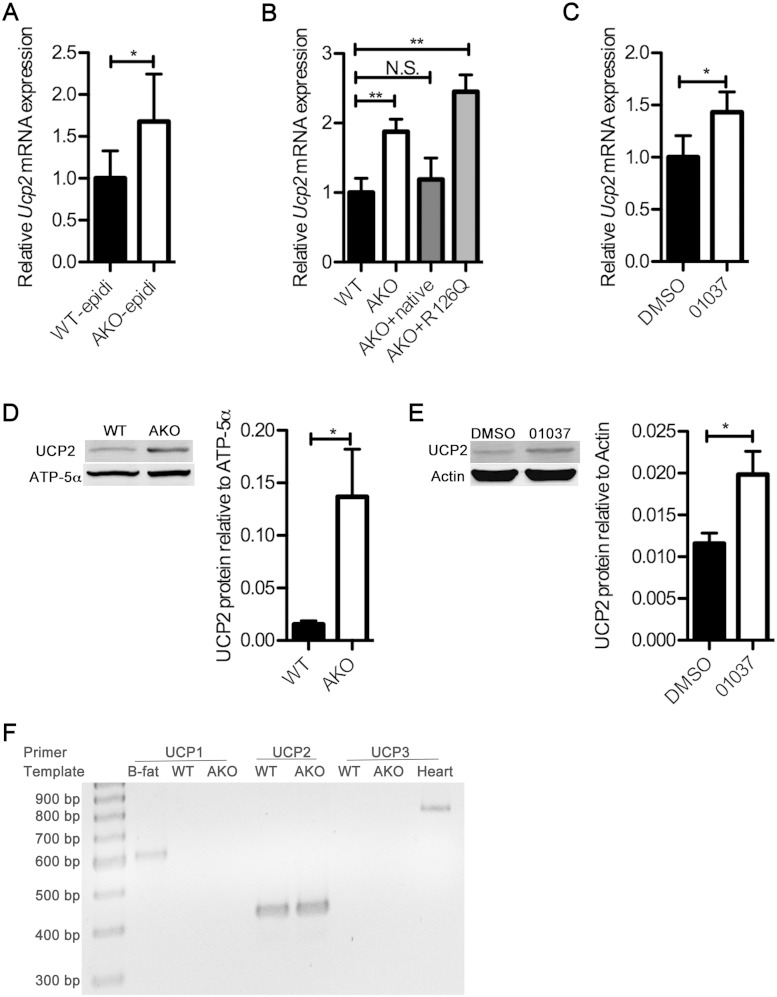
Previous reports have demonstrated that the intracellular fatty acid pool in FABP4/aP2-deficient macrophages and adipocytes is increased and has an altered composition (6, 20). Work in other systems, particularly in the liver, has shown that the expression of UCP2 mRNA and protein is increased with obesity and regulated by fatty acids and that lipid overload leads to increased expression of UCP2 as part of a counterregulatory cycle (14, 21). Since FABPs establish an intracellular equilibrium between bound and free fatty acids (FFAs), we hypothesized that loss of FABP4/aP2 would result in increased availability of lipid and upregulation of UCP2. To test this hypothesis, we evaluated the expression of uncoupling proteins using real-time qPCR and determined that UCP2 mRNA is increased approximately 60% in adipocyte FABP (AFABP)/aP2-deficient SVF cells compared to levels in wild-type mice (Fig. 1A). Similarly, UCP2 mRNA levels were increased ∼2-fold in cell lines derived from AFABP/aP2-deficient mice (referred to as AKO macrophages) compared to levels in control cells (Fig. 1B). The phenotype of FABP4/aP2 deficiency in macrophages can be mimicked by treatment of cultured macrophages with a previously identified and characterized chemical inhibitor of FABPs, HTS01037 (8). Treatment of Raw 264.7 macrophages with HTS01037 also significantly increased the UCP2 mRNA level (Fig. 1C). To further confirm that the upregulation of UCP2 is responsive to FABP4/aP2 deficiency, AFABP/aP2−/− macrophages were reconstituted with either a wild-type FABP4/aP2 or a non-fatty acid binding mutant of AFABP/aP2 (R126Q) (22) to a level comparable to that of FABP4/aP2 in wild-type macrophages (results not shown). Reexpression of wild-type FABP4/aP2 but not the R126Q mutant reduced UCP2 mRNA to a level comparable to that in wild-type macrophages (Fig. 1B), implying that the intracellular FFA-FABP equilibrium is a major control element for UCP2 expression. Analysis of UCP2 protein levels in FABP4/aP2-deficient macrophages showed that UCP2 is significantly increased compared to the wild-type level (Fig. 1D). Moreover, the treatment of Raw 264.7 macrophages with the FABP inhibitor HTS01037 also increased the UCP2 protein level (Fig. 1E). Furthermore, the increased expression of uncoupling protein was specific for UCP2 as there was no evidence of any expression of UCP1 or UCP3 (Fig. 1F). These results in sum indicate that FABP4/aP2-FFA equilibrium controls the expression of UCP2.
FIG 1.
Loss of FABP4/aP2 increases UCP2 expression. (A) UCP2 mRNA level normalized to macrophage F4/80 in the stromal vascular fraction of epididymal adipose tissue obtained from high-fat diet-fed wild-type (WT-epidi) and AFABP/aP2−/− (AKO-epidi) mice. (B) UCP2 mRNA level in wild-type, AKO, AKO+native (AFABP/aP2−/− macrophages reconstituted with WT AFABP/aP2), and AKO+R126Q (AFABP/aP2−/− macrophages reconstituted with a non-fatty acid binding mutant of AFABP/aP2) macrophages. (C) UCP2 mRNA level in Raw 264.7 macrophages treated with 30 μM HTS01037 for 24 h. DMSO, dimethyl sulfoxide. (D) UCP2 protein expression in AKO and WT macrophage mitochondrial fractions determined by Western blotting. (E) UCP2 expression in Raw 264.7 macrophages treated with 30 μM HTS01037 as determined by Western blotting. (F) RT-PCR amplification of UCP1, -2, and -3 from wild-type (WT) and AFABP/aP2−/− (AKO) macrophages. Amplification of UCP1 from brown fat (B-fat) was used as a positive control for UCP1 expression while a UCP3 expression control was amplified from a heart cDNA sample. *, P < 0.05; ** P < 0.01; NS, not significant.
Unsaturated fatty acids induce UCP2 expression in macrophages via PPARγ.
