Abstract
We investigated the oncogenic role of SETDB1 focusing on non-small cell lung cancer (NSCLC) having high expression of this protein. A total of 387 lung cancer cases were examined by immunohistochemistry, 72% of NSCLC samples were positive for SETDB1 staining, compared to 46% samples of normal bronchial epithelium (106 cases) (p<0.0001). Percent positive cells and intensity of staining increased significantly with increased grade of disease. Forced expression of SETDB1 in NSCLC cell lines enhanced their clonogenic growth in vitro and markedly increased tumor size in a murine xenograft model; while silencing (shRNA) SETDB1 in NSCLC cells slowed their proliferation. SETDB1 positively stimulated activity of the WNT/β-catenin pathway and diminished P53 expression resulting in enhanced NSCLC growth in vitro and in vivo. Our finding suggests therapeutic targeting SETDB1 may benefit patients whose tumors express high levels of SETDB1.
Keywords: SETDB1, WNT, tumorigenesis, lung
Introduction
Lung cancer is the most common cancer worldwide, accounting for 1.3 million deaths annually. Non-small cell lung cancer (NSCLC) represents about 80% of lung cancers [1]; the patients are often diagnosed at advanced stages when surgical cure is no longer possible [2]. Several oncogenic drivers have been identified for this cancer. However, currently only two categories of targeted therapies are available: one is gefitinib/erlotinib, tyrosine kinase inhibitors targeting the mutated epidermal growth factor receptor (EGFR) especially occurring in Asian (30~50%) [3,4]. The other is crizotinib, a kinase inhibitor targeting the echinoderm microtubule-associated protein-like 4 anaplastic lymphoma kinase (EML4-ALK) fusion protein (approximately 4–7% of patients) [5,6]. A large majority of NSCLC patients receive conventional chemotherapy, associated with toxicity and often only a marginal survival benefit. Searching for additional aberrant pathways in NSCLC is needed to identify novel “druggable” targets, leading to the development of new targeted therapeutic strategies.
Many biological processes, such as gene transcription, genome stability and recombination/DNA repair, are controlled by chromatin structures [7]. The primary architecture of chromatin is organized by histones, a class of small nuclear proteins which can be modified by acetylation, ubiquitination, phosphorylation or methylation [7,8]. These modifications, also referred to as the “histone code”, coordinate and help control genomic transcription and play a crucial role in maintaining genomic stability [9,10]. Overall, acetylation of histones usually marks transcriptional activating region; and the process is tightly controlled by histone acetyltransferases (HATs) and histone deacetylases (HDACs) [11]. In contrast, methylation of histones results in a much more complex and diverse regulatory pattern. Generally, methylation of histone H3 lysine 4 (H3K4), lysing 36 (H3K36) and lysing 79 (H3K79) is associated with gene activation, whereas methylation of histone H3K9, H3K27 and H4K20 usually lead to gene repression [12]. The process of histone methylation is, in part dynamically controlled by a variety of SET-domain-containing methyltransferases and demethylases [13].
SETDB1 is a histone H3 lysine 9 methyltransferase, which was initially identified as a binding partner of KRAB-associated protein-1 (KAP1) [14]. It transfers the methyl group(s) to histone H3K9, helping to control heterochromatin formation and chromatin organization [15]. SETDB1 is also required for efficient endogenous proviral silencing during early embryogenesis, which is indispensable for mammalian embryonic development [16].
Materials and Methods
Patient samples
Sixty primary NSCLC samples and their paired adjacent normal (for SETDB1 mRNA expression analysis by RT-PCR) were obtained from Shanghai Chest Hospital with consent obtained from patients [17,18]. Lung cancer tissue array used in immunohistochemical assay was described previously [IRB protocol (02-07-011-13; UCLA Institutional Medical Review Board)] [19,20].
Immunohistochemical assays
Polyclonal rabbit anti-SETDB1 antibody obtained from Sigma-Aldrich (HPA018142) was used for the IHC experiments after the specificity testing (Supplementary Fig. S1). Detailed IHC assays were described in the supplemental materials.
Cell lines and cell culture
Non-small cell lung cancer (NSCLC) cell lines (PC14, A549, HCC2279, H1299, H23, and HCC1975) were grown in RPMI 1640 plus 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37°C.
Generation of stably-transfected cell line
The entire coding region of SETDB1 was amplified from pCMV2-SETDB1, which was a generous gift from Dr. Frank J. Rauscher III. The lentiviral SETDB1 overexpression and shRNA vectors were generated as described in Supplementary Material and Method. The target sequences of these shRNA are listed in Supplementary Table S1. Stably transfected cell lines were obtained by selection with 2–5 μg/ml puromycin.
RNA and protein extraction
Total RNA was extracted from either cell lines or xenograft tissues using a QIAamp RNA kit (Qiagen). Cells were lysed with M-PER Mammalian Protein Extraction Reagent (Thermo scientific) containing protease inhibitor cocktail (Roche Life Science). Protein quantification was determined with BCA protein assay kit (Thermo scientific).
Microarray analysis
Microarray analyses were performed with H1299 cells expressing either GFP or SETDB1. Triplicate experimental and control samples were used for the analysis. The array hybridization was performed with the Human HT-12v4 chip Expression BeadChip (Illumina). Pathway analysis was accomplished with KEGG database. Real time PCR (RT-PCR) was performed to validate the selected genes.
Western blot analysis
Western blot analysis was performed with the primary antibodies against: SETDB1 (Sigma-Aldrich and Proteintech group), α-tubulin (Sigma-Aldrich), β-actin (Sigma-Aldrich), β-catenin (Cell Signaling Technology), Histone H3 (Cell Signaling Technology), Cyclin B1 (Cell Signaling Technology), Cyclin D1 (Santa Cruz Biotechnology), c-MYC (Santa Cruz Biotechnology) and H3K9me1/me2/me3 (Upstate Biotechnology).
Colony formation assays
Colony formation assays were performed in 1-ml cultures in 12-well flat-bottom plates: The base layer (0.5 ml) contained 0.25 ml of 1% low melting agarose and 0.25 ml 2 × RPMI with 20% FBS. After solidified, cells were added and mixed into the upper layer (0.5 ml) containing 0.25 ml of 0.7% low melting agarose and 0.25 ml 2×RPMI with 20% FBS. The plates were incubated for 21~35 days at 37°C in a fully humidified atmosphere of 5% CO2 in the air. Colony numbers were calculated after staining with crystal violet.
