Abstract
Long non-coding-RNAs are emerging as important regulators of cellular functions but little is known on their role in human immune system. Here we investigated long intergenic non-coding-RNAs (lincRNAs) in thirteen T and B lymphocyte subsets by RNA-seq analysis and de novo transcriptome reconstruction. Over five hundred new lincRNAs were identified and lincRNAs signatures were described. Expression of linc-MAF-4, a chromatin-associated TH1-specific lincRNA, was inversely correlated with MAF, a TH2-associated transcription factor. Linc-MAF-4 down-regulation skewed T cell differentiation toward TH2. We identified a long-distance interaction between linc-MAF-4 and MAF genomic regions, where linc-MAF-4 associates with LSD1 and EZH2, suggesting linc-MAF-4 regulated MAF transcription by recruitment of chromatin modifiers. Our results demonstrate a key role of lincRNAs in T lymphocyte differentiation.
Introduction
Lymphocytes enable us to fight and survive infections, but are also major drivers of immune-mediated diseases, such as allergy and autoimmunity. These different type of immune responses are mostly coordinated by distinct CD4+ T cell subsets through signals delivered both by cytokines and by cell-to-cell contacts1. Development and differentiation programs of CD4+ T lymphocytes subsets with distinct effector functions have been extensively studied in terms of signaling pathways and transcriptional networks, and a certain degree of functional plasticity between different subsets has been recently established2. Indeed, CD4+ T cell subset flexibility in the expression of genes coding for cytokines and transcription factors allows the immune system to dynamically adapt to the many challenges it faces3. As CD4+ T lymphocyte subsets are no longer considered stable and terminally differentiated cell lineages, the question arises as to how lymphocyte phenotype and functions can be modulated and whether these new findings offer new therapeutic opportunities.
Besides the well-established role of transcription factors as instructive signals for cell differentiation toward a given lineage, other cues, such as epigenetic modifications, can regulate maintenance of cellular states4. In this context non-coding RNAs (ncRNAs) are emerging as a new regulatory layer impacting on both the development and the functioning of the immune system5, 6. Among the several classes of ncRNAs that play a specific role in lymphocyte biology, microRNAs are the best characterized7-11. Although thousands of long intergenic non-coding RNAs (lincRNAs) have been identified in the mammalian genome by bioinformatics analyses of transcriptomic data12-14, their functional characterization is still largely incomplete. The functional studies performed to date have shown that lincRNAs contribute to the control of cell differentiation and to the maintenance of cell identity through different modes of action15. Nuclear lincRNAs act mainly through their association with chromatin-modifying complexes16-18. Whereas, cytoplasmic lincRNAs can modulate translational control19 and transcript stability20 directly by base pairing with specific targets or indirectly as competing endogenous RNAs21-23. Few examples of functional lincRNAs have been recently described in the mouse immune system. A broad analysis performed by interrogating naïve and memory CD8+ cells purified from mouse spleen with a custom array of lincRNAs reported the identification of 96 lymphoid-specific lincRNAs and suggested a role for lincRNAs in lymphocyte differentiation and activation24. The lincRNA NeST has been found to be downregulated during lymphocyte activation in a reciprocal manner to expression of interferon-γ (IFN-γ) and to control susceptibility to Theiler’s virus and Salmonella infection in mice through epigenetic regulation of the Ifng locus25, 26. More recently, mouse lincRNA-Cox2 has been reported to be induced downstream Toll-like receptor signaling and to mediate the activation and repression of distinct sets of immune target genes involved in inflammatory responses27. Another study on mouse thymocytes and mature peripheral T cells allowed the identification of lincRNAs with specific cell expression pattern during T cell differentiation and of a CD4+ TH2 specific lincRNA - LincR-Ccr2-5′AS - involved in the regulation of CD4+ TH2 lymphocytes migration28. Although these studies highlight the relevance of lincRNAs in regulating immune responses, a thorough analysis of their expression profile and functional role in the human immune system is still lacking.