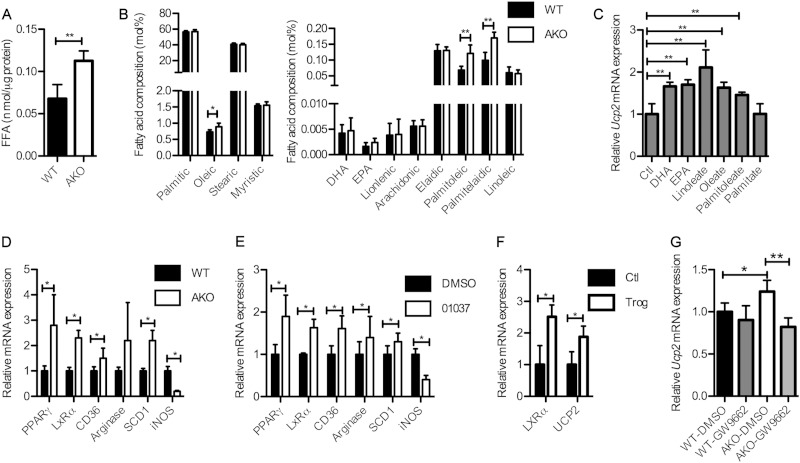
Previous reports have shown that in multiple systems UCP2 expression can be induced by unsaturated fatty acids (14, 23–25). Moreover, FABPs are identified as lipid chaperones involved in establishment of the bound-versus-free FA equilibrium (16, 26). Consistent with previous work, intracellular free fatty acids in FABP4/aP2−/− macrophages were increased ∼70% compared to the level of the wild type (Fig. 2A) (6). Moreover, fatty acid composition analysis revealed that monounsaturated fatty acid levels, such as those of oleic acid (18:1), palmitoleic acid (16:1 cis), and palmitelaidic acid (16:1 trans), were selectively elevated in FABP4/aP2−/− macrophages (Fig. 2B). To test which molecular species of fatty acid could induce UCP2 expression in macrophages, Raw 264.7 cells were treated for 24 h with different fatty acids (4:1, FFA/bovine serum albumin), and the expression of UCP2 was evaluated. The results demonstrate that polyunsaturated fatty acids, including docosahexaenoic acid (DHA; 22:6), eicosapentaenoic acid (EPA; 20:5), linoleate (18:2), and the monounsaturated fatty acids oleate (18:1) and palmitoleate (16:1) can each induce UCP2 expression in macrophages, while the saturated fatty acid palmitate (16:0) was unable to elicit any response (Fig. 2C). Unsaturated fatty acids have been shown to act as ligands of a family of transcription factors, peroxisomal proliferator-activated receptors (PPARs), which are involved in regulating the expression of a cohort of genes involved in lipid metabolism (27). PPARγ is the major form of PPARs expressed in macrophages and suppresses the expression of a large set of inflammatory genes (28). Real-time qPCR analysis showed that both PPARγ and its target genes, such as liver X receptor alpha (LXRα), cluster of differentiation 36 (CD36), arginase and stearoyl coenzyme A (stearoyl-CoA) desaturase 1 (SCD1), were upregulated in FABP4/aP2−/− macrophages compared to levels in wild-type macrophages. In contrast, the expression of the proinflammatory gene inducible nitric oxide synthase (iNOS) is downregulated in FABP4/aP2−/− macrophages (Fig. 2D). In addition, treatment of wild-type peritoneal macrophages with the FABP inhibitor HTS01037 showed similar results (Fig. 2E). In order to determine the role of PPARγ in macrophage UCP2 expression, Raw 264.7 cells were treated with the PPARγ agonist troglitazone. Troglitazone treatment increased the expression of UCP2 in macrophages, as well as LXRα (Fig. 2F). On the other hand, treatment of FABP4/aP2−/− macrophages with the PPARγ antagonist GW9662 reduced UCP2 expression to a level comparable to that in the wild type (Fig. 2G).
FIG 2.
Unsaturated fatty acids induce UCP2 expression via PPARγ. (A) Intracellular free fatty acids measured in WT (wild-type) and AKO (AFABP/aP2−/−) macrophages. (B) Fatty acid composition in WT and AKO macrophages. (C) UCP2 mRNA level in Raw 264.7 macrophages treated with 300 μM docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), linoleate, oleate, palmitoleate, or palmitate. Fatty acids were added in complex to bovine serum album at a molar ratio of 4:1. Ctl, control. (D) mRNA levels of PPARγ, liver X receptor alpha (LXRα), cluster of differentiation 36 (CD36), arginase, stearoyl-CoA desaturase 1 (SCD1), and inducible nitric oxide (iNOS) in WT and AKO peritoneal macrophages. (E) PPARγ, LXRα, CD36, arginase, SCD1, and iNOS mRNA levels in WT peritoneal macrophages treated with 10 μM HTS01037. (F) mRNA levels of LXRα and UCP2 in Raw 264.7 macrophages treated with 5 μM troglitazone for 24 h. (G) mRNA levels of UCP2 in WT and AKO macrophages treated with 5 μM GW9662 for 24 h. *, P < 0.05; ** P < 0.01.
UCP2 upregulation negates palmitate-induced ER stress in FABP4/aP2-deficient macrophages.
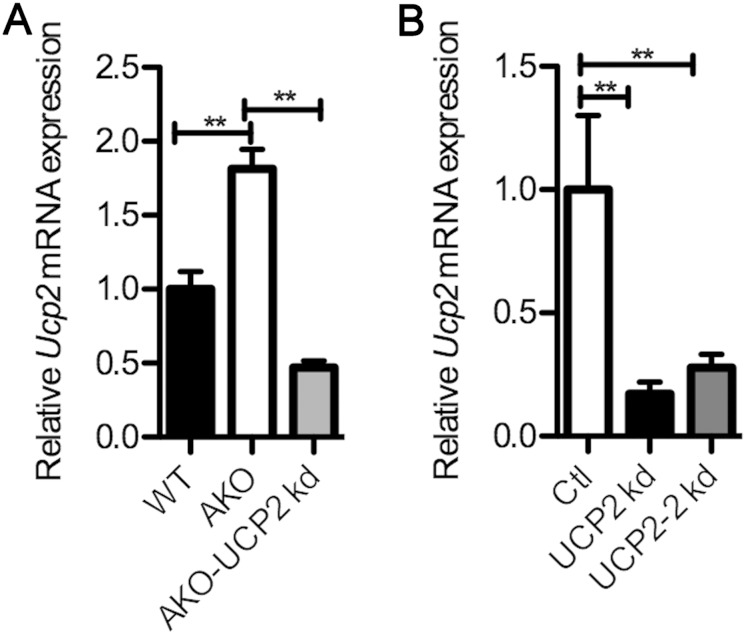
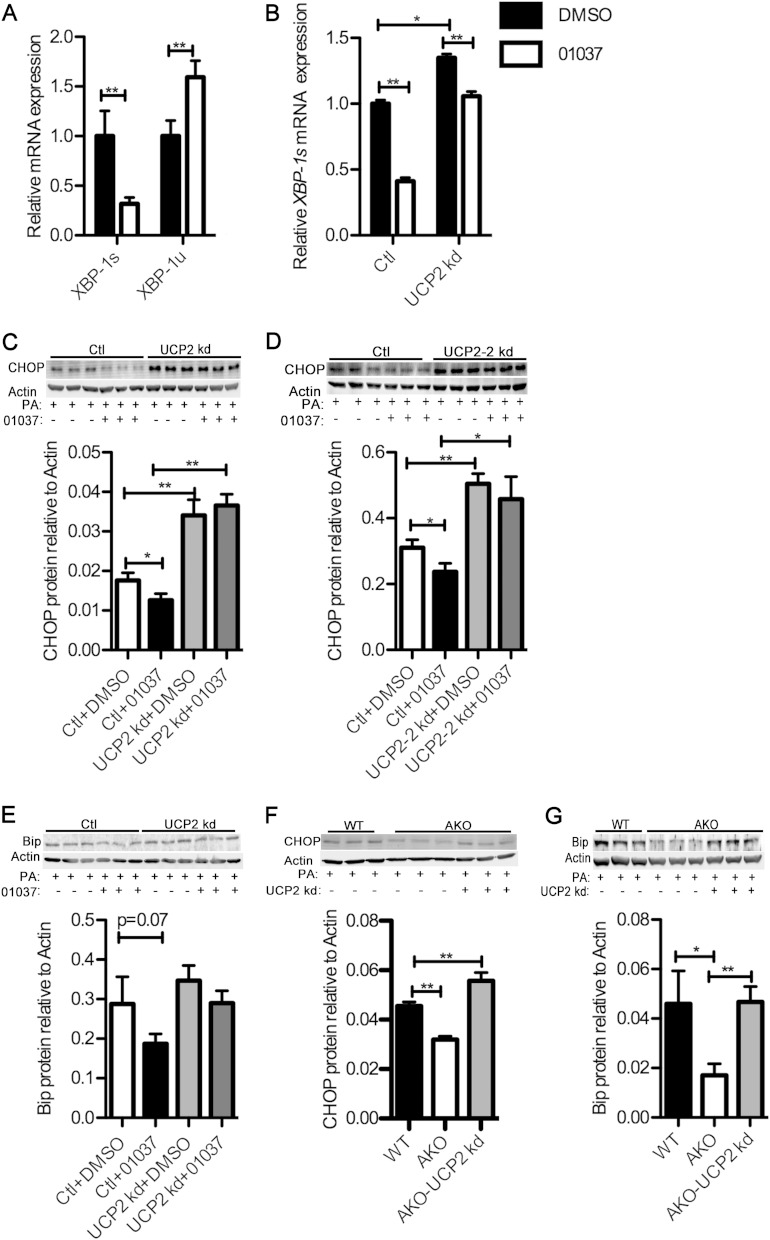
The alleviation of lipid-induced macrophage ER stress can be accomplished by either knocking out FABP4/aP2 genetically or inhibiting the FABP-fatty acid interaction with small-molecule inhibitors (6). In addition, the reduction in macrophage ER stress has been shown to offer protection against atherosclerosis (29). We therefore evaluated the role of UCP2 upregulation in FABP4/aP2−/− macrophages as mediating reduced ER stress in palmitate-treated macrophages by silencing UCP2 in AKO and Raw 264.7 macrophages (Fig. 3A and B). Spliced X-box binding protein 1 (XBP-1s) (30, 31) was markedly downregulated in Raw 264.7 cells treated with HTS01037, while the level of the unspliced form was increased, suggesting that loss of FABP4/aP2 protects macrophages from ER stress (Fig. 4A). Treatment of UCP2 knockdown and control cells with HTS01037 showed that UCP2-silenced macrophages have a basal increase of XBP-1s and a blunted response to HTS01037 (Fig. 4B).
