Abstract
DNA microarrays and specific reverse-transcription polymerase chain reaction assays were used to reveal transcriptional patterns in the blood of children presenting with dengue shock syndrome (DSS) and well-matched patients with uncomplicated dengue. The transcriptome of patients with acute uncomplicated dengue was characterized by a metabolically demanding “host-defense” profile; transcripts related to oxidative metabolism, interferon signaling, protein ubiquination, apoptosis, and cytokines were prominent. In contrast, the transcriptome of patients with DSS was surprisingly benign, particularly with regard to transcripts derived from apoptotic and type I interferon pathways. These data highlight significant heterogeneity in the type or timing of host transcriptional immune responses precipitated by dengue virus infection independent of the duration of illness. In particular, they suggest that, if transcriptional events in the blood compartment contribute to capillary leakage leading to hypovolemic shock, they occur before cardiovascular decompensation, a finding that has implications for rational adjuvant therapy in this syndrome.
Dengue viruses (DENVs) are arboviruses that can cause an acute, systemic febrile illness that varies in its severity. Dengue is endemic in many countries in tropical regions and is a major public health problem. As yet, no vaccine or specific antiviral therapy is available for the prevention or management of dengue.
Dengue hemorrhagic fever (DHF) is a severe presentation of DENV infection. The important clinical events in DHF are hemorrhagic manifestations and capillary leakage syndrome. At its most severe, capillary leak results in hypovolemic shock, or dengue shock syndrome (DSS); this requires urgent intravenous fluid resuscitation to restore intravascular volume. A risk factor for DHF/DSS is secondary infection with a DENV serotype distinct from the individual’s DENV infection history [1-3]. The basis for secondary infection as a risk factor for severe disease has been ascribed to the infection-enhancing potential of sub- or nonneutralizing antibodies from a previous infection [4], cross-reactive memory T cells that secrete vasodilatory cytokines [5-7], and host genetic predisposition [8, 9]. Studies to investigate the pathogenesis of dengue are made more challenging by the absence of a robust animal model of disease.
There remains considerable uncertainty as to how the DENV-host interaction results in vascular leak, the most important clinical characteristic of DHF. Early viral load in vivo appears to be important, given that patients with DHF have significantly higher serum viral loads and NS1 concentrations (possibly driven by antibody-dependent enhancement) than do patients with dengue fever (DF) [10, 11]. DHF has also been associated with a robust host inflammatory immune response—significantly greater plasma concentrations of inflammatory cytokines [12-14] and activated lymphocytes are found in patients with DHF than in those with DF [7, 15].
Type I interferons (IFN-α/β) are likely important in the host defense against DENV infection. Mice deficient in IFN-α/β receptors are more susceptible to DENV infection [16], and IFN-α/β can protect cells against DENV infection in vitro [17]. Once infected, however, DENV nonstructural proteins can functionally attenuate the activity of IFN-α/β–mediated antiviral mechanisms, suggesting an evolved viral strategy for escaping early innate immune defenses [18, 19]. Finally, the process of antibody-dependent enhancement of infection may result in attenuation of IFN-α/β–mediated up-regulation of transcription factors important to innate antiviral effector mechanisms [20].
Genomewide gene expression studies of dengue represent an opportunity to identify novel markers associated with immunity and disease pathogenesis. Ubol et al. [21] demonstrated an early difference in the transcriptome between children with DF and those with DHF, and we previously found that transcripts from type I IFN–stimulated genes (ISGs) were less abundant in adult patients with DSS than in those with less-severe dengue [22]. The present study describes global patterns of gene transcript abundance in whole blood from 2 groups of pediatric patients with dengue who exhibited different clinical phenotypes after identical illness durations. A key finding was the relatively low abundance of transcripts from ISGs among patients with DSS at the time of cardiovascular decompensation.
METHODS
Patients and clinical investigations
From July to November 2005, we prospectively enrolled 111 children with acute dengue into a study of transcriptional responses in whole blood at the Hospital for Tropical Diseases (HTD), Ho Chi Minh City, Vietnam. Written informed consent was obtained from the parents of each patient. The study protocol was approved by the Scientific and Ethical Committee of HTD and the Oxfordshire Tropical Research Ethical Committee. Clinical and hematological assessments were performed daily. Samples for RNA analysis were collected at the time of hospital admission and at a 1-month follow-up visit. Hemoconcentration was determined by comparing the maximum hematocrit recorded during hospitalization with the value recorded at convalescence. On the basis of World Health Organization classification criteria, 27 patients had DSS (pulse pressure of ≤20 mm Hg with poor peripheral perfusion and rapid, weak pulse) and were admitted to the intensive care unit and received appropriate resuscitation. The remaining 84 patients had dengue without evidence of cardiovascular compromise. We have used the term “uncomplicated dengue” to refer to these hospitalized patients because they did not require any significant clinical interventions and were managed throughout in the general dengue ward.
For the 27 patients with DSS, we selected RNA samples from 9 patients who presented to the hospital with DSS on the fourth day of illness (with the day of onset of symptoms taken as day 1) for microarray analysis. For each patient with DSS, an age- and (where possible) sex-matched control patient with uncomplicated dengue and an identical duration of illness was selected.
Dengue serology and polymerase chain reaction (PCR)
Serological testing of paired plasma samples was performed using a dengue IgM and IgG capture ELISA (Panbio). The interpretation of primary versus secondary acute serological profiles was performed as suggested by the manufacturer. An internally controlled and serotype-specific real-time reverse-transcription PCR (RT-PCR) assay was used to measure DENV loads in plasma, as described elsewhere [22].
RNA extraction
Whole blood (2.5 mL) was collected directly into PAXgene RNA tubes (Qiagen) at the bedside. RNA extraction was performed using PAXgene RNA kits (Qiagen).
Gene expression microarray
Biotinylated amplified cRNA was generated by in vitro transcription technology using the Illumina TotalPrep RNA Amplification Kit (Ambion), in accordance with the manufacturer’s instructions. After purification, 850 ng of cRNA was hybridized to an Illumina HumanRef-8 V1BeadChip (containing probes to 23,961 RefSeq gene sequences) at 55°C for 18 h, in accordance with the manufacturer’s instructions. This was followed by washing, blocking, and streptavidin-Cy3–staining steps. Finally, the chip was scanned with an Illumina Bead Array Reader confocal scanner.
Array normalization
Standard normalization procedures (GeneSpring GX software; version 7.3; Silicon Genetics) for 1-color array data were used. In brief, data transformation was corrected for low signal, with values recorded at <0.01 increased to the minimum (0.01). Per-chip (mean) normalization accounted for chip variability by dividing all of the measurements on each chip by a 50th percentile value. Per-gene normalization accounted for variability between probe sets for different genes. Only genes that were detected in at least 1 sample were included in further analysis (13,535 genes). Analysis of variance (ANOVA) t test (unequal variances, nonmultiple corrections, and false discovery rate <5%) was used for all comparisons.
TaqMan low-density array
Whole-blood total RNA (300 ng) was reversed transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems). PCR was performed using Microfluidic cards (Applied Biosystems), in accordance with the manufacturer’s instructions. Transcript abundance data (cycle threshold [CT] value) for each gene was manually checked to remove the false-positive values, using SDS software (version 2.2) and RQ Manager (version 2.2.2; Applied Biosystems). Relative transcript abundance was determined using the 2−ΔΔCT method [23]. In brief, this method uses a single sample, termed the “calibrator sample,” as a comparator for every unknown sample’s gene expression level. The calibrator can be any sample chosen to have all of the genes expressed (CT value of <36 cycles). The calibrator is analyzed on every assay plate with the unknown samples of interest. The relative fold difference is calculated using the following formula: fold induction = 2−ΔΔCT, where ΔΔCT = (CT of gene of interest in unknown sample – CT of 18S gene in unknown sample) – (CT of gene of interest in calibrator – CT of 18S gene in calibrator). The fold differences in transcript abundance in samples from patients with different clinical phenotypes were compared by the Mann-Whitney U test, using MultiExperiment Viewer software (version 4.1.01; available at: http://www.tm4.org) [24].
Pathway analysis
Ingenuity Pathway Analysis software (version 7; Ingenuity Systems) was used to analyze enriched or less-abundant gene lists. For canonical pathways, the significance of the association was measured using Fisher’s exact test to calculate a P value determining the probability that the association between the genes in the data set and those in the canonical pathway is explained by chance alone.