Luciferase reporter assay
Dual luciferase reporter assay system (Promega) was used according to the manufacturer’s instructions. GFP control or SETDB1 stably transfected cells were seeded in 0.5 ml medium in 12 well tissue culture plates. Cells were transfected with 1 μg of either TOP or FOP flash and 0.5 ng of pRL-TK vector. Seventy-two hours after transfection, the levels of WNT pathway activity were measured by the luciferase activity.
ChIP assay
ChIP assays were performed according to the protocol described previously (http://www.lbl.gov/LBL-Programs/lifesciences/BissellLab/labprotocols/chip.htm). Briefly, 7 million H1299 cells overexpressing SETDB1 were cross-linked with 1% formaldehyde for 10 min. Lysates were sonicated on ice to shear DNA to lengths between 500 and 800 bp. Chromatin was immunoprecipitated overnight at 4°C with either anti-SETDB1 antibody (Proteintech group) or normal Rabbit IgG (Santa Cruz Biotechnology). ChIP-enriched DNA was quantified by quantitative PCR and the PCR product was further examined by agarose gel electrophoresis and ethidium bromide staining.
Animal studies
Age-matched nude mice (5–6 weeks old) were used for the in vivo xenograft experiments. A total of 3 million H1299 cells either stably expressing SETDB1 or GFP (control) were suspended in a 1:1 mixture of fetal bovine serum and Matrigel (BD Labware) and injected subcutaneously into both flanks of nude mice.
Results
SETDB1 is recurrently amplified and highly expressed in NSCLC patients
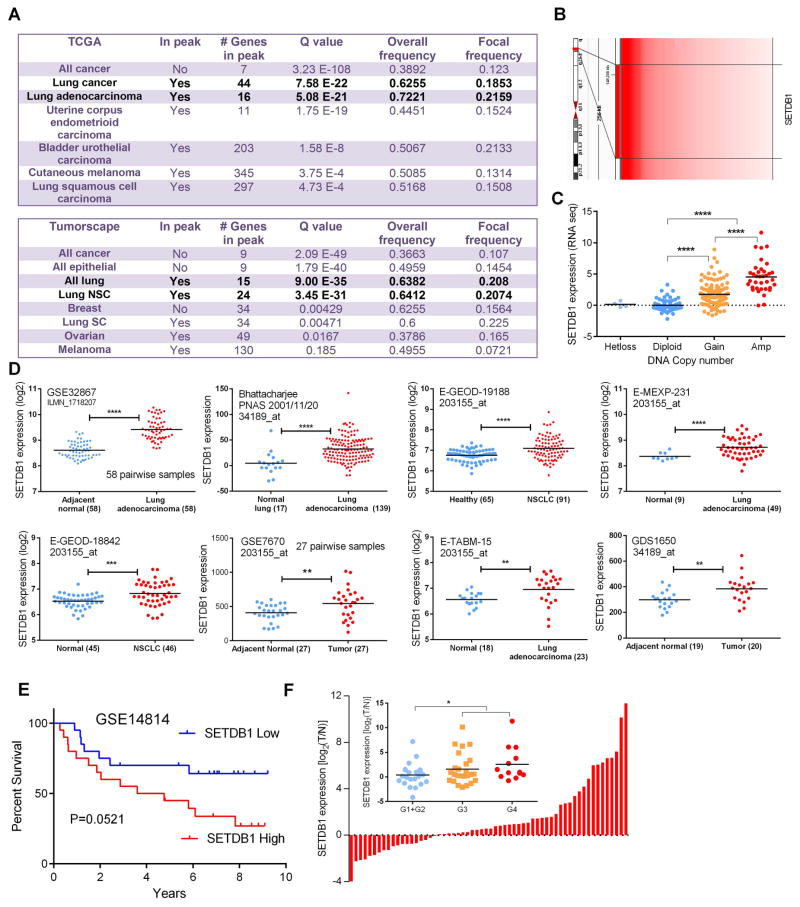
Gene copy number alterations were examined by 250k SNP-Chip analysis in nine NSCLC cell lines [17]. Gain or amplification (Amp) of chromosome 1q21 was identified as a common alteration among these cell lines. SETDB1 is one of the possible target genes located in this region which prompted further investigations. Tumors in TCGA (6,547 tumor samples) and Tumorscape (3,131 tumor samples) databases were examined for the SETDB1 copy number changes. SETDB1 was significantly amplified in ~20% of NSCLC samples in the TCGA compilation (with a focal frequency of 0.2159, Q value 5.08 E-21) and in Tumorscape dataset (with a focal frequency of 0.2074, Q value 3.45 E-31). The results are summarized in Figs. 1 A, B and Supplementary Fig. S2.
Fig. 1. Elevated expression of SETDB1 in NSCLC patients.
A. Copy number alterations of SETDB1 across a variety of cancer types (TCGA and Tumorscape database). SETDB1 is located at the peak region of the amplicon in lung cancer in both lung cancer sample collection. Significance was defined as Q value (calculation was performed on the database servers). B. Heatmap schematic illustration of copy number amplification of SETDB1 in tumorscape lung cancer samples which harbor 1q21 amplification. Position of SETDB1 is indicated. C. Gain of SETDB1 DNA copy number correlates with an increased mRNA expression in the NSCLC samples. D. Elevated expression of SETDB1 was noted in 8 different lung cancer patient cohorts. Data were retrieved from GEO and EBI Gene Expression Atlas database (*, P≤0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). E. Kaplan-Meier plots of overall survival: comparison of cases with highest (20 patients) versus lowest (20 patients) expression of SETDB1 in NSCLC patients (GSE14814). P value was calculated by log-rank test. F. mRNA levels of SETDB1 (examined by Realtime PCR) in a set of 60 paired samples of NSCLC. Inner picture, elevated expression of SETDB1 correlates with the increasement of tumor grade.
Next, we explored whether either the copy number gain or amplification in the genomic DNA translated into elevated SETDB1 expression. Microarray expression data were examined for each patient that had also been analyzed for SETDB1 copy number using the TCGA lung adenocarcinoma dataset. Compared to the samples grouped as either heterozygous loss (Hetloss) and Diploid cohorts, SETDB1 mRNA levels were significantly elevated in those grouped as either gain (Gain) or amplified (Amp) in copy number of the SETDB1 cohorts (Fig. 1 C), indicating that the copy number gain or amplification of SETDB1 loci resulted in elevated SETDB1 transcripts in lung cancer samples. Indeed, the elevation of SETDB1 transcripts were also noted in 8 independent expression microarray datasets of lung cancer which were done by different research groups [expression data were collected from GEO or Expression Atlas database (EMBL-EBI)] (Fig. 1 D). Furthermore, patients with elevated SETDB1 expression displayed a worse outcome compared with those with a lower SETDB1 expression (GSE14814) (Fig. 1 E).