FIG 3.
Knockdown of UCP2 in FABP4/aP2-deficient and Raw 264.7 macrophages. (A) UCP2 mRNA levels in wild-type (WT), AKO, and AKO-UCP2 kd (UCP2 knockdown AKO macrophages) macrophages. (B) UCP2 mRNA levels in control cells (Ctl) and in UCP2 kd and UCP2-2 kd (alternatively silenced UCP2 knockdown cells) Raw 264.7 macrophages. **, P < 0.01.
FIG 4.
UCP2 upregulation mediates decreased ER stress in FABP4/aP2-deficient macrophages. (A) XBP-1s and XBP-1u (unspliced XBP-1) mRNA levels in Raw 264.7 macrophages treated with 30 μM HTS01037. (B) XBP-1s mRNA levels in UCP2 knockdown and control macrophage cell lines treated with 30 μM HTS01037 for 24 h. (C to E) CHOP (C and D) and Bip (E) abundance as determined by Western blotting in UCP2 knockdown and control cells pretreated with vehicle (Ctl+DMSO) or HTS01037 (30 μM; 01037) for 3 h and treated with 500 μM palmitate (PA) for 16 h. (F) CHOP abundance as determined by Western blotting in WT, AKO, and AKO-UCP2 kd (UCP2 knockdown AKO macrophages) cells treated with 300 μM palmitate for 24 h. (G) Bip abundance as determined by Western blotting in WT, AKO, and AKO-UCP2 kd cells treated with 500 μM palmitate for 12 h. *, P < 0.05; **, P < 0.01.
Lipid loading, especially palmitate treatment, has been shown to induce expression of proteins involved in the ER stress response, such as CHOP (C/EBP homologous protein) and immunoglobulin heavy-chain binding protein (Bip) (6). To assess the role of UCP2 induction in regulating the ER stress response to palmitate, UCP2 knockdown and control cells were pretreated with HTS01037 or vehicle, and palmitate-induced ER stress was evaluated. Expression of CHOP and Bip with palmitate treatment was markedly reduced in response to HTS01037 treatment in macrophages, and this protection was negated in UCP2 knockdown macrophages (Fig. 4C to E). In order to further determine the role of UCP2 in inhibiting the ER stress response, UCP2 was knocked down in FABP4/aP2-deficient macrophages (AKO-UCP2 kd), and palmitate-induced ER stress was evaluated. Importantly, palmitate treatment of macrophages showed increased expression of CHOP and Bip in wild-type and AKO-UCP2 kd macrophages compared to levels in FABP4/aP2−/− (AKO) macrophages (Fig. 4F and G). Taken together, these results strongly indicate that UCP2 expression in FABP4/aP2-deficient macrophages plays an important role in mediating the reduced lipid-induced ER stress.
UCP2 mediates the decreased inflammatory signaling in FABP4/aP2-deficient macrophages.
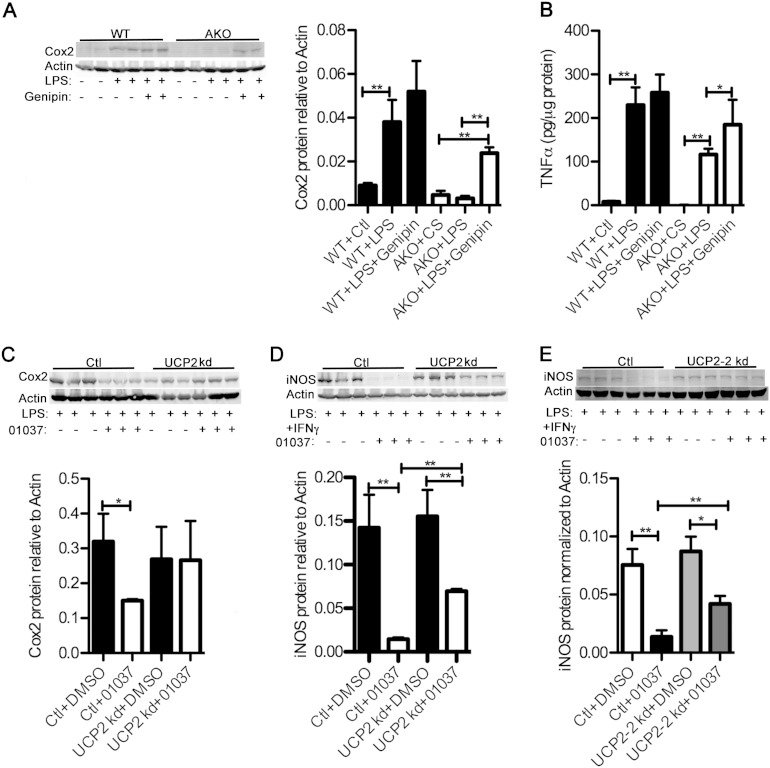
FABP4/aP2−/− macrophages have reduced expression of inflammatory proteins such as cyclooxygenase 2 (Cox2) and iNOS (7, 10). In order to determine if UCP2 is involved in the suppression of inflammatory signaling in FABP4/aP2 knockout macrophages, wild-type and FABP4/aP2−/− macrophages were treated with lipopolysaccharide (LPS) in the presence or absence of genipin, a UCP2 inhibitor (32), and the inflammatory response was profiled. Consistent with previous studies, LPS-induced Cox2 expression and TNF-α secretion were significantly lower in FABP4/aP2−/− macrophages (Fig. 5A and B). Moreover, LPS and genipin cotreatment of FABP4/aP2−/− macrophages significantly increased Cox2 expression and TNF-α secretion in FABP4/aP2−/− macrophages compared to levels with LPS treatment alone (Fig. 5A and B), suggesting that increased UCP2 expression may attenuate inflammatory responsiveness. To further confirm UCP2's role in suppressing inflammatory signaling, control cells and UCP2 knockdown Raw 264.7 macrophages were pretreated with HTS01037 and stimulated with LPS or LPS plus gamma interferon (IFN-γ). In control cells, HTS01037 treatment significantly reduced LPS-induced Cox2 expression as well as LPS-plus-IFN-γ-induced iNOS expression (Fig. 5C to E). However, the effect of HTS01037 was significantly reduced, if not totally abolished, in UCP2 knockdown macrophages (Fig. 5C to E). Taken together, these data indicate that UCP2 plays an important role in mediating the reduced inflammation in FABP4/aP2-deficient macrophages and that the key determinant of metabolic improvement in FABP-deficient mice is upregulation of UCP2.
FIG 5.