The present study is based on a RNA-seq analysis of thirteen highly purified primary human lymphocytes subsets. We performed a de novo transcriptome reconstruction, and discovered over five hundred new long intergenic non-coding RNAs (lincRNAs). We identified several lymphocyte subset-specific lincRNAs signatures, and found that linc-MAF-4, a chromatin associated CD4+ TH1 specific lincRNA, correlates inversely with the transcription factor MAF and that its down-regulation skews CD4+ T cell differentiation toward TH2 phenotype. We provide the first comprehensive inventory of human lymphocytes lincRNAs and demonstrate that lincRNAs can be key to lymphocyte differentiation. This resource will likely help a better definition of lincRNAs role in lymphocytes differentiation, plasticity and effector functions.
Results
LincRNAs discriminate human lymphocyte subsets
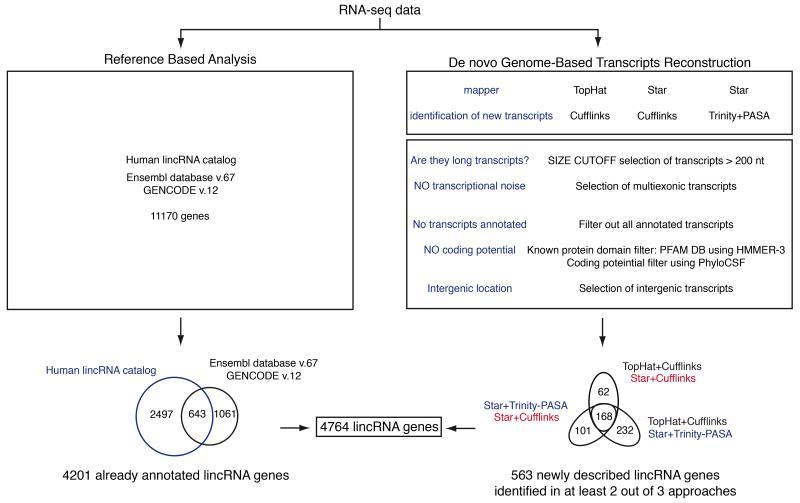
To assess lincRNA expression in human primary lymphocytes, RNA was extracted from thirteen lymphocyte cell subsets (Table 1) purified from peripheral blood mononuclear cells (PBMCs) of five healthy donors11. The polyadenylated RNA fraction was then analyzed by paired-end RNA sequencing obtaining about 1.7 billion mapped reads. To enrich for transcripts deriving from “bona fide” active genes, we applied an expression threshold (“0.21” fragments per kilobases of exons per million fragments mapped, FPKM) defined through the integration of RNAseq and chromatin state ENCODE project data29. We found a total of 31,902 expressed genes (including both protein coding and non-coding genes) in the 13 subsets (Table 1 and Supplementary Fig. 1a), of which 4,201 were lincRNAs annotated in public resources12, 30 (Fig. 1). To identify novel lincRNAs expressed in primary human lymphocytes, we used three de novo transcriptome reconstruction strategies that are based on the combination of two different sequence mappers, TopHat and Star31, 32, with two different tools for de novo transcripts assembly, Cufflinks and Trinity33, 34. LincRNAs were identified within the newly described transcripts exploiting the following process. We selected transcripts that were longer than 200 nucleotides and multiexonic, which did not overlap with protein coding genes (thus counting out unreliable single-exon fragments assembled from RNA-seq). Transcripts that contain a conserved protein-coding region and transcripts with ORFs that contain protein domains catalogued in Pfam protein family database35 were excluded. We utilized PhyloCSF, a comparative genomics method that assesses multispecies nucleotide sequence alignment based on a formal statistical comparison of phylogenetic codon models36, which efficiently identifies non-coding RNAs as demonstrated by ribosome profiling experiments37. Finally, we defined a stringent de novo lincRNA set including those genes for which at least one lincRNA isoform was reconstructed by two assemblers out of three. Through this conservatively multi-layered analysis we identified 563 novel lincRNAs genes, increasing by 11.8% the number of lincRNAs known to be expressed in human lymphocytes.
Table 1. Purification and RNA-sequencing of human primary lymphocyte subsets.