To confirm our in silico observations, we independently interrogated our own NSCLC patient sample collection (NSCLC verses normal lung matched pairs). Among 60 matched samples, 23 NSCLC tumors had upregulation of SETDB1 mRNA by greater than 2-fold (Fig. 1 F). In addition, elevated expression of SETDB1 was noted in the tumors which were either grade 3 or 4 tumors (Fig. 1 F).
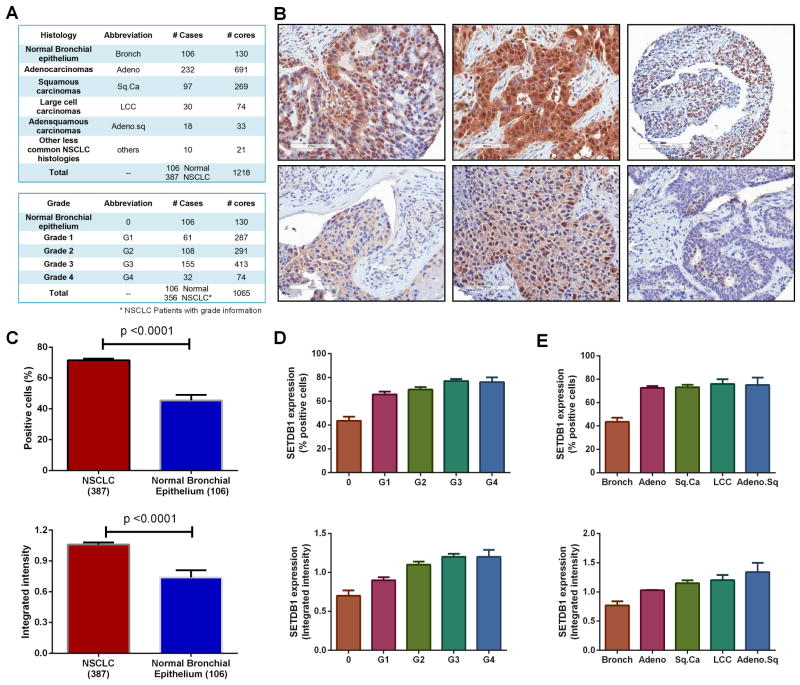
We then examined SETDB1 protein expression in 387 lung cancer samples and 106 normal lung samples by Tissue Microarray (TMA). Details of the patient information are listed in Fig. 2 A. Representative examples of tumor sections with either strong or weak nuclear staining of SETDB1 are shown in Fig. 2 B. A total of a mean 71.5% of NSCLC cells were positive for SETDB1, compared to 45.5% in normal bronchial epithelium cells (p<0.0001); the mean intensity value of SETDB1 staining was significantly higher in tumors than in the normal control samples (1.06 Vs 0.74) (Fig. 2 C). Both the percent positive cells and intensity of staining for SETDB1 was greater in the NSCLC cells than non-malignant bronchial epithelium control group (“bronch”); a significant association was consistently noted between SETDB1 expression and tumor grade (Figs. 2 D, E).
Fig. 2. SETDB1 protein expression is elevated in NSCLC samples.
A. Subtypes of lung cancer samples examined by immunohistochemistry (IHC). B. Representative examples of SETDB1 staining in NSCLC tumor sections. Upper panel: strong SETDB1 staining; lower panel: weak SETDB1 staining. C. Total of 387 NSCLC cases was analyzed by IHC. SETDB1 positive staining cells (upper row) and integrated intensities (lower row) in NSCLC samples compared to normal bronchial epithelium. D. SETDB1 positive staining cells (%) (upper row) and intensity of staining (lower row) correlates with the tumor grade. E. SETDB1 expression in different subtypes of lung cancer. Data are presented as percent positivity (upper row) and intensity of staining (lower row) is higher in each different subtype of cancer compared to normal.
SETDB1 promotes anchorage-independent growth in vitro and tumor growth in vivo
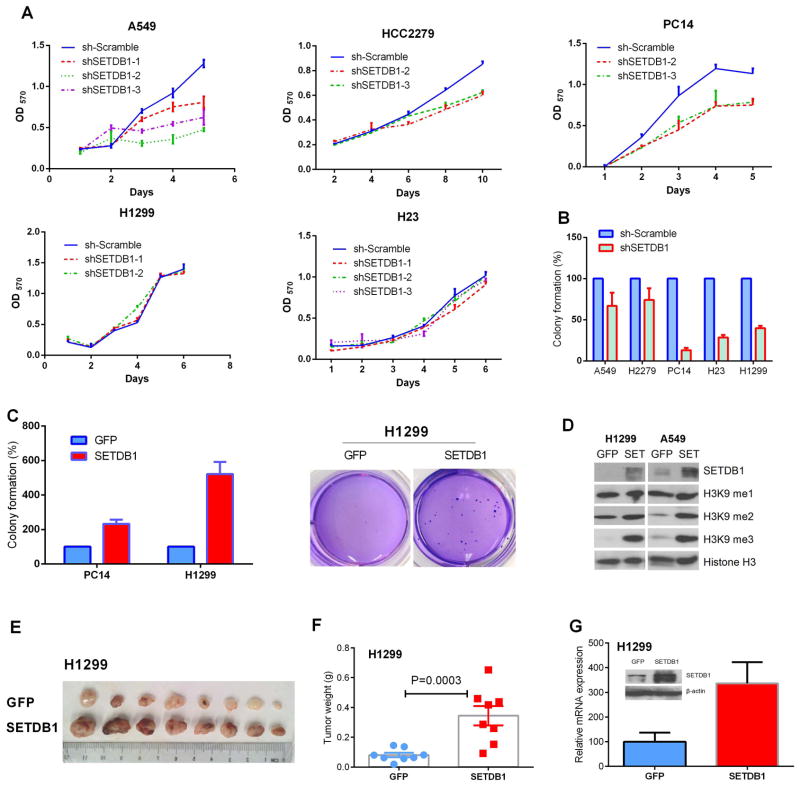
To test the functional importance of our findings, we silenced SETDB1 expression in several NSCLC cell lines using three shRNA targeting different coding regions of the gene. Silencing SETDB1 slowed proliferation in the cell lines A549, PC14 and HCC2279, whereas the changes were not significant in H1299 and H23 cells (Fig. 3 A). In contrast, SETDB1 silencing reduced the anchorage-independent clonogenic growth capacity of all five of these NSCLC cell lines when examined by soft agar clonogenic assays (Fig. 3 B). Reduction of clonogenic growth was especially prominent in TP53-deficient cell lines with decreased clonal growth of 86%, 71% and 60% in SETDB1 silenced PC14, H23 and H1299 cells, respectively (Detailed mutational information of cell lines is described in Supplementary Table S2). On the other hand, significantly elevated clonogenic growth was observed in cell lines with forced expression of SETDB1 (Fig. 3 C). This was especially dramatic (5-fold) in the H1299 cells having deletion of TP53 (Fig. 3 C). Consistent with the characterized role of SETDB1 as a histone H3K9 methyltransferase; elevated H3K9 di- and tri-methylation was noted in both H1299 and A549 cell lines when SETDB1 was overexpressed (Fig. 3 D).