UCP2 upregulation mediates decreased inflammation in FABP4/aP2-deficient macrophages. (A) Cyclooxygenae 2 (Cox2) abundance measured by Western blotting in WT and AKO macrophages cotreated with or without LPS (100 ng/ml) and genipin (40 μM) for 18 h. (B) Secreted TNF-α in cell culture medium determined by ELISA in WT and AKO macrophages cotreated with or without LPS (100 ng/ml) and genipin (40 μM) for 8 h. (C) Cox2 abundance as determined by Western blotting in UCP2 knockdown and control macrophages pretreated with vehicle or HTS01037 (30 μM) for 3 h and then treated with LPS (100 ng/ml) for 12 h. (D and E) iNOS abundance as determined by Western blotting in UCP2 knockdown and control macrophages pretreated with vehicle or HTS01037 (30 μM) for 3 h and then treated with LPS (100 ng/ml) and IFN-γ (10 U) for 12 h (UCP2 kd) and 4 h (UCP2-2 kd). *, P < 0.05; **, P < 0.01.
UCP2 decreases the hydrogen peroxide level and oxidative stress in FABP4/aP2-deficient macrophages.
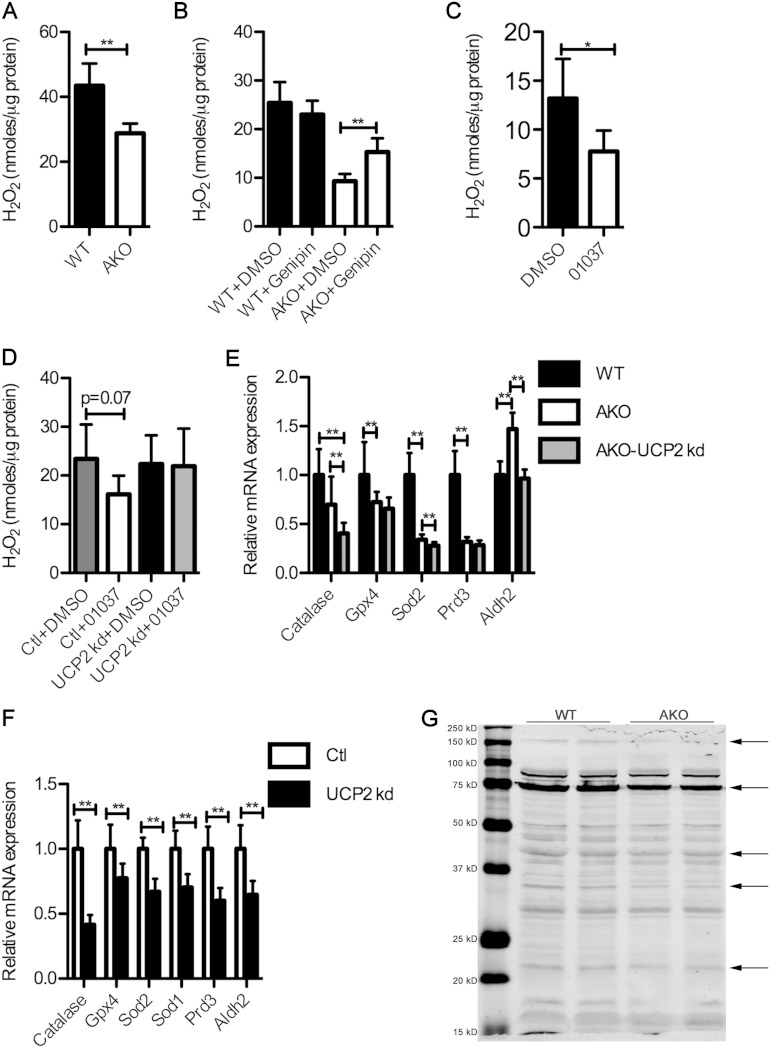
A well-defined role for UCP2 is suppression of reactive oxygen species (ROS) production (13, 14). In order to determine the effect of FABP4/aP2 loss on ROS level in macrophages, the intracellular hydrogen peroxide levels in both FABP4/aP2−/− and wild-type macrophages were determined. Figure 6A shows that FABP4/aP2−/− macrophages have a significantly lower level of intracellular hydrogen peroxide. Moreover, treatment of control and FABP4/aP2−/− macrophages with genipin to inhibit UCP2 attenuated the decreased ROS levels in FABP4/aP2-deficient cells but had little effect on control macrophages (Fig. 6B). Interestingly, HTS01037 treatment of Raw 264.7 macrophages, which mimics the knockdown of FABP4/aP2, also led to decreased intracellular hydrogen peroxide (Fig. 6C). Moreover, HTS01037 treatment of UCP2 knockdown cells was not able to reduce the intracellular hydrogen peroxide level (Fig. 6D), suggesting that the effect of HTS01037 treatment on hydrogen peroxide level is likely to be mediated by UCP2 expression as well. Taken together, the results indicate that genetic loss or chemical inhibition of FABP4/aP2 leads to the reduced level of intracellular hydrogen peroxide in a UCP2-dependent manner.
FIG 6.
UCP2 upregulation decreases intracellular hydrogen peroxide in FABP4/aP2-deficient macrophages. Intracellular hydrogen peroxide levels were measured by Amplex Red assay in WT and AKO macrophages (A), WT and AKO macrophages treated with vehicle or genipin (50 μM) for 6 h (B), Raw 264.7 macrophages treated with HTS01037 (30 μM) for 24 h (C), and UCP2 knockdown and control cells treated with HTS01037 (30 μM) for 6 h (D). Antioxidant gene mRNA levels were determined in WT, AKO, and AKO-UCP2 kd cells (E) and in UCP2 knockdown and control cells (F). (G) Protein carbonylation measured by Western blotting with anti-HNE antibody in WT and AKO mitochondrial proteins. *, P < 0.05; **, P < 0.01.
Oxidative stress is a key contributor to mitochondrial dysfunction and apoptosis (33, 34). Consistent with decreased hydrogen peroxide that is indicative of reduced oxidative stress, most of the antioxidants, if not all, are decreased in FABP4/aP2-deficient macrophages (Fig. 6E). Surprisingly, silencing of UCP2 also leads to a decrease in the mRNA expression levels of several antioxidant enzymes (Fig. 6E and F). One of the effects of oxidative stress is protein carbonylation, the covalent modification of proteins with reactive lipid aldehydes that is linked to mitochondrial dysfunction (35). Utilizing an antibody directed to carbonylated proteins, and consistent with reduced ROS levels, FABP4/aP2−/− macrophages exhibit reduced protein carbonylation (Fig. 6G).
FABP4/aP2−/− macrophages have higher mitochondrial respiration capacity.
