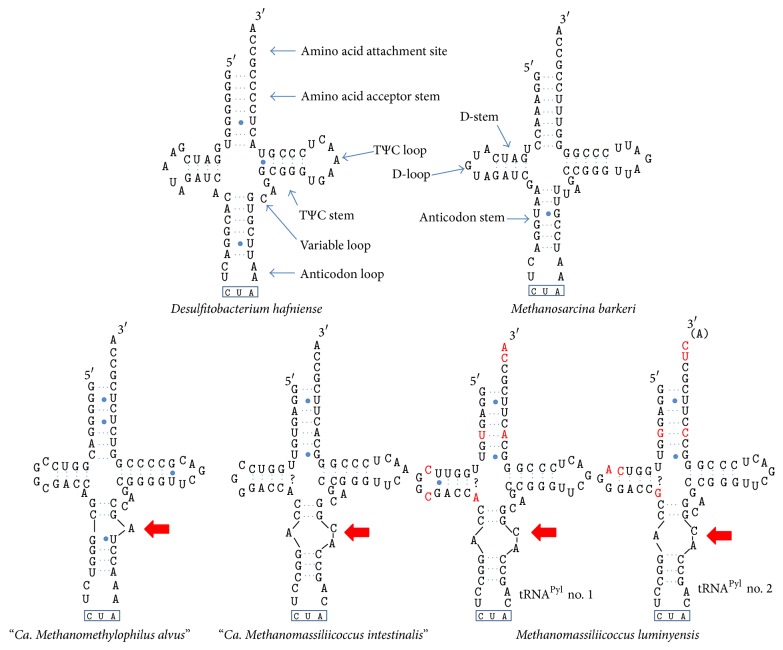
In the paper titled “Unique Characteristics of the Pyrrolysine System in the 7th Order of Methanogens: Implications for the Evolution of a Genetic Code Expansion Cassette,” a mistake arose in M. luminyensis tRNAPyl sequences shown in the lower part of Figure 2. In this erratum, we report the corrected Figure 2 with the correct tRNAPyl sequences.
Figure 2.
Secondary structure of the tRNAPyl in the Thermoplasmata-related 7th order of methanogens. The stem-loop structure of the tRNAPyl in the 7th order of methanogens is shown, deduced by comparison with the structure of this tRNA in bacteria (Desulfitobacterium hafniense, top left) and in Methanosarcinaceae (Methanosarcina barkeri, top right), adapted from [11, 13]. The name of each region of the tRNA is indicated in the upper part. The anticodon CUA (corresponding to the amber codon) is outlined in blue. The red arrows indicate the shortened anticodon stem in the 7th order of methanogens broken by unpaired bases that form an unusual loop in this region. The question marks represent other possible base pairing. For the two tRNAPyl in M. luminyensis, the modified bases between no. 1 and no. 2 are written in red.