Fig. 3. Aberrant expression of SETDB1 in NSCLC cells affects their proliferation in liquid culture, clonogenic growth in semi-soft cultures and tumor size in nude mice.
A. Effect of stably silencing SETDB1 on NSCLC cell growth in liquid culture. MTT assay was performed in 96 wells plate. Mean ± SD of 6 wells. B. Effect of silencing of SETDB1 on colony formation of NSCLC cells. Cells were seeded into soft agar in triplicate, and colonies were counted after 21~28 days of culturing. Mean ± SD (3 wells) are expressed as percent variation relative to scramble shRNA infected cells (control). C. Effect of forced expression of SETDB1 on colony formation of PC14 and H1299 NSCLC cells. Cells were seeded into soft agar in triplicate dishes; and colonies were counted on day 28 of culture. Values are expressed as fold variation relative to GFP overexpressing cells (control). Results are mean ± SD of 3 experiments. Right panel, representative pictures of colonies of H1299 overexpressing SETDB1. D. Histone H3K9 methylation status of NSCLC cells with stable overexpression of SETDB1 (H1299 and A549) was examined by western blot using antibodies specific for H3K9me1, H3K9me2, and H3K9me3 (mono-, di- and tri- methylation of H3K9, respectively). Expression levels of histone H3 protein were used as an internal control. E. SETDB1 stimulates NSCLC tumor growth in mice. Photograph of tumors dissected from nude mice which were injected with H1299 cells overexpressing either GFP (upper row) or SETDB1 (lower row). Tumor cells (3 × 106) were suspended in a 1:1 mixture of fetal bovine serum and Matrigel (BD Labware) and injected into both flanks of five week-old nude mice. Tumors were removed on day 21 from initiation of the experiment. F. Weights of tumors are shown in Fig. 3 E. The bars show differences in average weight of tumors in two groups (mean ± S.D., n = 8). Difference of mean weights between control (GFP) and SETDB1 overexpressing tumors was statistically significant (p-value of 0.0003). G. Expression level of SETDB1 in the tumors with forced expression of either GFP or SETDB1. Proteins and mRNA were isolated from xenografts. Tumor mRNA values were measured by RT-PCR and normalized with β-actin. Values are expressed as fold variation of SETDB1 overexpressing tumors relative to GFP control tumors. SETDB1 protein was measured by western blot (inner picture).
To analyze the effect of SETDB1 on cell growth in vivo, xenografts were established by subcutaneous injection of H1299 cells having forced expression of either SETDB1 or GFP into an immunodeficient murine model (Fig. 3 E). Tumor sizes and weights were significantly increased in mice injected with forced expression of SETDB1 H1299 cells than GFP control cells (p value of 0.0003) (Fig. 3 F). Total RNA and protein was extracted from the tumors, and the overexpressions of SETDB1 in these tumors were further confirmed by both RT-PCR and western blot (Fig. 3 G).
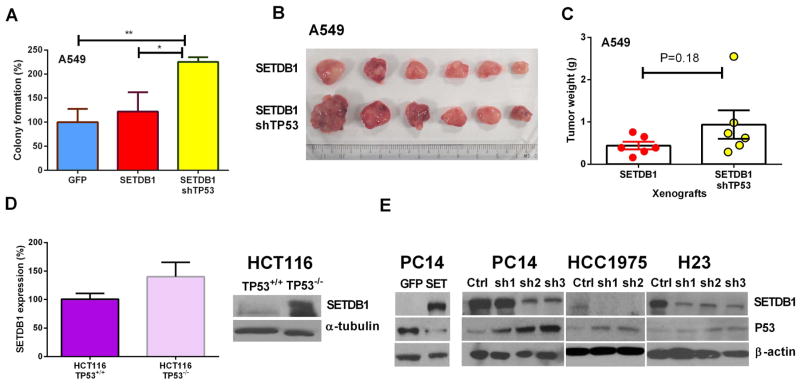
We also explored the possible interaction of P53 and SETDB1. Interestingly, forced silencing of TP53 in SETDB1 overexpressing A549 cells (TP53 wild type) resulted in a remarkable increase of their clonogenic growth in soft agar (Fig. 4 A), as well as the in vivo xenografts, albeit the latter was not statistically significant (Figs. 4 B, C) (p = 0.18). This suggests that a functional TP53 may antagonize the SETDB1 oncogenic effect. We made use of the isogenetic TP53−/− vs TP53+/+ HCT116 model to examine further the SETDB1 expression. Both the mRNA and protein levels of SETDB1 were increased in the TP53−/− compared to the TP53+/+ HCT116 cells. On the other hand, forced expression of SETDB1 significantly reduced the protein level of TP53. Likewise, silencing SETDB1 in three NSCLC lines increased their protein levels of P53 (Fig. 4 E). Taken together, SETDB1 and TP53 appear to modulate expression of each other.
Fig. 4. Inverse correlation between the protein levels of P53 and SETDB1.