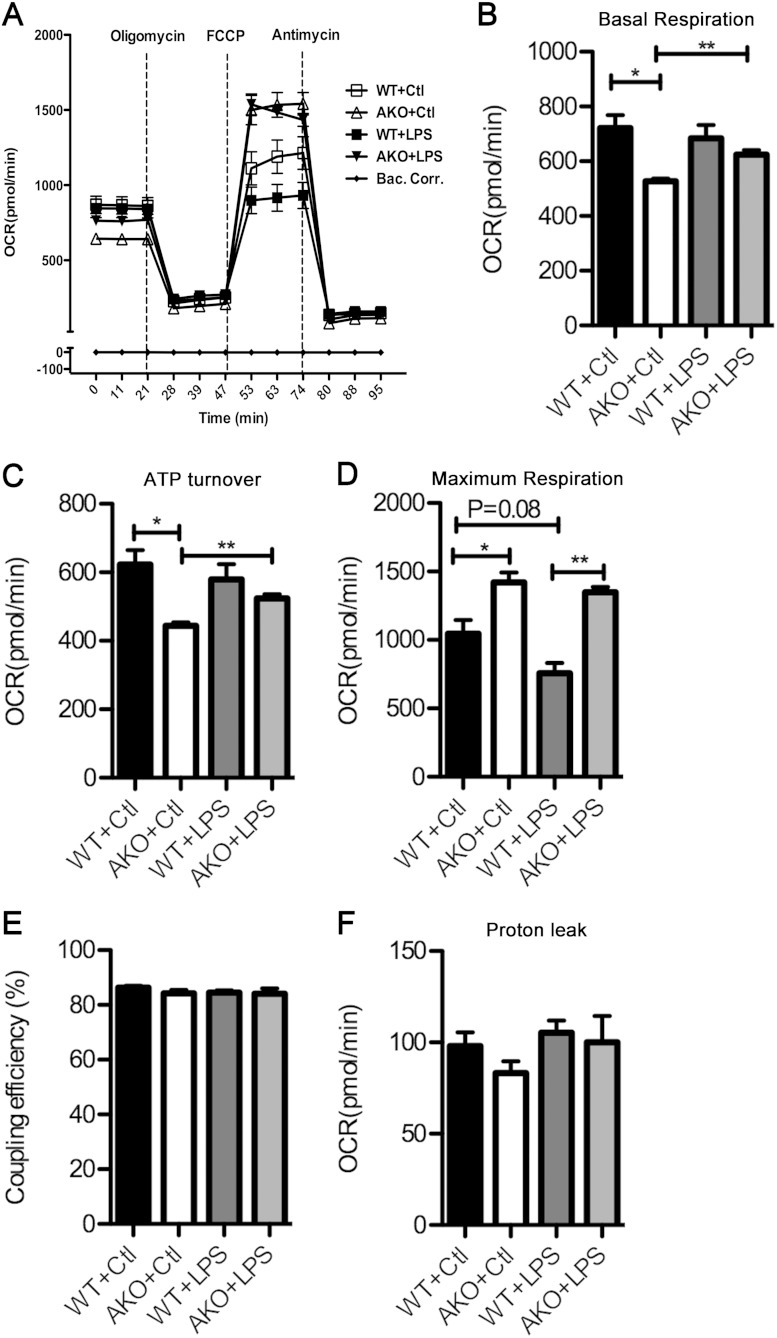
Reduced carbonylation is predictive of improved mitochondrial function, and to test this hypothesis, cellular respiration was evaluated both at the basal level and in response to an LPS challenge. As shown in Fig. 7, FABP4/aP2−/− macrophages exhibited a lower level of basal respiration and ATP turnover but a significantly higher level of maximum respiration. Upon LPS treatment, FABP4/aP2−/− macrophages have a significant increase in basal respiration and ATP turnover, while wild-type macrophages lose the maximum respiration capacity (Fig. 7B to D). However, no difference in coupling efficiency and proton leak between the two cell lines or treatments was observed (Fig. 7E and F).
FIG 7.
Cellular respiration of FABP4/aP2-deficient and wild-type macrophages. Oxygen consumption rates (OCRs) were determined in WT and AKO macrophages treated with or without LPS (100 ng/ml) for 4 h (oligomycin, 2 μM; FCCP, 0.4 μM; antimycin, 4 μM). *, P < 0.05; **, P < 0.01. Bac corr, background correction.
UCP2 mediates decreased basal respiration and lactate production of FABP4/aP2−/− macrophages but not increased fatty acid oxidation.
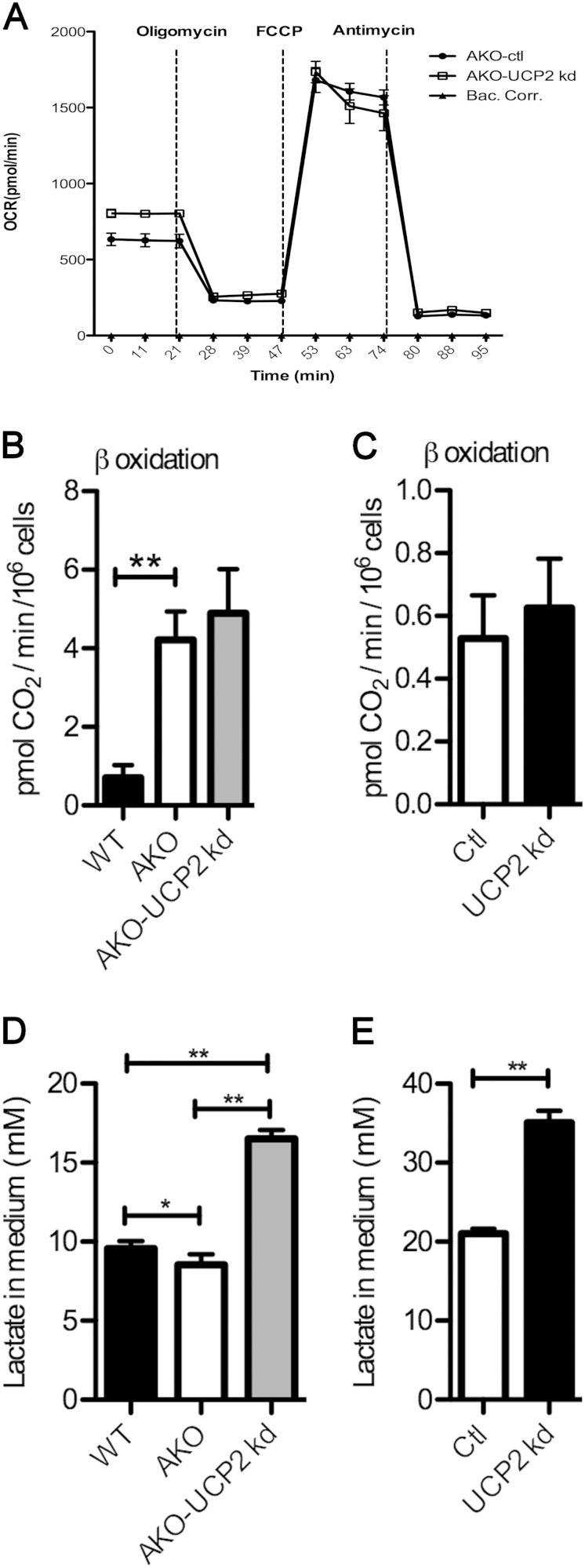
Consistent with a role of UCP2 in control of basal respiration, silencing of UCP2 in FABP4/aP2−/− macrophages increased basal, but not maximal, respiration compared to control cell levels (Fig. 8A). Additionally, fatty acid oxidation was increased in macrophages lacking FABP4/aP2 (Fig. 8B). However, no difference in fatty acid oxidation was observed in UCP2 knockdown FABP4/aP2−/− or Raw 264.7 macrophages (Fig. 8B and C), suggesting a UCP2-independent process. FABP4/aP2−/− macrophage cell culture medium had reduced lactate compared to wild-type macrophages (Fig. 8D). This is consistent with the lower basal respiration of FABP4/aP2−/− macrophages (Fig. 7B). In support of the role of UCP2 mediating the decreased lactate production in FABP4/aP2−/− macrophages, the lactate level was increased about 70% in the cell culture medium of UCP2 knockdown FABP4/aP2−/− cells and UCP2 knockdown Raw 264.7 cells compared with levels in control macrophages (Fig. 8D and E).
FIG 8.
Metabolic impact of UCP2 silencing on macrophages. (A) Oxygen consumption rate (OCR) in FABP4/aP2-deficient and AKO-UCP2 kd macrophages. (B) β-Oxidation measured in WT, AKO, and AKO-UCP2 kd cells. (C) β-Oxidation measured in UCP2 kd cells and control cells. (D) Cell culture medium lactate level measured in WT, AKO, and AKO-UCP2 kd cells. (E) Cell culture medium lactate level measured in UCP2 knockdown and control cells. *, P < 0.05; **, P < 0.01.