Statistical analysis
ANOVA was used to analyze microarray data, using GeneSpring GX software. The Mann-Whitney U test was used to analyze quantitative RT-PCR data, using MultiExperiment Viewer. Differences for which P < .05 were regarded as significant.
RESULTS
Patient population
The aim of the present study was to describe early whole-blood transcriptional responses in 2 groups of pediatric patients (n = 9 per group), comparing children presenting with uncomplicated dengue and children presenting with DSS. The clinical and hematological characteristics, including absolute counts of the major lymphocyte subsets, for the 18 patients with dengue are described in table 1. Patients with DSS had lower platelet nadirs and, on the basis of changes in hemoconcentration, significantly greater vascular leak than did patients with uncomplicated dengue (table 1). On the fourth day of illness, when blood samples for microarray analysis were collected, patients with DSS had significantly higher absolute counts of CD3+CD8+ T cells (table 1) but significantly lower plasma viremia levels than did patients with uncomplicated dengue (table 1).
Table 1. Characteristics of the patient population sampled for whole-blood RNA.
| Variable | Uncomplicated dengue (n = 9) | DSS (n = 9) | P a |
|---|---|---|---|
| Male sex, no. (%) | 5 (55.6) | 4 (44.4) | |
| Age, range, years | 9–14 | 10–14 | |
| Day of illness | 4 | 4 | |
| Febrile, no. (%) | 9 (100) | 0 (0) | |
| Fever dayb | −2.5 (−4 to −1) | 0 | |
| Infecting serotype, no. | |||
| DENV-1 | 4 | 1 | |
| DENV-2 | 0 | 7 | |
| DENV-3 | 5 | 0 | |
| DENV-4 | 0 | 1 | |
| Clinical severity, no. | |||
| DF | 7 | … | |
| DHFII | 2 | … | |
| DHFIII | … | 8 | |
| DHFIV | … | 1 | |
| Viremia, mean (range), cDNA copies/mL | 48,317,364 (19,775–163,000,000) | 819,735 (24–4,234,125) | .006 |
| Secondary infection, no. (%) | 9 (100) | 9 (100) | |
| Platelet nadir, cells/μL | 44,000 (34,000–110,000) | 24,000 (11,000–86,000) | |
| Hemoconcentration, % | 15.11 (−9 to 22) | 28 (13–53) | .001 |
| Cell counts, cells/μL | |||
| CD3+ T cells | 649 (262–1097) | 845 (599–1387) | |
| CD3+CD4+ T cells | 331 (161–649) | 371 (155–561) | |
| CD3+CD8+ T cells | 207 (67–309) | 417 (293–752) | .01 |
| CD19+ B cells | 196 (75–352) | 229 (109–502) | |
| CD16+CD56+ NK cells | 124 (37–424) | 148 (51–236) |
NOTE. Data are median (range) values, unless otherwise indicated. DENV, dengue virus; DF, dengue fever; DHF, dengue hemorrhagic fever; DSS, dengue shock syndrome.
Mann-Whitney U test.
The day of defervescence was regarded as fever day 0.
Microarray data analysis
Analysis was focused on identifying transcripts that were differentially abundant between acute (day 4) and convalescent (day 30) samples. For the resulting 13,535 gene transcripts, we applied the following criteria in all comparisons: (1) gene transcripts were detected in at least 50% of samples, (2) the mean fold change between 2 conditions was >1.5-fold, and (3) the false-discovery rate was <5%. By these criteria, microarray data analysis revealed 3092 gene transcripts that were differentially abundant (523 less abundant and 2569 enriched) in acute samples from patients with DSS compared with that in autologous convalescent samples and 2471 gene transcripts that were differentially abundant (267 less abundant and 2204 enriched) in acute samples from patients with uncomplicated dengue compared with that in autologous convalescent samples. There were 1197 genes common to the 2 gene lists. The 100 most differentially abundant transcripts in acute and convalescent samples from each patient group are listed in tables 2 and 3.
Table 2. Top 100 differentially abundant transcripts in samples from febrile patients with uncomplicated dengue collected on day 4 relative to autologous convalescent samples collected on day 30.
| GenBank | Symbol | Fold change* | Full name |
|---|---|---|---|
| NM_002982 | CCL2 | +771 | Chemokine (C-C motif) ligand 2 |
| NM_005623 | CCL8 | +580.4 | Chemokine (C-C motif) ligand 8 |
| NM_002391 | MDK | +133.2 | Midkine (neurite growth–promoting factor 2) |
| NM_023930 | KCTD14 | +120.5 | Potassium channel tetramerization domain containing 14 |
| NM_001565 | CXCL 10 | +91.25 | Chemokine (C-X-C motif) ligand 10 |
| NM_005532 | IFI27 | +74.97 | … |
| NM_003865 | HESX1 | +71.55 | Homeobox, ES cell expressed 1 |
| NM_052884 | SIGLEC11 | +66.11 | Sialic acid–binding Ig-like lectin 11 |
| NM_172369 | C1QG | +51.85 | Complement component 1, q subcomponent, gamma polypeptide |
| NM_176870 | MT1K | +48.84 | Metallothionein 1M |
| NM_194323 | OTOF | +37.7 | Otoferlin |
| NM_033220 | TRIM14 | +36.9 | Tripartite motif containing 14 |
| NM_016135 | ETV7 | +32.84 | ets variant gene 7 (TEL2 oncogene) |
| NM_002933 | RNASE1 | +31.65 | Ribonuclease, RNase A family, 1 (pancreatic) |
| NM_001699 | AXL | +26.13 | AXL receptor tyrosine kinase |
| NM_145060 | MGC10200 | +22 | Chromosome 18 open reading frame 24 |
| NM_017414 | USP18 | +21.14 | Ubiquitin-specific peptidase 18 |
| NM_004972 | JAK2 | +19.79 | Janus kinase 2 (a protein tyrosine kinase) |
| NM_015341 | BRRN1 | +18.99 | Barren homolog 1 (Drosophila) |
| NM_006547 | IMP-3 | +18.92 | IGF-II mRNA–binding protein 3 |
| NM_000491 | C1QB | +18.03 | Complement component 1, q subcomponent, beta polypeptide |
| NM_014736 | KIAA0101 | +15.66 | … |
| NM_000063 | C2 | +15.56 | Complement component 2 |
| NM_144590 | MGC22805 | +14.43 | Ankyrin repeat domain 22 |
| NM_020954 | KIAA1618 | +14.16 | KIAA1618 |
| NM_021991 | JUP | +14.03 | Junction plakoglobin |
| NM_015535 | DNAPTP6 | +13.41 | DNA polymerase–transactivated protein 6 |
| NM_015714 | G0S2 | +13.28 | G0/G1 switch 2 |
| NM_006281 | STK3 | +12.97 | Serine/threonine kinase 3 (STE20 homolog, yeast) |
| NM_003571 | BFSP2 | +12.81 | Beaded filament structural protein 2, phakinin |
| NM_003543 | HIST1H4H | +12.2 | Histone 1, H4h |
| NM_014398 | LAMP3 | +12.14 | Lysosomal-associated membrane protein 3 |
| NM_015149 | RGL1 | +11.93 | Ral guanine nucleotide dissociation stimulator–like 1 |
| NM_005082 | ZNF147 | +11.75 | Tripartite motif containing 25 |
| NM_012297 | G3BP2 | +11.41 | … |
| NM_025193 | HSD3B7 | +11.23 | Hydroxy delta 5-steroid dehydrogenase, 3 beta– and steroid delta–isomerase 7 |
| NM_033023 | PDGFA | +11.18 | Platelet-derived growth factor alpha polypeptide |