A. Silencing TP53 enhanced the colony formation of A549 cells (wild type P53) having forced expression of SETDB1. B. A549 NSCLC cells were either stably overexpressing SETDB1 (upper row) or stably overexpressing SETDB1 and silencing TP53 (lower row). These two cohorts of cells (5 × 106) were injected into the opposite flanks of nude mice. Tumors were photographed (Panel B) and weighed (Panel C) on day 35 after initiation of the study. D. SETDB1 expression levels in the HCT116 TP53+/+ and TP53−/− HCT116 cells. Left panel, SETDB1 mRNA expression; right panel, western blot of the same cells. E. Protein levels (western blot) of P53 and SETDB1 in NSCLC cells (PC14, HCC1975, H23) with either stable overexpression or silencing of SETDB1.
SETDB1 regulates the WNT pathway
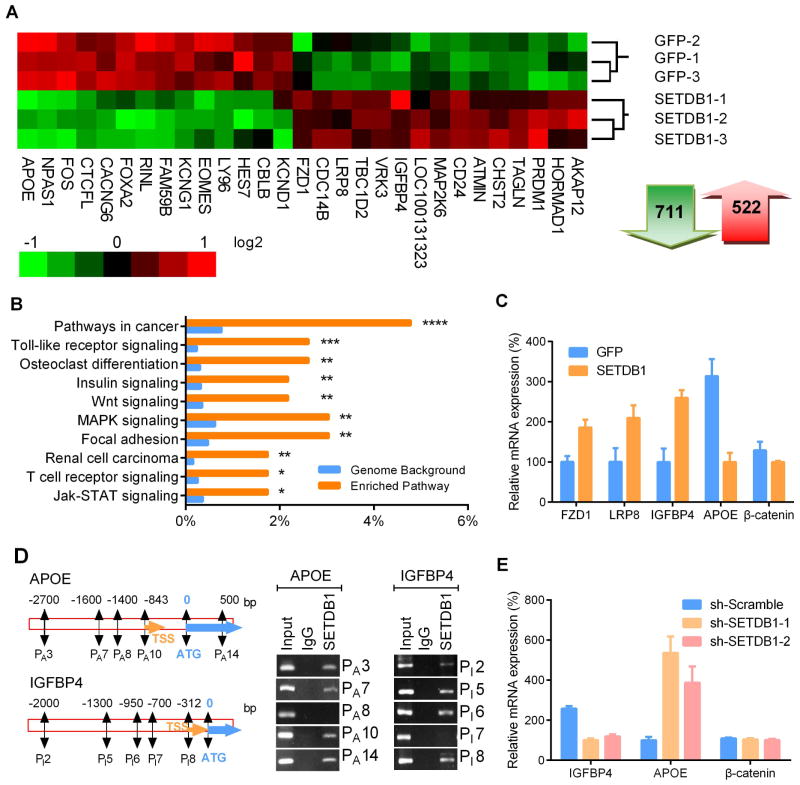
To investigate the molecular mechanism underlying the SETDB1 contribution to enhanced growth of lung cancer, we utilized cDNA microarray to profile gene expression changes in H1299 cells (Fig. 5 A). A total of 711 genes were down-regulated and 522 genes were up-regulated after forced expression of SETDB1. Pathway analysis showed cancer-related genes were significantly altered in SETDB1 overexpressing cells (p value <0.0005). In addition, genes related to signaling by Toll-like receptors, insulin, WNT, MAPK, focal adhesion, as well as JAK-STAT pathways were also significantly enriched (Fig. 5 B). The WNT pathway was selected for further study because both SETDB1 and WNT pathways are critical to the maintenance of self-renewal of stem cells [21–23]. Four WNT relevant genes identified by RNA array were validated by RT-PCR: APOE (4-fold down-regulation), IGFBP4 (3-fold up-regulation), FZD1 (1.7-fold up-regulation) and LRP8 (1.7-fold up-regulation) (Fig. 5 C).
Fig. 5. Altered gene expression in NSCLC cells with forced expression of SETDB1.
A. RNA array data was transformed into heat-map of gene expression in H1299 NSCLC cells stably expressing SETDB1. Green and red represents down-regulation and up-regulation of gene expression, respectively. Total 711 genes were down-regulated and 522 genes were upregulated after overexpression of SETDB1. B. Pathway enrichment analysis between SETDB1 and control GFP overexpressing H1299 cells using the KEGG database. Top 10 significantly enriched pathways are presented in the graph (*, P≤0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). C. RT-PCR validation of expression of genes related to the WNT signaling pathway in SETDB1 overexpressing H1299 cells. Relative mRNA amounts were normalized to β-actin. D. Recruitment of SETDB1 to the promoter region of APOE and IGFBP4 was examined by ChIP. Nucleotide segments 2 ~ 2.3 kb upstream and 600 ~ 1000 bp downstream from the starting site (ATG) were divided into 14 (APOE) and 10 (IGFBP4) regions, enrichment of each fragment (250~300 bp) were examined by PCR. Enrichment occurred in the promoter regions of PA 3, PA 7, PA 10 and PA 14 of the APOE promoter (left panel) and regions of PI 2, PI 5, PI 6 and PI 8 of the IGFBP4 promoter (right panel). TSS, transcriptional start site; ATG, protein start codon. Negative results of promoter regions PA 8 (APOE) and PI 7 (IGFBP4) were shown as controls for respective genes. E. mRNA levels of IGFBP4, APOE and β-catenin in SETDB1 silenced H1299 cells. Relative mRNA amounts were normalized with β-actin. Error bars indicate SD (n = 4).
We examined whether SETDB1 regulated these genes by binding directly to their promoter regions. Each promoter was divided into 10~14 sections (~ 200 bp), and ChIP-PCR was performed to determine the SETDB1 binding regions (Supplementary Table S2). Three SETDB1 enrichment sites were identified in the promoter region of APOE at −850 bp, −1600 bp, −2700 bp upstream from the start codon ATG, as well as 500 bp downstream of the start codon (Fig. 5 D, upper and left panels). In addition, three regions of enrichment occurred in the promoter of IGFBP4 (up-regulated gene by SETDB1) (Fig. 5 D, lower and right panels); but no enrichment was detected in the promoter region of the other two SETDB1 upregulated genes, FZD1 and LRP8 (data not shown), suggesting that upregulation of these gene may not be directly controlled by SETDB1. Consistent with SETDB1 directly binding to the promoter region of APOE and IGFBP4, silencing of SETDB1 significantly downregulated IGFBP4 and upregulated APOE mRNA, but did not affect the mRNA levels of β-catenin (Fig. 5 E).