DISCUSSION
Chronic activation of ER stress and inflammation in macrophages contributes to the pathogenesis of various metabolic disarrangements such as atherosclerosis and type 2 diabetes (4, 29). The stressors that can lead to macrophage ER stress and inflammation include oxidative stress and high levels of intracellular cholesterol and saturated fatty acids (29). Prolonged elevation of macrophage ER stress and inflammation has been proposed to contribute to macrophage apoptosis and lead to plaque necrosis and rupture (2, 36). Therefore, understanding the biological processes involved in counteracting macrophage ER stress and inflammation is crucial for development of specific strategies to improve metabolic poise. Interestingly, FABP4/aP2 has been shown to play an important role in mediating both ER stress and inflammation in macrophages (6, 37). Genetic ablation or chemical inhibition of FABP4/aP2 alleviates macrophage inflammation and ER stress (6, 8, 10). The increase in monounsaturated fatty acids, PPARγ, and LXRα activity have all been suggested as mediating the anti-inflammatory and anti-ER stress effects of FABP4/aP2 deficiency (6, 10). However, the molecular relationship of FABP deficiency to inflammation and ER stress outcome is still unclear. Increased expression of UCP2 in FABP4/aP2−/− macrophages provides a mechanistic basis for the anti-inflammatory, anti-ER stress outcomes. Studies in a number of systems have implicated UCP2 as a major control element in macrophage ER stress, inflammation, and diet-induced atherosclerosis (38–40). Interestingly, a UCP2 promoter region -866G→A polymorphism which decreases UCP2 expression has been associated with increased risk of obesity, decreased insulin level, and type 2 diabetes (14). Moreover, type 2 diabetes patients bearing the G allele have a higher inflammatory status (41). In a second study, a UCP2 -866G→A polymorphism is associated with multiple chronic inflammatory diseases, including Crohn's disease, ulcerative colitis, and psoriasis (42).
It has been proposed that unsaturated fatty acids (or their metabolites) are potential ligands for LXRα and PPARγ (27). Fatty acids and the expression of LXRα and PPARγ are increased in FABP4/aP2-deficient macrophages and have all been previously shown to upregulate UCP2 expression (14). Interestingly, a previous study by Erbay et al. showed that the unsaturated fatty acid pool is increased in FABP4/aP2−/− macrophages (6). Results herein show that monounsaturated fatty acids are increased in FABP4/aP2−/− macrophages and that unsaturated, but not saturated, fatty acids induce UCP2 expression in macrophages, consistent with a report on fatty acid induction of UCP2 in liver cells (43). Moreover, given the increase in the intracellular free fatty acid pool in FABP4/aP2−/− macrophages, it is very likely that they directly or indirectly activate PPARγ, thereby inducing UCP2 expression. However, this does not exclude the possibility that fatty acids also directly regulate UCP2 activity.
Paradoxically, despite the existence of the other FABP in macrophages (FABP5/mal1), the deficiency of FABP4 leads to an anti-inflammatory and antiatherosclerotic phenotype (7). One explanation is that FABP4 has a higher affinity for unsaturated fatty acids than FABP5 (44). Therefore, the loss of FABP4 will affect the pool of available free unsaturated fatty acids, especially monounsaturated fatty acids. An additional possibility is that the pools of fatty acids bound to FABP4 are distinct from those bound to FABP5 such that loss of FABP4 could exhibit a unique phenotype. Experiments to test these hypotheses are under way.
The reduced ER stress and inflammation demonstrated in both FABP4/aP2-deficient and FABP4/aP2-inhibited macrophages were markedly abolished or compromised by knockdown or inhibition of UCP2. This observation places UCP2 as a modulator between the FABP-FFA equilibrium and ER stress and inflammation, which is a role for UCP2 that has previously not been appreciated. It is noteworthy that the findings do not rule out contributions from other parallel pathways that reduce ER stress and inflammation. However, the results imply that the UCP2-mediated pathway is likely the major determinant of the FABP4/aP2-deficient macrophage phenotype since UCP2 knockdown or inhibition greatly compromised the protection from loss of FABP4/aP2 function.
The prominent and well-defined role of UCP2 is to suppress ROS (13). The data herein show that FABP4/aP2−/− macrophages have a lower level of intracellular hydrogen peroxide that is dependent on UCP2 expression, as evidenced by restoration via genipin inhibition of UCP2. Hydrogen peroxide has been shown to directly oxidize the side chains of several amino acids, particularly cysteine, thereby affecting the cellular redoxome. Alternatively, if not detoxified by antioxidants such as catalase, peroxiredoxin, or glutathione peroxidase, hydrogen peroxide could react with iron to form hydroxyl radicals and oxidize lipids. 4-Hydroxynonenal (4-HNE) and 4-oxononenal (4-ONE) are the two most well-studied lipid peroxidation products and have been shown to covalently modify proteins, a process termed as protein carbonylation. This frequently leads to loss or alteration of protein activity and mitochondrial dysfunction (35, 45). Interestingly, a decreased level of mitochondrial protein carbonylation was observed in FABP4/aP2−/− macrophages, consistent with improved mitochondrial function (35). The results shown in Fig. 7 demonstrating that FABP4/aP2−/− macrophages were protected from LPS-induced loss of mitochondrial respiration capacity support this concept. Interestingly, it has been shown that LPS-induced mitochondrial dysfunction mainly relies on the increased ROS production and that suppression of ROS not only protects cells from LPS-induced mitochondrial dysfunction but also greatly attenuates LPS-induced inflammatory responses (46, 47). Further work is still required to determine if the decreased hydrogen peroxide level in FABP4/aP2−/− macrophages is responsible for the protection from LPS-induced mitochondrial dysfunction.
Both hydrogen peroxide and oxidized lipids have also been shown to activate ER stress and inflammatory pathways (48, 49). It is tempting to speculate that the UCP2 suppression of ROS in FABP4/aP2−/− macrophages broadly impacts not only mitochondrial function but also endoplasmic reticulum function and inflammatory pathway activity (Fig. 9). Interestingly, knockdown of UCP2 in macrophages increases lactate production. The increased lactate production indicates increased energy production through glycolysis, a common indicator of electron transport chain dysfunction (50). Additionally, this may also explain UCP2 dependency of the lower basal respiration of FABP4/aP2-deficient macrophages. As a suppressor of ROS production, higher UCP2 expression in FABP4/aP2−/− macrophages leads to lower cellular oxidative stress. Since oxidative stress drives the upregulation of antioxidants, it is reasonable that most genes linked to antioxidant biology are expressed at lower levels in FABP4/aP2−/− cells than in wild-type macrophages (Fig. 6E). The reduced UCP2 expression in macrophages, which leads to increased oxidative stress, would be predicted to lead to increased antioxidant protein expression. Paradoxically, in the FABP4/aP2−/− UCP2 kd macrophages, expression of antioxidant enzymes is downregulated. This suggests that UCP2 is involved in a more complex regulation of antioxidant expression. Nevertheless, the loss of antioxidant capacity of UCP2 knockdown is consistent with a previous report showing that UCP2-deficient mice have reduced antioxidant capacity (38).
FIG 9.