| NM_006101 | KNTC2 | +11.13 | Kinetochore associated 2 |
| NM_080416 | PNUTL2 | +10.52 | Septin 4 |
| NM_181791 | GPR141 | +10.36 | G protein–coupled receptor 141 |
| NM_004336 | BUB1 | +10.12 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| NM_000355 | TCN2 | +9.911 | Transcobalamin II, macrocytic anemia |
| NM_003965 | CCRL2 | +9.651 | … |
| NM_000359 | TGM1 | +9.519 | Transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyftransferase) |
| NM_002053 | GBP1 | +9.327 | Guanylate binding protein 1, interferon inducible, 67 kDa |
| NM_145918 | CTSL | +9.251 | Cathepsin L |
| NM_152899 | IL4I1 | +9.109 | Interleukin 4 induced 1 |
| NM_176797 | P2RY6 | +8.92 | Pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| NM_015589 | SAMD4 | +8.824 | Sterile alpha motif domain containing 4 |
| NM_001775 | CD38 | +8.729 | CD38 antigen (p45) |
| NM_002692 | POLE2 | +8.686 | Potymorase (DNA directed), epsilon 2 (p59 subunit) |
| NM_021105 | PLSCR1 | +8.353 | Phospholipid scramblase 1 |
| NM_030928 | CDT1 | +8.299 | DNA replication factor |
| NM_000062 | SERPING1 | +8.229 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) |
| NM_001735 | C5 | +8.171 | Complement component 5 |
| NM_001827 | CKS2 | +8.033 | CDC28 protein kinase regulatory subunit 2 |
| NM_002164 | INDO | +8.011 | Indoleamine-pyrrole 2,3 dioxygenase |
| NM_020367 | C12orf6 | +7.946 | Poly (ADP-ribose) polymerase family, member 11 |
| NM_147150 | AKAP2 | +7.799 | PALM2-AKAP2 protein |
| NM_174938 | FRMD3 | +7.765 | FERM domain containing 3 |
| NM_002189 | IL 15RA | +7.651 | Interleukin 15 receptor, alpha |
| NM_021063 | HIST1H2BD | +7.559 | Histone 1, H2bd |
| NM_003745 | SOCS1 | +7.499 | Suppressor of cytokine signaling 1 |
| NM_005101 | G1P2 | +7.325 | Interferon, alpha-inducible protein (clone IFI-15K) |
| NM_005502 | ABCA1 | +7.309 | ATP-binding cassette, subfamily A (ABC1), member 1 |
| NM_080657 | cig5 | +7.278 | Radical S-adenosyl methionine domain containing 2 |
| NM_012456 | TIMM10 | +7.146 | Translocase of inner mitochondrial membrane 10 homolog (yeast) |
| NM_001444 | FABP5 | +7.141 | Fatty acid–binding protein 5 (psoriasis associated) |
| NM_022147 | IFRG28 | +6.954 | 28-kDa interferon responsive protein |
| NM_181782 | NCOA7 | +6.927 | … |
| NM_005953 | MT2A | +6.896 | Metallothionein 2A |
| NM_003283 | TNNT1 | +6.857 | Troponin T type 1 (skeletal, slow) |
| NM_012081 | ELL2 | +6.764 | Elongation factor, RNA polymorase II, 2 |
| NM_032804 | C10orf22 | +6.66 | Chromosome 10 open reading frame 22 |
| NM_003878 | GGH | +6.465 | Gamma glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| NM_021170 | Hes4 | +6.46 | Hairy and enhancer of split 4 (Drosophila) |
| NM_152792 | FLJ25084 | +6.448 | Skin aspartic protease |
| NM_004419 | DUSP5 | +6.423 | Dual specificity phosphatase 5 |
| NM_138720 | HIST1H2BD | +6.318 | Histone 1, H2bd |
| NM_138801 | GALM | +6.316 | Galactose mutarotase (aldose 1 epimerase) |
| NM_144573 | NEXN | +6.227 | Nexilin (F actin–binding protein) |
| NM_004030 | IRF7 | +6.219 | Interferon regulatory factor 7 |
| NM_006634 | VAMP5 | +6.21 | Vesicle-associated membrane protein 5 (myobrevin) |
| NM_003504 | CDC45L | +6.202 | CDC45 cell division cycle 45–like (S. cerevisiae) |
| XM_209643 | LOC285510 | +6.184 | … |
| NM_006820 | C1orf29 | +6.129 | Interferon induced protein 44–like |
| NM_175622 | MT1J | +6.104 | Metallothionein 1J |
| NM_004856 | KIF23 | +5.95 | Kinesin family member 23 |
| NM_018438 | FBXO6 | +5.95 | F-box protein 6 |
| NM_016817 | OAS2 | +5.887 | 2′–5′-oligoadenylate synthetase 2, 69/71 kDa |
| NM_002638 | PI3 | −5.917 | Peptidase inhibitor 3, skin derived (SKALP) |
| NM_000478 | ALPL | −16.52 | Alkaline phosphatase, liver/bone/kidney |
| NM_002001 | FCER1A | −8.06 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| NM_177417 | KLC2L | −8.06 | Kinesin light chain 3 |
| NM_003944 | SELENBP1 | −7.63 | Selonium binding protein 1 |
| NM_000347 | SPTB | −7.29 | Spectrin, beta, erythrocytic (includes spherocytosis, dmical type I) |
| NM_005332 | HBZ | −7.09 | Hemoglobin, zeta |
| NM_030758 | OSBP2 | −6.99 | Oxysterol binding protein 2 |
| NM_024748 | FLJ11539 | −6.06 | … |
| NM_004778 | GPR44 | −5.98 | G protein–coupled receptor 44 |
Fold charges reflect greater (positive fold change) or lesser (negative fold change) abundance in samples from patients with acute uncomplicated dengue
Table 3. Top 100 differentially abundant transcripts in samples from patients with dengue shock syndrome (DSS) collected on day 4 relative to autologous convalescent samples collected on day 30.
| GenBank | Symbol | Fold changea | Full name |
|---|---|---|---|
| NM_002417 | MKI67 | +185.30 | Antigen identified by monoclonal antibody Ki-67 |
| NM_012112 | TPX2 | +161.90 | … |
| NM_024032 | MGC3130 | +156.50 | Chromosome 17 open reading frame 53 |
| NM_002933 | RNASE1 | +103.70 | Ribonuclease, RNase a family, 1 (pancreatic) |
| NM_152562 | CDCA2 | +64.90 | Cell division cycle associated 2 |
| NM_004181 | UCHL 1 | +60.71 | Ubiquitin carboxyl-terminal esterase I1 (ubiquitin thiolesterase) |
| NM_002997 | SDC1 | +59.92 | Syndecan 1 |
| NM_005480 | TROAP | +59.35 | Trophinin associated protein (tastin) |
| NM_052884 | SIGLEC11 | +41.31 | Sialic acid–binding Ig–like lectin 11 |
| NM_030928 | CDT1 | +39.79 | DNA replication factor |
| NM_003088 | FSCN1 | +39.29 | Fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| NM_015341 | BRRN1 | +37.47 | Barren homolog 1 (Drosophila) |
| NM_023930 | KCTD14 | +34.03 | Potassium channel tetramerization domain containing 14 |
| NM_001505 | GPR30 | +29.95 | G protein–coupled receptor 30 |
| NM_005532 | IFI27 | +29.76 | … |
| NM_016354 | SLCO4A1 | +28.6 | … |
| NM_003504 | CDC45L | +25.63 | CDC45 cell division cycle 45–like (S. cerevisiae) |
| NM_021000 | PTTG3 | +25.56 | … |
| NM_004237 | TRIP13 | +25.06 | Thyroid hormone receptor interactor 13 |
| NM_004336 | BUB1 | +24.53 | Bub1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| NM_080668 | CDCA5 | +23.13 | Cell division cycle associated 5 |
| NM_145060 | MGC10200 | +22.91 | Chromosome 18 open reading frame 24 |
| NM_001905 | CTPS | +21.54 | CTP synthase |
| NM_002466 | MYBL2 | +21.18 | v-myb myeloblastosis viral oncogene homoiog (avian)–like 2 |
| NM_003173 | SUV39H1 | +2.05 | Suppressor of variegation 3–9 homolog 1 (Drosophila) |