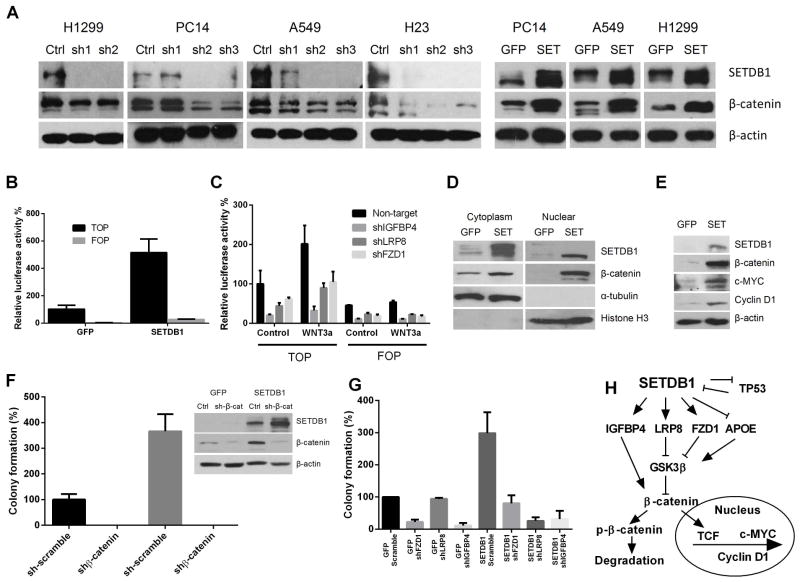
Since APOE, FZD1, LRP8 and IGFBP4 all participated in the WNT/β-catenin signaling by inhibiting the degradation of β-catenin protein [24–28], we speculated that alteration of these genes may lead to accumulation of β-catenin protein and activation of the WNT pathway. As anticipated, western blot demonstrated a prominent elevation of β-catenin protein in SETDB1 overexpressing cells and a decreased level of β-catenin protein when silencing SETDB1 in NSCLC (Fig. 6 A). As SETDB1 caused no significant change of β-catenin mRNA in either SETDB1 overexpressed or silenced cells (Figs. 5 C, E), the pathway of accumulation of β-catenin probably is post-transcriptional.
Fig. 6. Protein levels of β-catenin in the NSCLC cells with either stably overexpressed or silenced SETDB1.
A. Western analysis of β-catenin accumulation in NSCLC cell lines with either silenced or overexpressed SETDB1. Ctrl, sh-Scramble; sh1,2,3, three shRNA targeting to SETDB1. B. Relative TOP/FOP activities when overexpressing SETDB1 in H1299 cells. The stably forced expression of either SETDB1 or GFP containing cells were transfected with either pGL-TOP or pGL-FOP luciferase-reporter constructs. Luciferase activities were measured 72 hours after the transfection and normalized to the corresponding co-transfected Renilla luciferase activity. Data are shown as the ratio between TOP/FOP and Renilla. Error bars represent SD of three independent experiments. C. Silencing of IGFBP4, LRP8 or FZD1 reduced the WNT activity in H1299 NSCLC cells. Cells stably infected with either IGFBP4, LRP8 or FZD1 were transfected with either TOP or FOP, as well as Renilla control vector, and the levels of WNT pathway activity were determined by TOP/FOP assay. D. Cytoplasmic and nuclear fraction of H1299 cells stably overexpressing either GFP or SETDB1. Cells were grown on 10 cm dishes to 50% ~ 70% confluence, and the cytoplasmic and nuclear proteins were prepared as described in Materials and Methods. Cytoplasmic α-tubulin and nuclear histone H3 was used as controls of protein fractionation. E. Western analysis of alternations of downstream genes (c-MYC, Cyclin D1) of the WNT pathway in SETDB1 stably overexpressing H1299 NSCLC cells. F. Effect of stable silencing of β-catenin on clonogenic growth of H1299 cells overexpressing either GFP (control) or SETDB1. Values are expressed as % of GFP control cells, mean ± SD (three independent experiments). The silencing efficiency of β-catenin was determined by Western blot (right side). G. Clonogenic growth of either GFP or SETDB1 forced expressing H1299 cells after stable silencing of FZD1, LRP8 or IGFBP4. Values are expressed as % of GFP control cells. Experiments were performed in triplicate, and results are presented as mean ± SD. H. Schematic illustration of the proposed signaling interaction of SETDB1 and WNT pathway.
Activation of WNT pathway is required for SETDB1 induced tumorigenesis
To prove further that SETDB1 increased the activity of the WNT pathway, the TOP/FOP reporter system was employed to measure the activity of the WNT pathway [29]. Overexpression of SETDB1 in H1299 cells significantly increased the value of TOP luciferase activity (Fig. 6 B). Consistently, silencing of either FZD1, LRP8 or IGFBP4 significantly reduced TOP luciferase activity in H1299 cells (Fig. 6 C). Since the hallmark of activation of WNT signaling pathway is nuclear localization of β-catenin [30–32], the total proteins of transfected H1299 cells were separated into cytoplasmic and nuclear fractions to examine the nuclear β-catenin accumulation. Lysates were collected, blotted and probed with β-catenin antibody. Examining of cytoplasmic protein α-tubulin and nuclear protein Histone H3 (compartment specific controls) confirmed good nuclear/cytoplasmic protein separation. A significant increase of β-catenin protein migrated into the nucleus of SETDB1 overexpressing H1299 cells (Fig. 6 D), consistent with the activation of WNT pathway in SETDB1 overexpressing cells. When examining the downstream pathway of WNT, the elevated protein levels of β-catenin stimulated by SETDB1 was associated with increased levels of c-MYC and Cyclin D1 (Fig. 6 E).
Rescue experiments were carried out to test the concept that activation of WNT pathway is required for transformation induced by SETDB1. Depletion of β-catenin abolished the colony-forming ability of SETDB1 overexpressing H1299 cells (Fig. 6 F). Furthermore, silencing of either FZD1, LRP8 or IGFBP4 concomitantly with forced expression of SETDB1 also suppressed the enhanced clonogenic and liquid culture growth of SETDB1 overexpressing cells (Fig. 6 G) (Supplementary Fig. S3). Taken together, these results highlight the critical role of the β-catenin pathway in SETDB1 mediating transformation.