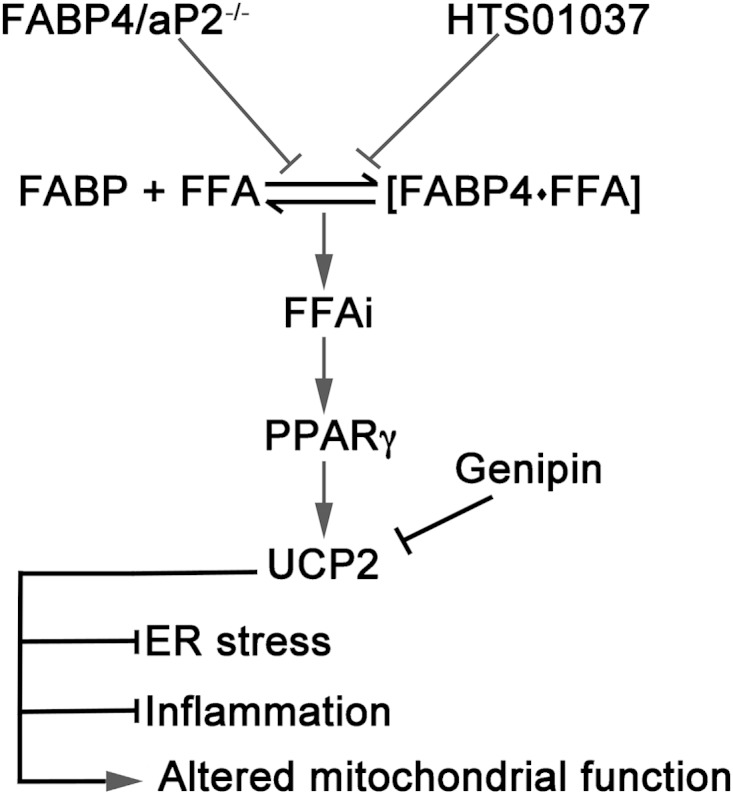
Schematic model of the role of UCP2 upregulation in macrophages. Loss or inhibition of FABP4/aP2 increases the intracellular free fatty acid levels and induces expression of UCP2 via a PPARγ-mediated pathway. Increased expression of UCP2 reduces oxidative stress in the mitochondrion, alleviates ER stress and inflammation, and alters mitochondrial function in macrophages. FFAi, intracellular free fatty acids.
Overall, the studies herein provide a mechanistic basis for the metabolic improvement in FABP4/aP2-deficient cells generated by either genetic or pharmacologic means. Loss of FABP-FFA equilibrium is likely to increase the bioavailability of fatty acids, particularly unsaturated lipids that have the potential to increase the expression of UCP2. Whether this mechanism exists in nonmacrophage cells and would pertain to the metabolic changes observed in FABP1, FABP2, or FABP3 null mice remains unknown (51). However, it may be that FABP-dependent modulation of intracellular FFA levels and therefore of UCP2 expression is a common property in many cell types.
ACKNOWLEDGMENTS
We thank the members of the Bernlohr laboratory for helpful discussions during the study and preparation of the manuscript. We also thank Edward McFalls, VA Medical Center, Minneapolis, MN, for helpful discussions and reagents in the preliminary phase of the study as well as Nicholas Kvalheim for assistance with real-time PCR measurements. The support of the Minnesota Supercomputer Institute is gratefully acknowledged.
This study was supported by NIH R01 DK053189 to D.A.B., NIH T32 AG029796 to K.A.S., and the Minnesota Obesity Center (NIH P30 DK050456).
REFERENCES
- 1.Hummasti S, Hotamisligil GS. 2010. Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ Res 107:579–591. doi: 10.1161/CIRCRESAHA.110.225698. [DOI] [PubMed] [Google Scholar]
- 2.Hotamisligil GS. 2010. Endoplasmic reticulum stress and atherosclerosis. Nat Med 16:396–399. doi: 10.1038/nm0410-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J. 2013. Adipose tissue macrophages in the development of obesity-induced inflammation, insulin resistance and type 2 diabetes. Arch Pharm Res 36:208–222. doi: 10.1007/s12272-013-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cnop M, Foufelle F, Velloso LA. 2012. Endoplasmic reticulum stress, obesity and diabetes. Trends Mol Med 18:59–68. doi: 10.1016/j.molmed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen A, Tao H, Metrione M, Hajri T. 2014. Very low density lipoprotein receptor (VLDLR) expression is a determinant factor in adipose tissue inflammation and adipocyte-macrophage interaction. J Biol Chem 289:1688–1703. doi: 10.1074/jbc.M113.515320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erbay E, Babaev VR, Mayers JR, Makowski L, Charles KN, Snitow ME, Fazio S, Wiest MM, Watkins SM, Linton MF, Hotamisligil GS. 2009. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat Med 15:1383–1391. doi: 10.1038/nm.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makowski L, Boord JB, Maeda K, Babaev VR, Uysal KT, Morgan MA, Parker RA, Suttles J, Fazio S, Hotamisligil GS, Linton MF. 2001. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat Med 7:699–705. doi: 10.1038/89076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertzel AV, Hellberg K, Reynolds JM, Kruse AC, Juhlmann BE, Smith AJ, Sanders MA, Ohlendorf DH, Suttles J, Bernlohr DA. 2009. Identification and characterization of a small molecule inhibitor of fatty acid binding proteins. J Med Chem 52:6024–6031. doi: 10.1021/jm900720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan H, Cheng CC, Kowalski TJ, Pang L, Shan L, Chuang CC, Jackson J, Rojas-Triana A, Bober L, Liu L, Voigt J, Orth P, Yang X, Shipps GW Jr, Hedrick JA. 2011. Small-molecule inhibitors of FABP4/5 ameliorate dyslipidemia but not insulin resistance in mice with diet-induced obesity. J Lipid Res 52:646–656. doi: 10.1194/jlr.M012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. 2005. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and IκB kinase activities. J Biol Chem 280:12888–12895. doi: 10.1074/jbc.M413788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuncman G, Erbay E, Hom X, De Vivo I, Campos H, Rimm EB, Hotamisligil GS. 2006. A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci U S A 103:6970–6975. doi: 10.1073/pnas.0602178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diano S, Horvath TL. 2012. Mitochondrial uncoupling protein 2 (UCP2) in glucose and lipid metabolism. Trends Mol Med 18:52–58. doi: 10.1016/j.molmed.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Krauss S, Zhang CY, Lowell BB. 2005. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol 6:248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 14.Donadelli M, Dando I, Fiorini C, Palmieri M. 2014. UCP2, a mitochondrial protein regulated at multiple levels. Cell Mol Life Sci 71:1171–1190. doi: 10.1007/s00018-013-1407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long EK, Hellberg K, Foncea R, Hertzel AV, Suttles J, Bernlohr DA. 2012. Fatty acids induce leukotriene C4 synthesis in macrophages in a fatty acid binding protein-dependent manner. Biochim Biophys Acta 1831:1199–1207. doi: 10.1016/j.bbalip.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertzel AV, Smith LA, Berg AH, Cline GW, Shulman GI, Scherer PE, Bernlohr DA. 2006. Lipid metabolism and adipokine levels in fatty acid-binding protein null and transgenic mice. Am J Physiol Endocrinol Metab 290:E814–E823. doi: 10.1152/ajpendo.00465.2005. [DOI] [PubMed] [Google Scholar]
- 17.Curtis JM, Grimsrud PA, Wright WS, Xu X, Foncea RE, Graham DW, Brestoff JR, Wiczer BM, Ilkayeva O, Cianflone K, Muoio DE, Arriaga EA, Bernlohr DA. 2010. Downregulation of adipose glutathione S-transferase A4 leads to increased protein carbonylation, oxidative stress, and mitochondrial dysfunction. Diabetes 59:1132–1142. doi: 10.2337/db09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. 2006. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 19.Wiczer BM, Bernlohr DA. 2009. A novel role for fatty acid transport protein 1 in the regulation of tricarboxylic acid cycle and mitochondrial function in 3T3-L1 adipocytes. J Lipid Res 50:2502–2513. doi: 10.1194/jlr.M900218-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coe NR, Simpson MA, Bernlohr DA. 1999. Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. J Lipid Res 40:967–972. [PubMed] [Google Scholar]
- 21.Chavin KD, Yang S, Lin HZ, Chatham J, Chacko VP, Hoek JB, Walajtys-Rode E, Rashid A, Chen CH, Huang CC, Wu TC, Lane MD, Diehl AM. 1999. Obesity induces expression of uncoupling protein-2 in hepatocytes and promotes liver ATP depletion. J Biol Chem 274:5692–5700. doi: 10.1074/jbc.274.9.5692. [DOI] [PubMed] [Google Scholar]
- 22.Smith AJ, Thompson BR, Sanders MA, Bernlohr DA. 2007. Interaction of the adipocyte fatty acid-binding protein with the hormone-sensitive lipase: regulation by fatty acids and phosphorylation. J Biol Chem 282:32424–32432. doi: 10.1074/jbc.M703730200. [DOI] [PubMed] [Google Scholar]