Purity achieved (mean ± SD) by sorting 13 human lymphocyte subsets (isolated from peripheral blood lymphocytes) by various surface marker combinations (sorting phenotype) and number of expressed genes (FPKM> 0.21). Cells were sorted from 4-5 different individuals for each lymphocyte subset and RNA sequencing carried out for each sample separately.
| Subset | Purity (%) | Sorting phenotype | Genes |
|---|---|---|---|
| CD4+ naïve | 99.8 ± 0.1 | CD4+ CCR7+ CD45RA+ CD45RO− | 20061 |
| CD4+ TH1 | 99.9 ± 0.05 | CD4+ CXCR3+ | 20855 |
| CD4+ TH2 | 99.7 ± 0.3 | CD4+ CRTH2+ CXCR3− | 19623 |
| CD4+ TH17 | 99.1 ± 1 | CD4+ CCR6+ CD161+ CXCR3− | 20959 |
| CD4+ Treg | 99.0 ± 0.8 | CD4+ CD127− CD25+ | 21435 |
| CD4+ TCM | 98.4 ± 2.8 | CD4+ CCR7+ CD45RA− CD45RO+ | 20600 |
| CD4+ TEM | 95.4 ± 5.5 | CD4+ CCR7− CD45RA− CD45RO+ | 19800 |
| CD8+ TCM | 98.3 ± 0.8 | CD8+ CCR7+ CD45RA− CD45RO+ | 20901 |
| CD8+ TEM | 96.8 ± 0.9 | CD8+ CCR7− CD45RA− CD45RO+ | 21813 |
| CD8+ naïve | 99.3 ± 0.2 | CD8+ CCR7+ CD45RA+ CD45RO− | 20611 |
| B naïve | 99.9 ± 0.1 | CD19+ CD5− CD27− | 21692 |
| B memory | 99.1 ± 0.8 | CD19+ CD5− CD27+ | 21239 |
| B CD5+ | 99.1 ± 0.8 | CD19+ CD5+ | 22499 |
Figure 1. Identification of lincRNAs expressed in human lymphocyte subsets.
RNA-seq data generated from 63 lymphocyte samples were processed according to two different strategies: quantification of lincRNAs already annotated in public resources and de novo Genome Based Transcripts Reconstruction for the quantification of new lincRNAs expressed in human lymphocytes. Three methods for the identification of new transcripts were adopted: Reference Annotation Based assembly by Cufflinks with two different aligners (TopHat and STAR) and an approach that integrates Trinity and PASA software. Only transcripts reconstructed by at least two assemblers were considered. Novel transcripts were filtered with a computational analysis pipeline to select for lincRNAs. The number of lincRNA genes and transcripts identified in lymphocytes subsets is indicated.
The different classes of RNAs were evenly distributed among different lymphocytes subsets (Supplementary Fig. 1b) and the ratio of already annotated and newly identified lincRNAs was similar across different chromosomes (Supplementary Fig. 1c) and across various lymphocyte subsets (Supplementary Fig. 1d). As previously observed in different cell types12, 33, also in human lymphocytes lincRNAs were generally expressed at lower abundance than protein coding genes (Supplementary Fig. 1e). However, when transcripts were divided based on their expression in cell-specific and non specific (Supplementary Fig. 1f), we found that cell specific lincRNAs and cell specific protein coding genes, displayed similar expression levels (Supplementary Fig. 1e-g).