| NM_000170 | GLDC | +19.57 | Glycine dehydrogenase |
| NM_194323 | OTOF | +19.28 | Otoferlin |
| NM_015671 | DKFZP434J046 | +18.88 | Synonyms c19orf14, flj33298, dkfzp434j046, dkfzp686g1024; chromosome 19 open reading frame 14 |
| NM_003500 | ACOX2 | +18.86 | Acyl-coenzyme a oxidase 2, branched chain |
| NM_031299 | CDCA3 | +18.63 | Cell division cycle associated 3 |
| NM_020804 | PACSIN1 | +17.22 | Protein kinase C and casein kinase substrate in neurons 1 |
| XM_377087 | LOC401627 | +16.93 | … |
| NM_018101 | CDCA8 | +15.81 | Cell division cycle associated 8 |
| NM_017636 | TRPM4 | +15.41 | Transient receptor potential cation channel, subfamily M, member 4 |
| NM_172369 | C1QG | +14.97 | Complement component 1, q subcomponent, gamma polypeptide |
| NM_014900 | COBLL1 | +14.83 | … |
| NM_018410 | DKFZp762E1312 | +14.82 | Hypothetical protein dkfzp762e1312 |
| NM_182513 | Spc24 | +14.66 | Spindle pole body component 24 homolog (S. cerevisiae) |
| NM_003579 | RAD54L | +14.63 | Rad54-like (S. cerevisiae) |
| NM_007268 | Z39IG | +14.62 | V-set and immunoglobulin domain containing 4 |
| NM_012447 | STAG3 | +14.54 | … |
| NM_001255 | CDC20 | +13.57 | Cell division cycle 20 homolog (S. cerevisiae) |
| NM_001067 | TOP2A | +13.4 | Topoisomerase (DNA) II alpha 170 kDa |
| NM_001237 | CCNA2 | +13.39 | Cyclin a2 |
| NM_006623 | PHGDH | +13.28 | Phosphoglycerate dehydrogenase |
| XM_352965 | KIFC1 | +13.04 | … |
| NM_003258 | TK1 | +12.94 | Thymidine kinase 1, soluble |
| NM_004925 | AQP3 | +12.04 | Aquaporin 3 |
| NM_144988 | MGC19780 | +11.66 | … |
| NM_014152 | HSPC054 | +11.63 | … |
| NM_001168 | BIRC5 | +11.44 | Baculoviral iap repeat-containing 5 (survivin) |
| NM_005879 | TRIP | +11.35 | … |
| NM_015714 | G0S2 | +11.27 | G0/g1 switch 2 |
| NM_004260 | RECQL4 | +11.24 | RecQ protein–like 4 |
| NM_006845 | KIF2C | +11.15 | Kinesin family member 2c |
| NM_145249 | FAM14B | +10.67 | Family with sequence similarity 14, member b |
| NM_004701 | CCNB2 | +10.62 | Cydin b2 |
| NM_014488 | RAB30 | +10.59 | Rab30, member ras oncogene family |
| NM_024111 | MGC4504 | +10.56 | ChaC, cation transport regulator–like (E. coli) |
| NM_012140 | SLC25A10 | +10.23 | Solute earner family 25, member 10 |
| NM_021953 | FOXM1 | +10.1 | … |
| NM_024053 | C22orf18 | +9.548 | Chromosome 22 open reading frame 18 |
| NM_001775 | CD38 | +9.417 | CD38 antigen (p45) |
| NM_016459 | PACAP | +9.21 | Proapoptotic caspase adaptor protein |
| NM_003686 | EXO1 | +9.13 | … |
| NM_020120 | UGCGL 1 | +9.06 | UDP-glucose ceramide glucosyltransferase–like 1 |
| NM_006461 | SPAG5 | +9.03 | Sperm-associated antigen 5 |
| NM_005693 | NR1H3 | +8.98 | Nuclear receptor subfamily 1, group H, member 3 |
| NM_000491 | C1QB | +8.95 | Complement component 1, q subcomponent, beta poly peptide |
| NM_001192 | TNFRSF17 | +8.94 | Tumor necrosis factor receptor superfamily, member 17 |
| NM_001151 | SLC25A4 | +8.92 | Solute earner family 25 |
| NM_022346 | NCAPG | +8.90 | Non-SMC condensin I complex, subunit G |
| NM_003486 | SLC7A5 | +8.88 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 |
| NM_014736 | KIAA0101 | +8.82 | … |
| NM_014383 | TZFP | +8.807 | Zinc finger and BTB domain containing 32 |
| NM_015949 | C7orf20 | +8.74 | … |
| NM_003571 | BFSP2 | +8.71 | Beaded filament structural protein 2, phakinin |
| NM_152227 | SNX5 | +8.54 | Sorting nexin 5 |
| NM_004217 | AURKB | +8.44 | Aurora kinase B |
| NM_015426 | DKFZP434C245 | +8.43 | WD repeat domain 51a |
| NM_016359 | NUSAP1 | +8.43 | Nucleolar and spindle associated protein 1 |
| NM_002885 | RAP1GAI | −38.46 | RAP1, GTPase activating protein 1 |
| NM_000347 | SPTB | −26.17 | Spectrin, beta, erythrocytic (includes spherocytosis, clinical type 1) |
| NM_000345 | SNCA | −22.17 | Synuclein, alpha (non-A4 component of amyloid precursor) |
| NM_144665 | SESN3 | −17.88 | Sestrin 3 |
| NM_002638 | PI3 | −17.45 | Peptidase inhibitor 3, skin derived (SKALP) |
| NM_004667 | SDPR | −17.36 | Serum deprivation response (phosphatidylserine binding protein) |
| XM_353706 | LOC378204 | −16.05 | … |
| NM_024748 | FLJ11539 | −14.53 | … |
| NM_005629 | SLC6A8 | −14.32 | Solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| NM_003944 | SELENBP1 | −12.22 | Selenium binding protein 1 |
| XM_291438 | LOC343171 | −11.38 | Olfactory receptor, family 2, subfamily W, member 3 (OR2W3), mRNA |
| NM_182973 | LOC360200 | −10.40 | Transmembrane protease, serine 9 |
| NM,007308 | SNCA | −9.80 | Synuclein, alpha (non-A4 component of amyloid precursor) |
| NM_004778 | GPR44 | −9.43 | G protein-coupled receptor 44 |
| NM_006887 | ZFP36L2 | −9.34 | Zinc finger protein 36, C3H type–like 2 |
| NM_000342 | SLC4A1 | −9.25 | Solute earner family 4, anion exchanger |
| NM_006121 | KRT1 | −9.25 | Keratin 1 (epidermolytic hyperkeratosis) |
| NM_000559 | HBG1 | −8.84 | Hemoglobin, gamma A |
| NM_000184 | HBG2 | −8.54 | Hemoglobin, gamma G |
Fold changes reflect greater (positive fold change) or lesser (negative fold change) abundance in acute samples from patients with DSS
Uncomplicated dengue: acute versus convalescent samples
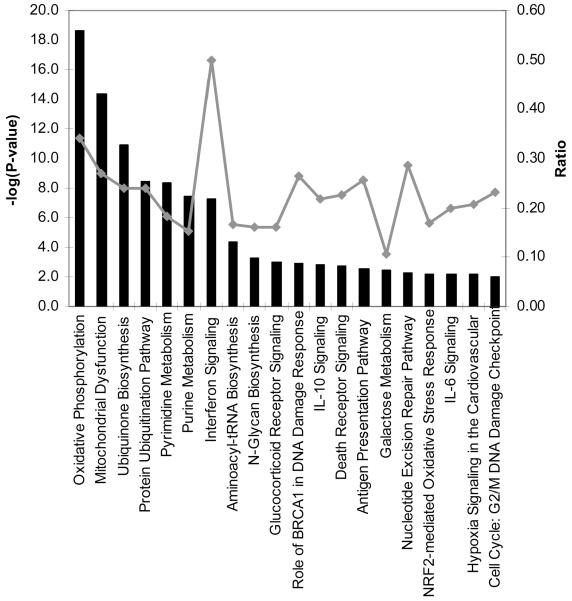
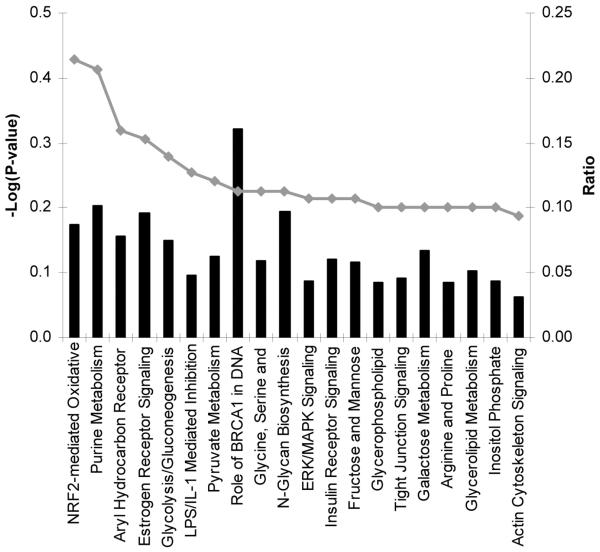
Unsupervised pathway analysis of transcripts either significantly enriched (n = 2204) or underrepresented (n = 267) in acute samples (relative to that in autologous convalescent samples) was used to identify biological themes in the transcriptome of patients with uncomplicated dengue. Transcripts significantly enriched in acute samples were derived from genes in pathways associated with oxidative metabolism, mitochondrial dysfunction, protein ubiquitination, and nucleotide metabolism, suggesting a highly active metabolic state consistent with a host response to infection (figure 1). Of immune-response pathways, the IFN pathway was most prominent, followed by interleukin (IL)–10, antigen presentation, and IL-6 signaling pathways. In contrast, among transcripts that were significantly less abundant in acute samples, there were no significant molecular themes arising from pathway analysis, most likely because of the small number (n = 267) of elements for analysis. Thus, the biological processes highlighted in the enriched population of transcripts are consistent with a metabolically demanding host response in which IFN-driven immune processes and cytokine networks are prominent.