- 23.Reilly JM, Thompson MP. 2000. Dietary fatty acids Up-regulate the expression of UCP2 in 3T3-L1 preadipocytes. Biochem Biophys Res Commun 277:541–545. doi: 10.1006/bbrc.2000.3705. [DOI] [PubMed] [Google Scholar]
- 24.Aubert J, Champigny O, Saint-Marc P, Negrel R, Collins S, Ricquier D, Ailhaud G. 1997. Up-regulation of UCP-2 gene expression by PPAR agonists in preadipose and adipose cells. Biochem Res Commun 238:606–611. doi: 10.1006/bbrc.1997.7348. [DOI] [PubMed] [Google Scholar]
- 25.Medvedev AV, Robidoux J, Bai X, Cao W, Floering LM, Daniel KW, Collins S. 2002. Regulation of the uncoupling protein-2 gene in INS-1 beta-cells by oleic acid. J Biol Chem 277:42639–42644. doi: 10.1074/jbc.M208645200. [DOI] [PubMed] [Google Scholar]
- 26.Coe NR, Bernlohr DA. 1998. Physiological properties and functions of intracellular fatty acid-binding proteins. Biochim Biophys Acta 1391:287–306. doi: 10.1016/S0005-2760(97)00205-1. [DOI] [PubMed] [Google Scholar]
- 27.Thompson MP, Kim D. 2004. Links between fatty acids and expression of UCP2 and UCP3 mRNAs. FEBS Lett 568:4–9. doi: 10.1016/j.febslet.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Olefsky JM, Glass CK. 2010. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol 72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 29.Tabas I. 2010. The role of endoplasmic reticulum stress in the progression of atherosclerosis. Circ Res 107:839–850. doi: 10.1161/CIRCRESAHA.110.224766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, Sun S, Sha H, Liu Z, Yang L, Xue Z, Chen H, Qi L. 2010. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr 15:13–25. doi: 10.3727/105221610X12819686555051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mailloux RJ, Adjeitey CN, Harper ME. 2010. Genipin-induced inhibition of uncoupling protein-2 sensitizes drug-resistant cancer cells to cytotoxic agents. PLoS One 5:e13289. doi: 10.1371/journal.pone.0013289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brune B, Dehne N, Grossmann N, Jung M, Namgaladze D, Schmid T, von Knethen A, Weigert A. 2013. Redox control of inflammation in macrophages. Antioxid Redox Signal 19:595–637. doi: 10.1089/ars.2012.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rath E, Haller D. 2012. Mitochondria at the interface between danger signaling and metabolism: role of unfolded protein responses in chronic inflammation. Inflamm Bowel Dis 18:1364–1377. doi: 10.1002/ibd.21944. [DOI] [PubMed] [Google Scholar]
- 35.Frohnert BI, Bernlohr DA. 2013. Protein carbonylation, mitochondrial dysfunction, and insulin resistance. Adv Nutr 4:157–163. doi: 10.3945/an.112.003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiwari RL, Singh V, Barthwal MK. 2008. Macrophages: an elusive yet emerging therapeutic target of atherosclerosis. Med Res Rev 28:483–544. doi: 10.1002/med.20118. [DOI] [PubMed] [Google Scholar]
- 37.Hui X, Li H, Zhou Z, Lam KS, Xiao Y, Wu D, Ding K, Wang Y, Vanhoutte PM, Xu A. 2010. Adipocyte fatty acid-binding protein modulates inflammatory responses in macrophages through a positive feedback loop involving c-Jun NH2-terminal kinases and activator protein-1. J Biol Chem 285:10273–10280. doi: 10.1074/jbc.M109.097907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moukdar F, Robidoux J, Lyght O, Pi J, Daniel KW, Collins S. 2009. Reduced antioxidant capacity and diet-induced atherosclerosis in uncoupling protein-2-deficient mice. J Lipid Res 50:59–70. doi: 10.1194/jlr.M800273-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Lu M, Sun XL, Qiao C, Liu Y, Ding JH, Hu G. 2014. Uncoupling protein 2 deficiency aggravates astrocytic endoplasmic reticulum stress and nod-like receptor protein 3 inflammasome activation. Neurobiol Aging 35:421–430. doi: 10.1016/j.neurobiolaging.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 40.Bai Y, Onuma H, Bai X, Medvedev AV, Misukonis M, Weinberg JB, Cao W, Robidoux J, Floering LM, Daniel KW, Collins S. 2005. Persistent nuclear factor-kappa B activation in Ucp2−/− mice leads to enhanced nitric oxide and inflammatory cytokine production. J Biol Chem 280:19062–19069. doi: 10.1074/jbc.M500566200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapice E, Pinelli M, Pisu E, Monticelli A, Gambino R, Pagano G, Valsecchi S, Cocozza S, Riccardi G, Vaccaro O. 2010. Uncoupling protein 2 G(-866)A polymorphism: a new gene polymorphism associated with C-reactive protein in type 2 diabetic patients. Cardiovasc Diabetol 9:68. doi: 10.1186/1475-2840-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X, Wieczorek S, Franke A, Yin H, Pierer M, Sina C, Karlsen TH, Boberg KM, Bergquist A, Kunz M, Witte T, Gross WL, Epplen JT, Alarcon-Riquelme ME, Schreiber S, Ibrahim SM. 2009. Association of UCP2-866 G/A polymorphism with chronic inflammatory diseases. Genes Immun 10:601–605. doi: 10.1038/gene.2009.29. [DOI] [PubMed] [Google Scholar]
- 43.Armstrong MB, Towle HC. 2001. Polyunsaturated fatty acids stimulate hepatic UCP-2 expression via a PPARα-mediated pathway. Am J Physiol Endocrinol Metab 281:E1197–E1204. [DOI] [PubMed] [Google Scholar]
- 44.Simpson MA, LiCata VJ, Ribarik Coe N, Bernlohr DA. 1999. Biochemical and biophysical analysis of the intracellular lipid binding proteins of adipocytes. Mol Cell Biochem 192:33–40. doi: 10.1023/A:1006819715146. [DOI] [PubMed] [Google Scholar]
- 45.Schaur RJ. 2003. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Mol Aspects Med 24:149–159. doi: 10.1016/S0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 46.Kruzel ML, Actor JK, Radak Z, Bacsi A, Saavedra-Molina A, Boldogh I. 2010. Lactoferrin decreases LPS-induced mitochondrial dysfunction in cultured cells and in animal endotoxemia model. Innate Immun 16:67–79. doi: 10.1177/1753425909105317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo SK, Yi HS, Yun HJ, Ko CH, Choi JW, Park SD. 2010. Ethylacetate extract from Draconis Resina inhibits LPS-induced inflammatory responses in vascular smooth muscle cells and macrophages via suppression of ROS production. Food Chem Toxicol 48:1129–1136. doi: 10.1016/j.fct.2009.06.043. [DOI] [PubMed] [Google Scholar]
- 48.Haberzettl P, Hill BG. 2013. Oxidized lipids activate autophagy in a JNK-dependent manner by stimulating the endoplasmic reticulum stress response. Redox Biol 1:56–64. doi: 10.1016/j.redox.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lisanti MP, Martinez-Outschoorn UE, Lin Z, Pavlides S, Whitaker-Menezes D, Pestell RG, Howell A, Sotgia F. 2011. Hydrogen peroxide fuels aging, inflammation, cancer metabolism and metastasis: the seed and soil also needs “fertilizer.” Cell Cycle 10:2440–2449. doi: 10.4161/cc.10.15.16870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishisaka A, Kawabata K, Miki S, Shiba Y, Minekawa S, Nishikawa T, Mukai R, Terao J, Kawai Y. 2013. Mitochondrial dysfunction leads to deconjugation of quercetin glucuronides in inflammatory macrophages. PLoS One 8:e80843. doi: 10.1371/journal.pone.0080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuhashi M, Hotamisligil GS. 2008. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]