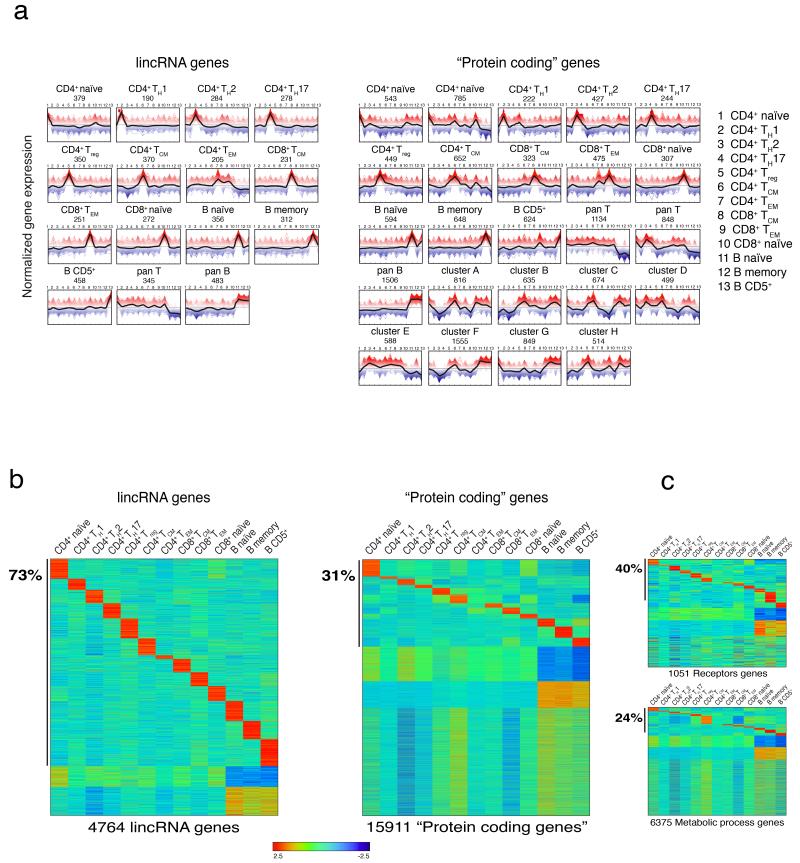
Lymphocytes subsets display very different migratory abilities and effector functions, yet they are very closely related from the differentiation point of view. As lincRNAs are generally more tissue specific than protein coding genes12, 38, we assessed the lymphocyte cell-subset specificity of lincRNAs. We therefore classified genes according to their expression profiles by unsupervised K-means clustering and found that lincRNAs were defined by 15 clusters and protein-coding genes by 24 clusters (Fig. 2a and Supplementary Fig. 2a). Remarkably, the percentage of genes assigned to the clusters specific for the different lymphocyte subsets was higher for lincRNAs (71%) than for protein-coding genes (34%) (Fig. 2b). This superiority stands out even when lincRNAs were compared with membrane receptor coding genes (40%) (Fig. 2c), which are generally considered the most accurate markers of different lymphocyte subsets. Similar results were obtained also using the heuristic expression threshold of FPKM>1 (Supplementary Fig. 2b). Altogether, based on RNA-seq analyses of highly purified primary T and B lymphocyte subsets, we provide a comprehensive landscape of lincRNAs expression in human lymphocytes. Exploiting a de novo transcriptome reconstruction we discovered 563 new lincRNAs, and found that lincRNAs are very effective in marking lymphocyte cell identity.
Figure 2. Definition of transcript clusters in human lymphocytes.
(a) Expression profiles of lincRNA and protein coding genes across 13 human lymphocyte subsets according to K-Means clusters definition. The black line represents the mean expression of the genes belonging to the same cluster. The peaks of expression profiles refer to the populations reported in legend according to numbering. (b) Specificity of lincRNAs and protein coding genes. Rows and columns are ordered based on a K-Means clustering of lincRNAs and protein coding genes across 13 human lymphocyte populations. Color intensity represents the Z-score log2-normalized raw FPKM counts estimated by Cufflinks. 79% of lincRNAs genes and 39% of protein coding genes are assigned to specific clusters. See also Supplementary Fig. 2a. (c) As in (b), performed on receptors and metabolic processes genes.