Figure 1. Pathway analysis of transcripts enriched in acute samples from patients with uncomplicated dengue.
Shown are the top 20 canonical pathways identified by unsupervised pathway analysis of filtered microarray data representing transcripts enriched (n = 2204) in acute samples (day 4 of illness) from patients with uncomplicated dengue relative to autologous convalescent samples (day 30). The strength of the statistical association is indicated by the length of the bars. The ratio value reflects the proportion of gene elements on the enriched gene list that belong to one of these canonical pathways.
DSS: acute versus convalescent samples
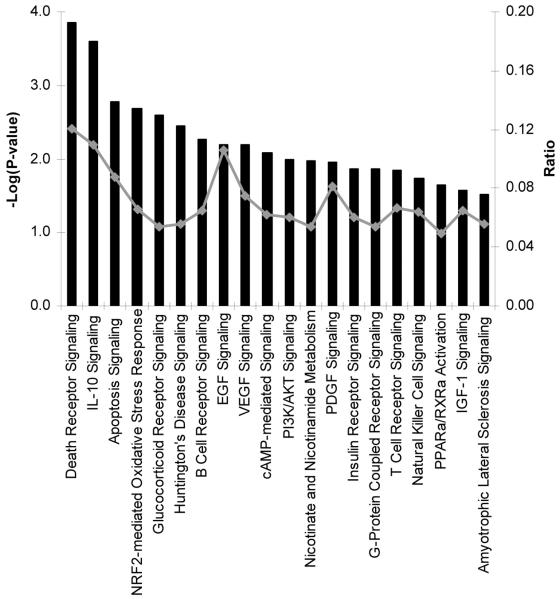
Surprisingly, unsupervised pathway analysis of transcripts significantly enriched (n = 2569) in acute samples from children with DSS did not identify any significant pathway or biological process (figure 2). Unsupervised analysis of underrepresented transcripts in acute samples (n = 523) implicated the death receptor, apoptotic, and IL-10 signaling pathways as being features of this “downregulated” transcriptome (figure 3). However, the strength of the association between these pathways and samples was not as robust as that observed between acute and convalescent samples in children with uncomplicated dengue (figure 1). Remarkably then, at a time when capillary leakage had precipitated hypovolemic shock in these afebrile patients with DSS, the whole-blood transcriptome appeared relatively muted from an immune-defense perspective.
Figure 2. Pathway analysis of transcripts enriched in acute samples from patients with dengue shock syndrome (DSS).
Figure 3. Pathway analysis of transcripts less abundant in acute samples from patients with dengue shock syndrome (DSS).
Shown are the top 20 canonical pathways identified by unsupervised pathway analysis of filtered microarray data representing transcripts less abundant (n = 523) in acute samples (day 4 of illness) from patients with DSS relative to autologous convalescent samples (day 30). The strength of the statistical association is indicated by the length of the bars. The ratio value reflects the proportion of gene elements on the enriched gene list that belong to one of these canonical pathways.
The absence of a strong transcriptional signature in the blood of children with DSS is in stark contrast with the prominent molecular themes identified in acute samples in febrile children with uncomplicated dengue (figure 1). These differences between acute and convalescent samples did not result from intrinsic differences between convalescent samples in the 2 patient groups; unsupervised comparison of these convalescent samples identified just 138 differentially abundant transcripts among 13,535 transcripts analyzed. Collectively, these data suggest fundamental differences in the evolution of the transcriptional response between these 2 groups of children with identical durations of illness but different clinical phenotypes.
Uncomplicated dengue versus DSS
The transcriptional profiles of acute samples from patients with uncomplicated dengue and those with DSS were compared directly to identify differentially abundant gene transcripts. The rationale for this comparison was to compare the host response in these 2 groups of patients with identical durations of illness but distinct clinical phenotypes and disease evolution. In unsupervised analysis, 1749 transcripts were differentially abundant between patients with acute uncomplicated dengue and those with DSS; transcripts from 1030 genes were enriched and 719 were less abundant in patients with uncomplicated dengue (table 4).
Table 4. Top 100 differentially abundant transcripts in samples from patients with uncomplicated dengue collected on day 4 relative to samples from patients with dengue shock syndrome collected on day 4.
| GenBank | Symbol | Fold changea | Function |
|---|---|---|---|
| NM_005623 | CCL8 | +170.64 | Chemokino (C-C motil) ligand 8 |
| NM_002982 | CCL2 | +86.95 | Chemokine (C-C motif) ligand 2 |
| NM_199204 | DHRS9 | +32.46 | Dehydrogenase/reductase (SDR family) member 9 |
| NM_004972 | JAK2 | +21.92 | Janus kinase 2 (a protein tyrosine kinase) |
| NM_002164 | INDO | +17.36 | Indoleamine-pyrrole 2,3 dioxygenase |
| NM_000067 | CA2 | +16.66 | Carbonic anhydrase II |
| NM_001565 | CXCL 10 | +16.26 | Chemokine (C-X-C motif) ligand 10 |
| NM_003865 | HESX1 | +12.45 | HESX homeobox 1 |
| NM_004657 | SDPR | +11.5 | Serum deprivation response (phosphatidylserine-binding protein) |
| NM_002053 | GBP1 | +10.68 | Guanylate binding protein 1, interferon inducible, 67 kDa |
| NM_002024 | FMR1 | +10.27 | Fragile X mental retardation 1 |
| NM_033238 | PML | +9.61 | Promyelocytic leukemia |
| XM_038864 | KIAA1632 | +8.13 | Homo sapiens KIAA1632 (KIAA1632), mRNA |
| XM_293276 | LOC347348 | +8.13 | … |
| NM_002729 | HHEX | +8.06 | Homeobox, hematopoietically expressed |
| NM_006079 | CITED2 | +7.87 | Cbp/p300-interacting transactivator with Glu/Asp rich carboxy-terminal domain, 2 |
| NM_014398 | LAMP3 | +7.57 | Lysosomal-associated membrane protein 3 |
| NM_030881 | DOX17 | +6.84 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 |
| NM_012329 | MMD | +6.71 | Monocyte to macrophage differentiation associated |
| NM_014016 | SACM1L | +6.62 | … |
| NM_001547 | IFIT2 | +6.62 | Interferon-induced protein with tetratricopeptide repeats 2 |
| XM_209643 | LOC285510 | +6.49 | … |
| NM_018682 | MLL5 | +6.02 | Myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila) |
| NM_181291 | WDR20 | +5.88 | Wd repeat domain 20 |
| NM_144590 | MGC22805 | +5.78 | Ankyrin repeat domain 22 |
| NM_006887 | ZFP36L2 | +5.71 | Zinc finger protein 36, C3H type–like 2 |
| NM_017719 | SNRK | +5.64 | SNF-related kinase |
| NM_174938 | FRMD3 | +5.61 | Ferm domain containing 3 |
| NM_000161 | GCH1 | +5.61 | GTP cyclohydrolase 1 (dopa-responsive dystonia) |
| NM_032860 | C6orf93 | +5.55 | Chromosome 6 open reading frame 93 |
| NM_003489 | NRIP1 | +5.37 | Nuclear receptor-interacting protein 1 |
| XM_059368 | LOC129607 | +5.23 | … |
| NM_021170 | Hes4 | +5.2 | Hairy and enhancer of split 4 (Drosophila) |
| NM_175862 | CD86 | +5.12 | CD86 antigen (CD28 antigen ligand 2, B7–2 antigen) |