Discussion
SETDB1 is within the 1q21 amplicon, a region recurrently amplified in a variety of solid tumors including lung, breast and ovarian cancers, as well as melanomas. SETDB1 is a methyltransferase transfering a methyl group to the histone 3 lysine 9 (H3K9) which is generally believed to suppress gene expression associated with heterochromatin formation [16,33]. Recently, SETDB1 was identified as a novel oncogene in a zebrafish melanoma model [34]. During the preparation and submission of our manuscript, another investigative group also noted an oncogenic effect of SETDB1 in human lung cancers [35]. Our study confirms their observation and further explores the mechanism by which SETDB1 has tumorigenic activity.
SETDB1 maintains the embryonic stemness by controlling the expression of stem cell-related factors (OCT4 and Nanog) [21]. However, SETDB1 did not affect the expression of these genes in our study of NSCLC cells (data not shown), but it had a profound effect on the WNT pathway. We examined, for the first time, expression of SETDB1 by IHC in a large cohort of well-annotated lung cancer samples (387 lung cancers vs 106 normal cases). SETDB1 was statistically higher (p < 0.0001) in NSCLC tumors than normal lung tissue as measured by both percent positive cells and their staining intensity for SETDB1. This occurred in adeno-, squamous, large cell carcinomas and adenosquamous carcinomas of the lung; and the levels of SETDB1 increased with increased grade of NSCLC.
WNT/β-catenin signaling plays a well-defined oncogenic role in colon and skin cancers [36]. Studies of both normal intestinal cells and colorectal cancer cells have shown that inhibition of WNT signaling exhausts normal intestinal cells and blocks the growth of colorectal cancer cells [37,38]. In addition, the WNT signaling pathway including β-catenin can also regulate the length of telomeres by directly controlling the transcription of telomerase reverse transcriptase, which further emphasizes the critical role that the WNT pathway has in regulating the cell cycle, cell division and “stemness” [39–41]. WNT/β-catenin pathway play an oncogenic role in NSCLC [36]. However, unlike colorectal cancer which has high levels of β-catenin as a result of mutational loss of the adenomatous polyposis coli (APC) gene, mutation of either APC or β-catenin is infrequent in lung cancers [42]. Our studies suggest that activation of WNT pathway by SETDB1 in lung cancer results in accumulation of nuclear β-catenin causing a transformation phenotype (Fig. 6 H).
As a H3K9 methyltransferase, SETDB1 has been characterized as a classic transcriptional repressor [21]. However, our microarray data showed that after induction of expression of SETDB1 in NSCLC, 711 genes transcriptionally decreased and another 522 genes increased in their expression. This up-regulation may represent secondary events as a result of down-regulation of repressive genes by SETDB1; or SETDB1 may enhance expression of select genes by unexplored mechanisms. Providing weight for the latter view, clear enrichment of SETDB1 on the promoter of a SETDB1 induced gene (IGFBP4) was noted. Indeed, studies by other researchers found that H3K9me3 marks are also presented in transcriptionally active genes [43–45]. For example, in a study of histone modification associated with human X chromosome, H3K9me3 was found prominently within the actively transcribed genes, and the highest levels of H3K9me3 occurred within the highly expressed genes [43]. In addition, evidence from Drosophila showed that SETDB1 could both repress, as well as activate genes; and this dual function was determined by the binding position of SETDB1 on the chromatin [46]. Notably, G9A, another H3K9 methyltransferase, also can enhance gene expression by acting as a transcriptional coactivator of the hormone receptor signaling pathways [47,48]. A similar activation effect also has been observed in the H3K27 methytransferase EZH2 [49].
SETDB1 does not contain a DNA binding motif, but can form a complex with KAP1 (Trim28) and HP1 or certain Zinc finger proteins, which contain a DNA binding motif resulting in DNA binding of the complex [14]. Adding to the complexity, SETDB1 can also bind to Suv39H1, G9a and GLP, which together can recruit additional factors to modulate transcription [50]. Further experiments are ongoing to elucidate the detailed mechanism of activation of genes by SETDB1. In addition, we made an unexpected observation, SETDB1 can lower expression of P53; likewise, P53 can decrease expression of SETDB1. Silencing of either one of these genes can reciprocally enhance the expression of another gene. Therefore, SETDB1 may further enhance cellular growth when silencing P53. This novel interaction requires further study.
In summary, we showed that some of the NSCLC samples have elevated SETDB1 expression associated with an increased grade of tumor. The increased levels of SETDB1 produced an increased clonogenic growth, associated with the activation of the WNT pathway and tumor growth. Our findings suggest inhibitors that therapeutically target SETDB1 may benefit the large population of NSCLC patients whose tumors have high expression of SETDB1.
Supplementary Material
Acknowledgments
This work was funded by the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award to H. Phillip Koeffler, and NIH grant R01CA026038-35, as well as the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence Initiatives, and Early Detection Research Network (NCI CA86366) Award to Lee Goodglick and David Chia. We thank Professor Frank J. Rauscher, Leonard Zon and Sudhakar Jha for their extremely helpful suggestions, insights and constructs. We thank members of Prof Lorenz Poellinger’s laboratory for advice and discussions.
Footnotes
No conflict of interests was declared.
Author contributions
Q-Y S, L-W D conceived, carried out the experiments, analyzed the data and wrote the manuscript, J-F X carried out the experiments, WC, S-L L, NH, XYL, LX, TH, and D-C L analyzed the data, LG, DC, VM, MA, SRK, NBD and JWS carried out the IHC experiments, SS, DX carried out the RT-PCR experiment in lung cancer patient samples, L-Z L and HY analyzed the microarray expression data, HPK conceived the experiment, analyzed the data, wrote and reviewed the manuscript.
SUPPLEMENTARY MATERIAL ON THE INTERNET
The following supplementary material may be found in the online version of this article: Supplementary materials and methods
Table S1. List of the short hairpin RNA (shRNA) target sequences used in this study.
Table S2. Mutational genetic background of the NSCLC cells.
Table S3. Primers used for the CHIP experiments.
Fig. S1. Testing the specificity of SETDB1 antibody.
Fig. S2. SETDB1 copy number alternations and mutation status in different types of cancer.
Fig. S3. Effect of silencing β-catenin, FZD1, LRP8 or IGFBP4 on the proliferation of H1299 with overexpression of either GFP or SETDB1.