| NM_014857 | HHL | +4.97 | RAB GTPase activating protein 1–like |
| NM_017631 | FLJ20035 | +4.97 | Hypothetical protein FLJ20035 |
| NM_006276 | SFRS7 | +4.95 | … |
| NM_016135 | ETV7 | +4.92 | ets variant gene 7 (TEL2 oncogene) |
| NM_000584 | IL8 | +4.9 | Interleukin 8 |
| NM_006022 | TSC22 | +4.73 | TSC22 domain family, member 1 |
| NM_003473 | STAM | +4.65 | Signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| MM_153045 | C9orf91 | +4.62 | Chromosome 9 open reading frame 91 |
| NM_004099 | STOM | +4.58 | Stomatin |
| NM_147128 | LOC223082 | +4.52 | … |
| NM_021105 | PLSCR1 | +4.48 | Phospholipid scramblase 1 |
| NM_033220 | TRIM14 | +4.46 | Tripartite motif containing 14 |
| NM_000689 | ALDH1A1 | +4.42 | Aldehyde dehydrogenase 1 family, member A1 |
| NM_145804 | ABTB2 | +4.42 | Ankyrin repeat and BTB (POZ) domain containing 2 |
| NM_018847 | KIAA1354 | +4.38 | Kelch-like 9 (Drosophila) |
| NM_178237 | SEC3L1 | +4.36 | … |
| NM_181782 | NCOA7 | +4.31 | … |
| NM_037817 | FLJ31033 | +4.31 | Hypothetical protein FLJ31033 |
| NM_000076 | CDKN1C | +4.31 | Cyclin dependent kinase inhibitor 1C (p57, Kip2) |
| NM_0O4842 | AKAP7 | +4.27 | A kinase (PRKA) anchor protein 7 |
| NM_003745 | SOCS1 | +4.25 | Suppressor of cytokine signaling 1 |
| NM_017837 | FLJ20477 | +4.21 | Phosphatidylinositol glycan, class V |
| NM_002835 | PTPN12 | +4.21 | Protein tyrosine phosphatase, nonreceptor type 12 |
| NM_173842 | IL 1RN | +4.2 | Interleukin 1 receptor antagonist |
| NM_022168 | MDA5 | +4.16 | Interferon induced with helicase C domain 1 |
| MM_199324 | HSHIN1 | +4.14 | OTU domain containing 4 |
| NM_001981 | EPS15 | +4.14 | Epidermal growth factor receptor pathway substrate 15 |
| NM_001735 | C5 | +4.13 | Complement component 5 |
| NM_002145 | HOXB2 | +4.11 | Homeobox 82 |
| NM_014314 | RIG-1 | +4.08 | Dead (AspGIu-Ala-Asp) box polypeptide 58 |
| NM_001186 | BACH1 | +4.04 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| NM_052941 | GBP4 | +4.03 | Guanylate binding protein 4 |
| NM_014358 | CLECSF9 | +4.03 | C-type lectin domain family 4, member E |
| NM_001712 | CEACAM1 | +3.92 | Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| NM_006074 | TRIM22 | +3.92 | Tripartite motif-containing 22 |
| NM_153341 | IBRDC3 | +3.9 | IBR domain containing 3 |
| NM_006415 | SPTLC1 | +3.87 | Serine palmitoyltransferase, long chain base subunit 1 |
| NM_001548 | IFIT1 | +3.83 | Interferon-induced protein with tetratnucleopeptide repeats 1 |
| NM_152522 | MGC33864 | +3.83 | ADP-ribosylation–like factor 6 interacting protein 6 |
| NM_020423 | PACE-1 | +3.81 | … |
| NM_013352 | SART2 | +3.81 | Squamous cell carcinoma antigen recognized by T cells 2 |
| NM_006018 | HM74 | +3.78 | G protein–coupled receptor 109B |
| NM_018993 | RIN2 | +3.77 | Ras and Rab interactor 2 |
| XM_353706 | LOC378204 | +3.74 | … |
| NM_017414 | USP18 | +3.74 | Ubiquitin-specific peptidase 18 |
| NM_015589 | SAMD4 | +3.71 | Sterile alpha motil domain containing 4 |
| NM_004241 | TRIP8 | +3.67 | Jumonji domain containing 1C |
| NM_006667 | PGRMC1 | +3.64 | Progesterone receptor membrane component 1 |
| NM_052942 | GBP5 | +3.63 | Guanylate binding protein 5 |
| NM_004863 | SPTLC2 | +3.63 | Serine palmitoyltransferase, long chain base subunit 2 |
| NM_153374 | MGC35274 | +3.62 | LysM, putative peptidoglycan binding, domain containing 2 |
| NM_021127 | PMAIP1 | +3.61 | Phorbol-12-myristate-13-acetate–induced protein 1 |
| NM_007268 | Z39IG | −15.82 | V-set and immunoglobulin domain containing 4 |
| NM_002417 | MKI67 | −9.91 | Antigen identified by monoclonal antibody Ki-67 |
| NM_005480 | TROAP | −8.27 | Trophinin associated protein (tastin) |
| NM_003088 | FSCN1 | −6.93 | Fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| NM_000478 | ALPL | −5.4 | Alkaline phosphatase, liver/bone/kidney |
| NM_002343 | LTF | −5.37 | Lactotransferrin |
| NM_015671 | DKFZP434J046 | −5.17 | Chromosome 19 open reading frame 14, homo sapiens WD repeat domain 62 (WDR62), mRNA |
| NM_005091 | PGLYRP1 | −4.79 | Peptidoglycan recognition protein 1 |
| NM_001925 | DEFA4 | −4.48 | Defensin, alpha 4, corticostatin |
| NM_005217 | DEFA3 | −4.28 | Defensin, alpha 3, neutrophil-specific |
| NM_004260 | RECQL4 | −4.04 | RecQ protein–like 4 |
| NM_021953 | FOXM1 | −3.77 | … |
| NM_002466 | MYBL2 | −3.73 | v-myb myeloblastosis viral oncogene homolog (avian)–like 2 |
| NM_002997 | SDC1 | −3.61 | Syndecan 1 |
Fold changes reflect greater (positive fold change) or lesser (negative fold change) abundance in acute samples from uncomplicated dengue patients.
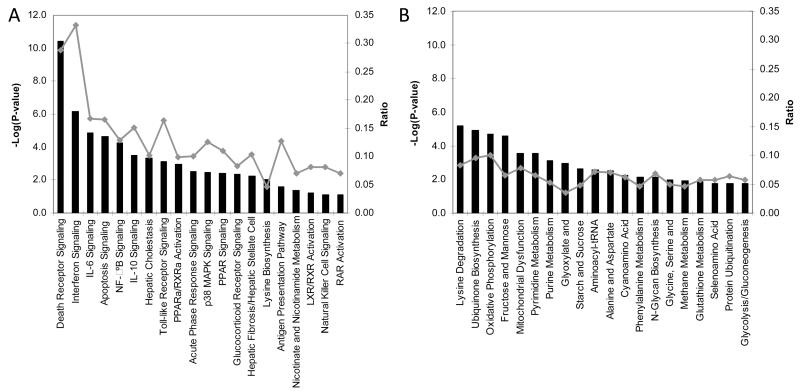
Unsupervised pathway analysis of transcripts significantly enriched in patients with uncomplicated dengue implicated the death receptor, IFN, apoptosis, IL-6, NF-κB, and IL-10 signaling pathways as being immune function–related processes distinguishing uncomplicated dengue from DSS (figure 4A). Unsupervised pathway analysis of transcripts less abundant in patients with uncomplicated dengue identified a diverse set of pathways (figure 4B), with the majority associated with cellular metabolic activities and not immune function. Collectively and in the context of the samples available, these data identify apoptotic and immune function–related molecular pathways as defining the transcriptional difference between these 2 groups of children with identical durations of illness but different clinical outcomes.
Figure 4. Pathway analysis of transcripts enriched or less abundant in acute samples from patients with uncomplicated dengue relative to that in acute samples from patients with dengue shock syndrome (DSS).
Shown are the top 20 canonical pathways identified by unsupervised pathway analysis of filtered microarray data representing transcripts significantly enriched (n = 1030) (A) or less abundant (n = 719) (B) in acute samples (day 4 of illness) from patients with uncomplicated dengue relative to that in acute samples from patients with DSS. The strength of the statistical association is indicated by the length of the bars. The ratio value reflects the proportion of gene elements on the enriched gene list that belong to one of these canonical pathways.
RT-PCR validation of gene transcript abundance
TaqMan RT-PCR assays were used to validate the relative abundance of transcripts identified as being differentially expressed between (1) acute versus convalescent samples from children with uncomplicated dengue and (2) acute samples from children with DSS versus those from children with uncomplicated dengue. The selection of gene transcripts to validate was influenced by 2 guiding themes: (1) they were biologically replicated in the array (i.e., they were relatively consistent between patient samples) and (2) genes related to immune function should be prominent, because these pathways distinguished uncomplicated dengue from DSS and, in general, a focus on the immune transcriptome would serve the wider purpose of informing models of immunopathogenesis. Validation of gene transcript abundance was performed with acute RNA samples from 8 patients with DSS (6 of whom were included in the microarray analysis) and 8 matched patients with uncomplicated dengue (6 of whom were included in the microarray analysis).
RT-PCR: acute versus convalescent samples among patients with uncomplicated dengue
Sixty-nine transcripts related to immune response were selected for validation by RT-PCR assays from among all transcripts significantly enriched in acute samples from patients with uncomplicated dengue relative to convalescent samples. Emphasis was placed on validating canonical ISGs, because these were highly enriched in the microarray (figure 1). Of the 69 transcripts selected, 64 (93%) were also differentially abundant when measured by RT-PCR, including those from 34 ISGs (figure 5A-5C). In addition to ISGs, transcripts more abundant in acute samples and relevant to pathogenesis included those from genes for complement factors (C2, C1qA, C1qB, C1qG, and C3aR), chemokines (CCL2, CCL8, and CXCL10), Toll-like receptor (TLR) 2 and 7, and cig5/viperin.
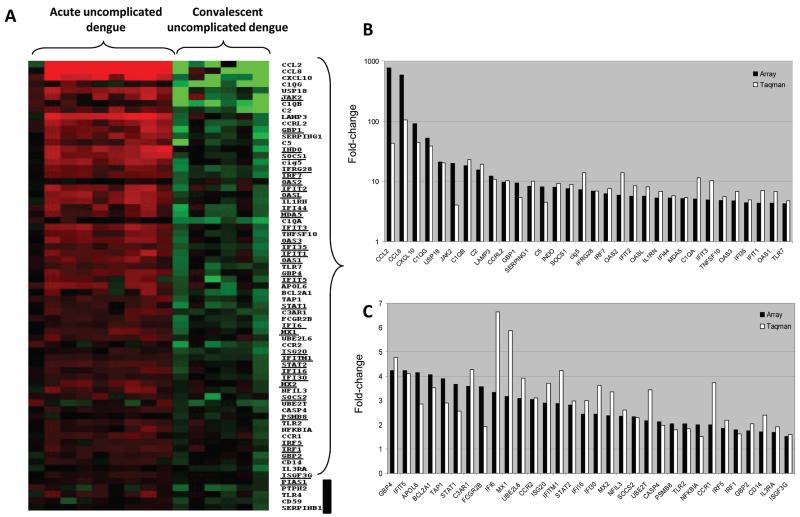
Figure 5. Reverse-transcription polymerase chain reaction (RT-PCR) validation of transcripts enriched in acute samples from patients with uncomplicated dengue relative to autologous convalescent samples.
Shown in panel A is a heat map of individual patient samples filtered on those transcripts enriched in acute samples (day 4 of illness) from patients with uncomplicated dengue relative to autologous convalescent samples (day 30) (n = 69) that were selected for RT-PCR validation. The gene symbols of canonical type I interferon–stimulated genes are underlined next to the heat map. Shown in panels B and C are graphs representing the mean fold difference in abundance for 64 (93%) of the 69 transcripts that were validated by RT-PCR. The fold difference is shown for both microarray analysis (black bars) and RT-PCR analysis (white bars), with values >1 indicating greater abundance in acute samples. Five of the 69 transcripts were not validated by RT-PCR (shown by the black vertical line next to the heat map).
RT-PCR: acute versus convalescent samples among patients with DSS
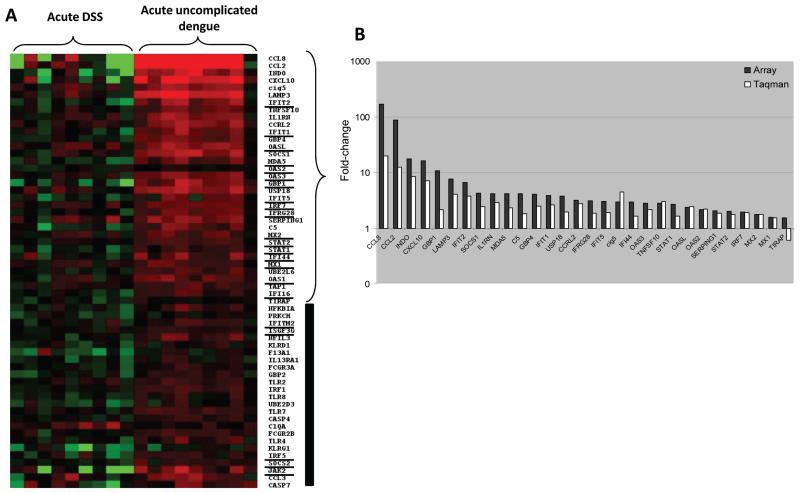
Nineteen transcripts related to immune response were selected for validation by RT-PCR assays from among transcripts differentially abundant in acute samples from patients with DSS relative to convalescent samples; of these, 11 transcripts were validated by RT-PCR (figure 6), with transcripts from the complement pathway being most prominent.
Figure 6. Reverse-transcription polymerase chain reaction (RT-PCR) validation of transcripts enriched or less abundant in acute samples from patients with dengue shock syndrome (DSS) relative to that in convalescent samples.
RT-PCR: acute uncomplicated dengue versus acute DSS
Fifty-nine transcripts related to immune response were selected for validation by RT-PCR assays from among transcripts significantly enriched in acute samples from patients with uncomplicated dengue relative to those from patients with DSS; this included 47 transcripts also investigated in the comparison of acute versus convalescent samples from patients with uncomplicated dengue (figure 5A-5C). Of these 59 transcripts, 30 (51%) were also differentially abundant when measured by RT-PCR (figure 7A and 7B), including those from 17 ISGs. In addition to ISGs, transcripts from genes for chemokines (CCL2, CCL8, and CXCL10) were prominently more abundant in acute samples from patients with uncomplicated dengue.
Figure 7. Reverse-transcription polymerase chain reaction (RT-PCR) validation of transcripts enriched in acute samples from patients with uncomplicated dengue relative to that in acute samples from patients with dengue shock syndrome (DSS).
Shown in panel A is a heat map of individual patient samples filtered on those transcripts (n = 59) enriched in acute samples from patients with uncomplicated dengue relative to that in acute samples from patients with DSS that were selected for RT-PCR validation. The gene symbols of canonical type I interferon–stimulated genes are underlined next to the heat map. Shown in panel B is a graph representing the mean fold difference in the abundance of 30 (51%) of the 59 transcripts that were validated by RT-PCR. The fold difference is shown for both microarray analysis (black bars) and RT-PCR analysis (white bars), with values >1 indicating greater abundance in acute samples. Twenty-nine of the 59 transcripts were not validated by RT-PCR (shown by the black vertical line next to the heat map).
Collectively, these RT-PCR data serve to validate one of the central findings of the microarray analysis: the prominence of IFN-related transcripts in the host-defense profile of the transcriptome of acute uncomplicated dengue versus convalescent samples and, conversely, their relative underabundance in children at the time of hypovolemic shock.
DISCUSSION
The present study attempted to better understand DSS by defining the whole-blood transcriptome at the time of hypovolemic shock and comparing it with that in autologous convalescence samples and to that in samples from well-matched children with acute uncomplicated dengue. A key and unexpected finding was the immunologically muted transcriptional profile in children with DSS relative to that in their own convalescent samples and to that in children with acute uncomplicated dengue but an identical duration of illness. In particular, transcripts belonging to pathways associated with apoptosis, cytokine signaling (IL-6 and IL-10), NF-κB, and IFN were less abundant in patients with DSS at the time of cardiovascular compromise, despite the presence of a measurable viremia. These data suggest that significant heterogeneity exists in the type or timing of host immune responses precipitated by DENV infection. Indeed, the present study suggests that, if whole-blood transcriptional events contribute to capillary leakage and DSS, then it occurs well before hypovolemic shock manifests.