References
- 1.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 2.Crino L, Weder W, van Meerbeeck J, et al. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21 (Suppl 5):v103–v115. doi: 10.1093/annonc/mdq207. [DOI] [PubMed] [Google Scholar]
- 3.Bell DW, Brannigan BW, Matsuo K, et al. Increased prevalence of EGFR-mutant lung cancer in women and in East Asian populations: analysis of estrogen-related polymorphisms. Clin Cancer Res. 2008;14:4079–4084. doi: 10.1158/1078-0432.CCR-07-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology. J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mano H, Takeuchi K. EML4-ALK fusion in lung. Am J Pathol. 2010;176:1553–1554. doi: 10.2353/ajpath.2010.091057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 8.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 9.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 10.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 12.Miao F, Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Mol Cell Biol. 2005;25:4650–4661. doi: 10.1128/MCB.25.11.4650-4661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 14.Schultz DC, Ayyanathan K, Negorev D, et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loyola A, Tagami H, Bonaldi T, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsui T, Leung D, Miyashita H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 17.Hayano T, Garg M, Yin D, et al. SOX7 is down-regulated in lung cancer. J Exp Clin Canc Res. 2013;32:17. doi: 10.1186/1756-9966-32-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi S, Deng YZ, Zhao JS, et al. RACK1 promotes non-small-cell lung cancer tumorigenicity through activating sonic hedgehog signaling pathway. J Biol Chem. 2012;287:7845–7858. doi: 10.1074/jbc.M111.315416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mah V, Seligson DB, Li A, et al. Aromatase expression predicts survival in women with early-stage non small cell lung cancer. Cancer Res. 2007;67:10484–10490. doi: 10.1158/0008-5472.CAN-07-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huerta-Yepez S, Yoon NK, Hernandez-Cueto A, et al. Expression of phosphorylated raf kinase inhibitor protein (pRKIP) is a predictor of lung cancer survival. BMC Cancer. 2011;11:259. doi: 10.1186/1471-2407-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilodeau S, Kagey MH, Frampton GM, et al. SETDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland JD, Klaus A, Garratt AN, et al. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254–264. doi: 10.1016/j.ceb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 24.Caruso A, Motolese M, Iacovelli L, et al. Inhibition of the canonical Wnt signaling pathway by apolipoprotein E4 in PC12 cells. J Neurocytol. 2006;98:364–371. doi: 10.1111/j.1471-4159.2006.03867.x. [DOI] [PubMed] [Google Scholar]
- 25.Pencheva N, Tran H, Buss C, et al. Convergent Multi-miRNA Targeting of ApoE Drives LRP1/LRP8-Dependent Melanoma Metastasis and Angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueno K, Hirata H, Majid S, et al. IGFBP-4 activates the Wnt/beta-catenin signaling pathway and induces M-CAM expression in human renal cell carcinoma. Int J Cancer. 2011;129:2360–2369. doi: 10.1002/ijc.25899. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Zhang X, Zhang L, et al. LRP8 mediates Wnt/β-catenin signaling and controls osteoblast differentiation. J Bone Miner Res. 2012;27:2065–2074. doi: 10.1002/jbmr.1661. [DOI] [PubMed] [Google Scholar]
- 28.Flahaut M, Meier R, Coulon A, et al. The Wnt receptor FZD1 mediates chemoresistance in neuroblastoma through activation of the Wnt/β-catenin pathway. Oncogene. 2009;28:2245–2256. doi: 10.1038/onc.2009.80. [DOI] [PubMed] [Google Scholar]
- 29.Barolo S. Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene. 2006;25:7505–7511. doi: 10.1038/sj.onc.1210057. [DOI] [PubMed] [Google Scholar]
- 30.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 31.Dvory-Sobol H, Sagiv E, Kazanov D, et al. Targeting the active β-catenin pathway to treat cancer cells. Mol Cancer Ther. 2006;5:2861–2871. doi: 10.1158/1535-7163.MCT-06-0122. [DOI] [PubMed] [Google Scholar]
- 32.Nicholas S, Tolwinski, Wieschaus E. A Nuclear Function for Armadillo/β-Catenin. PLoS Biol. 2004;2:8. doi: 10.1371/journal.pbio.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karimi MM, Goyal P, Maksakova IA, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Paredes M, de Paz AM, Simó-Riudalbas L, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2013;33:2807–2813. doi: 10.1038/onc.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Chem Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Hirata A, Utikal J, Yamashita S, et al. Dose-dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development. 2013;140:66–75. doi: 10.1242/dev.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Sousa EM, Vermeulen L, Richel D, et al. Targeting Wnt signaling in colon cancer stem cells. Clin Cancer Res. 2011;17:647–653. doi: 10.1158/1078-0432.CCR-10-1204. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmeyer K, Raggioli A, Rudloff S, et al. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Toh L, Lau P, et al. Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/beta-catenin pathway in human cancer. J Biol Chem. 2012;287:32494–32511. doi: 10.1074/jbc.M112.368282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X, Malhotra GK, Lele SM, et al. Telomerase-immortalized human mammary stem/progenitor cells with ability to self-renew and differentiate. Proc Natl Acad Sci U S A. 2010;107:14146–14151. doi: 10.1073/pnas.1009030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He B, Barg RN, You L, et al. Wnt signaling in stem cells and non-small-cell lung cancer. Clin Lung Cancer. 2005;7:54–60. doi: 10.3816/CLC.2005.n.022. [DOI] [PubMed] [Google Scholar]
- 43.Brinkman AB, Roelofsen T, Pennings SW, et al. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vakoc CR, Mandat SA, Olenchock BA, et al. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Hahn MA, Wu X, Li AX, et al. Relationship between gene body DNA methylation and intragenic H3K9me3 and H3K36me3 chromatin marks. PLoS One. 2011;6:e18844. doi: 10.1371/journal.pone.0018844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lundberg LE, Stenberg P, Larsson J. HP1a, Su(var)3-9, SETDB1 and POF stimulate or repress gene expression depending on genomic position, gene length and expression pattern in Drosophila melanogaster. Nucleic Acids Res. 2013;41:4481–4494. doi: 10.1093/nar/gkt158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bittencourt D, Wu D-Y, Jeong KW, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2012;109:19673–19678. doi: 10.1073/pnas.1211803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purcell DJ, Jeong KW, Bittencourt D, et al. A distinct mechanism for coactivator versus corepressor function by histone methyltransferase G9a in transcriptional regulation. J Biol Chem. 2011;286:41963–41971. doi: 10.1074/jbc.M111.298463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee ST, Li Z, Wu Z, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 50.Fritsch L, Robin P, Mathieu JR, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.