Capillary leakage leading to DSS is the most common life-threatening complication of dengue in children living in settings of endemicity. DSS typically occurs around the time of defervescence and at a time when the DENV burden is in steep decline. Host immune responses, driven at least in part by overall viral antigenic mass, have been repeatedly nominated as being important in promoting clinically significant capillary leakage (reviewed in [25]). The rationale for the present study was to better understand the host response at the time of vascular decompensation in pediatric patients with dengue. The patients with DSS in this study were afebrile at the time of hypovolemic shock (day 4) and on average had significantly lower viremias than did the comparison group of children with uncomplicated dengue, who, in contrast, were all still febrile. In essence then, although matched by duration of illness, overall serological response, age, and sex, these 2 groups of children had contrasting disease evolution profiles when samples for microarray analysis were collected.
Consistent with the concept that the patients in this study had different disease evolution profiles, we observed starkly different whole-blood transcriptional profiles between afebrile children with DSS and febrile children with uncomplicated dengue. Transcripts associated with type I IFN—beginning with the dsRNA-sensing molecule melanoma differentiation–associated protein 5 and extending to canonical ISGs plus chemokines (CCL2, CCL8, and CXCL10), complement subunits/receptors, and transcription factors and TLRs—were significantly more abundant in samples from febrile children with uncomplicated dengue than in autologous convalescent samples. Several of the ISGs (OAS1, OAS2, OAS3, IRF7, SOCS1, STAT1, and viperin) identified in this study were also abundant in the transcriptome of febrile Singaporean adults with uncomplicated dengue [26] and in experimentally DENV-infected macaque monkeys [27]. These prominent genes and pathways may serve as candidate genes in future host genetic studies of dengue susceptibility. In contrast, the transcriptome of patients with DSS at the time of decompensation was, from an immune-defense perspective, highly muted and instead contained a greater abundance of transcripts from metabolic pathways than that in patients with uncomplicated dengue and the same duration of illness. A possible explanation for this muted, or even repressed, immune-response signature is that the host-defense transcriptional profile is observed well before the point of defervescence and cardiovascular decompensation in patients with DSS. In this scenario, a very high initial viral antigen burden drives a rapid and robust host-defense transcriptional response that wanes by the time of defervescence and cardiovascular decompensation as viral antigen is cleared by secondary humoral and cellular immune responses. Paradoxically, the rapidity and strength of this antigen-driven secondary response, along with other risk factors, may largely account for the extent of capillary leakage in children with secondary dengue [12].
A conceivable but perhaps less likely explanation for the missing host-defense profile at the time of decompensation in patients with DSS is that it is attenuated throughout the course of disease. This attenuated response might be related to intrinsic host-genetic influences on gene expression, or it might be a virus-mediated phenomenon—either could result in higher initial viral burdens in vivo. Relatedly, the capacity of DENV non-structural proteins to inhibit the activation of ISGs in vitro has been well documented [18, 19]. Finally, the absence of an inflammatory transcriptome at the time of cardiovascular decompensation may explain why adjuvant corticosteroid use in patients with established DSS has little efficacy [28]—there is essentially little in the way of an inflammatory transcriptional response to modulate.
This present study extends our previous observations in adults with DSS, albeit using a different chip technology [22]. In contrast to our previous study, the present study was more comprehensive in that it sampled RNA from pediatric patients matched by age, sex, and duration of illness and validated a wider panel of transcripts by RT-PCR. A limitation of our present study is the relatively small sample size and lack of samples collected very early during the evolution of disease. In addition, the relative lack of known pathways associated with the many transcripts more abundant in patients with DSS may also reflect our incomplete understanding of transcriptional networks. Future studies will focus on the very early host viral events in febrile children who subsequently progress to DSS. Ultimately, a better understanding of the molecular evolution of DSS could help identify novel biomarkers that have prognostic utility and form the rationale for new clinical interventions for this important disease.
Acknowledgments
We thank Ling Ling (Genome Institute, Singapore) for expert advice on microarray analysis. We also thank the doctors, nurses, and participating patients at the Hospital for Tropical Diseases for contributing to the study.
Financial support: Wellcome Trust (Vietnam; grant B9RJIX0); Singapore Agency for Science Technology and Research (Singapore).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Thein S, Aung MM, Shwe TN, et al. Risk factors in dengue shock syndrome. Am J Trop Med Hyg. 1997;56:566–72. doi: 10.4269/ajtmh.1997.56.566. [DOI] [PubMed] [Google Scholar]
- 2.Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–80. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 3.Sangkawibha N, Rojanasuphot S, Ahandrik S, et al. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–69. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 4.Halstead SB, Lan NT, Myint TT, et al. Dengue hemorrhagic fever in infants: research opportunities ignored. Emerg Infect Dis. 2002;8:1474–9. doi: 10.3201/eid0812.020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangada MM, Endy TP, Nisalak A, et al. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J Infect Dis. 2002;185:1697–703. doi: 10.1086/340822. [DOI] [PubMed] [Google Scholar]
- 6.Loke H, Bethell DB, Phuong CX, et al. Strong HLA class I–restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J Infect Dis. 2001;184:1369–73. doi: 10.1086/324320. [DOI] [PubMed] [Google Scholar]
- 7.Mongkolsapaya J, Dejnirattisai W, Xu XN, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–7. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 8.Loke H, Bethell D, Phuong CX, et al. Susceptibility to dengue hemorrhagic fever in Vietnam: evidence of an association with variation in the vitamin D receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67:102–6. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- 9.Stephens HA, Klaythong R, Sirikong M, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–18. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 10.Libraty DH, Endy TP, Houng HS, et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185:1213–21. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 11.Vaughn DW, Green S, Kalayanarooj S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 12.Green S, Vaughn DW, Kalayanarooj S, et al. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J Infect Dis. 1999;179:755–62. doi: 10.1086/314680. [DOI] [PubMed] [Google Scholar]
- 13.Juffrie M, Meer GM, Hack CE, et al. Inflammatory mediators in dengue virus infection in children: interleukin-6 and its relation to C-reactive protein and secretory phospholipase A2. Am J Trop Med Hyg. 2001;65:70–5. doi: 10.4269/ajtmh.2001.65.70. [DOI] [PubMed] [Google Scholar]
- 14.Azeredo EL, Zagne SM, Santiago MA, et al. Characterisation of lymphocyte response and cytokine patterns in patients with dengue fever. Immunobiology. 2001;204:494–507. doi: 10.1078/0171-2985-00058. [DOI] [PubMed] [Google Scholar]
- 15.Green S, Pichyangkul S, Vaughn DW, et al. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis. 1999;180:1429–35. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 16.Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J Virol. 2004;78:2701–10. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol. 2000;74:4957–66. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones M, Davidson A, Hibbert L, et al. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol. 2005;79:5414–20. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, et al. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol. 2005;79:8004–13. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahalingam S, Lidbury BA. Suppression of lipopolysaccharide-induced antiviral transcription factor (STAT-1 and NF-kappa B) complexes by antibody-dependent enhancement of macrophage infection by Ross River virus. Proc Natl Acad Sci USA. 2002;99:13819–24. doi: 10.1073/pnas.202415999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ubol S, Masrinoul P, Chaijaruwanich J, Kalayanarooj S, Charoensirisuthikul T, Kasisith J. Differences in global gene expression in peripheral blood mononuclear cells indicate a significant role of the innate responses in progression of dengue fever but not dengue hemorrhagic fever. J Infect Dis. 2008;197:1459–67. doi: 10.1086/587699. [DOI] [PubMed] [Google Scholar]
- 22.Simmons CP, Popper S, Dolocek C, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 25.Rothman AL. Dengue: defining protective versus pathologic immunity. J Clin Invest. 2004;113:946–51. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fink J, Gu F, Ling L, et al. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl Trop Dis. 2007;1:e86. doi: 10.1371/journal.pntd.0000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sariol CA, Munoz-Jordan JL, Abel K, et al. Transcriptional activation of interferon-stimulated genes but not of cytokine genes after primary infection of rhesus macaques with dengue virus type 1. Clin Vaccine Immunol. 2007;14:756–66. doi: 10.1128/CVI.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panpanich R, Sornchai P, Kanjanaratanakorn K. Corticosteroids for treating dengue shock syndrome. Cochrane Database Syst Rev. 2006;3:CD003488. doi: 10.1002/14651858.CD003488.pub2. [DOI] [PubMed] [Google Scholar]