Identification of lincRNA signatures in lymphocytes
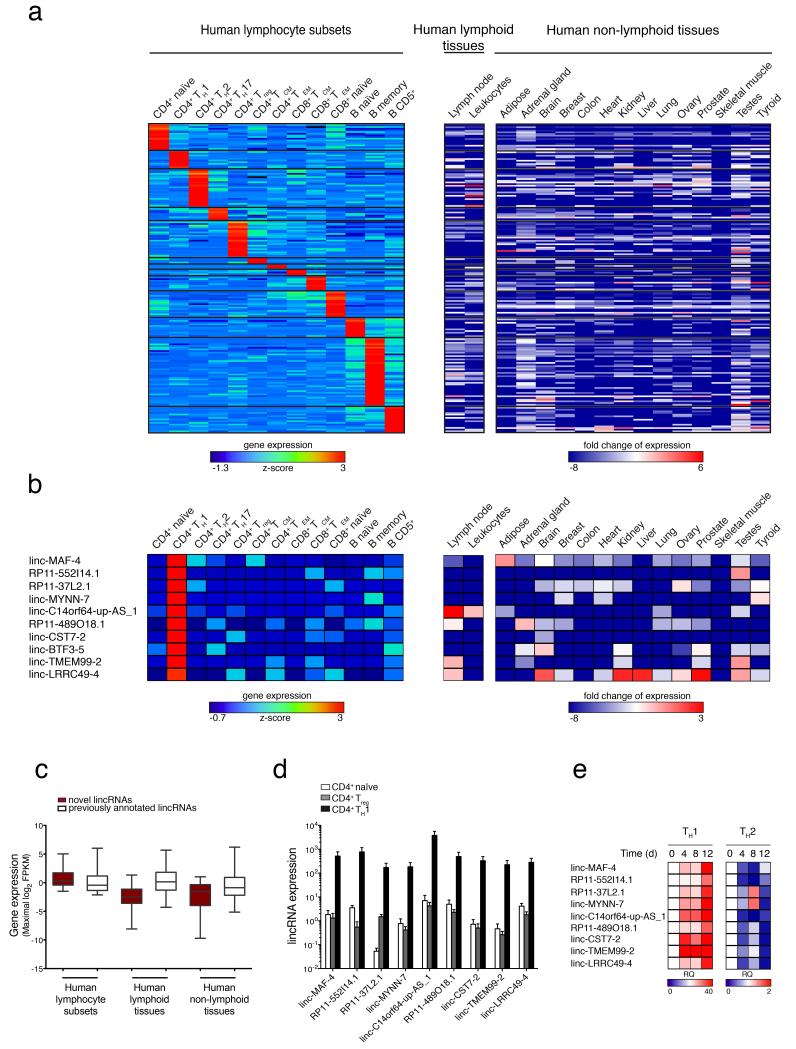
Next, we interrogated our dataset for the presence of lincRNAs signatures in the different lymphocyte subsets. We therefore looked for lincRNAs differentially expressed (P < 0.05; non-parametric Kruskal-Wallis test) that had more than 2.5-fold expression difference in a given cell subset compared to all the other subsets and that were expressed in at least 3 out of 5 individuals and found 172 lincRNAs that met these criteria (Fig. 3a and Supplementary Table 3). We integrated the human transcriptome database with our newly identified transcripts and thus created a new reference to assess more thoroughly expression of new transcripts, in other human tissues. Looking at lincRNAs signatures in a panel of sixteen human tissues (Human BodyMap 2.0 project) we found that lymphocytes signature lincRNAs were not only very poorly expressed in non-lymphoid tissues (Fig. 3a), but also that most signature lincRNAs were not detectable even in lymphoid tissues. These findings underscore the importance of assessing expression of lincRNAs (as well as of any highly cell-specific transcripts) in purified primary cells rather than in total tissues where a given cell subset-specific transcript is diluted by the transcripts of all the other cell types of the tissue. It is important to note that, the newly identified lincRNAs defined as signatures were more abundant (Fig. 3c) and more cell-specific (Supplementary Table 3) than the already annotated lincRNAs defined as signatures. Representative data obtained from the CD4+ TH1 cell subset are depicted in Fig. 2b; similar results were obtained for all the other subsets (Supplementary Table 3).
Figure 3. LincRNA signatures of human lymphocyte subsets.
(a) Heatmap of normalized expression values of lymphocytes signature lincRNAs selected on the basis of fold change (>2.5 with respect to all the other subsets), intrapopulation consistency (expressed in at least 3 out of 5 samples) and non parametric Kruskal-Wallis test (P < 0.05). Signature lincRNAs relative expression values were calculated as log2 ratios between lymphocyte subsets and a panel of human lymphoid and non-lymphoid tissues of the Human BodyMap 2.0 project (See also Supplementary Table 3). (b) CD4+ TH1 signature lincRNAs extracted from panel (a). The barcode on the left indicates already annotated lincRNAs (white) and novel lincRNAs (brick red). For novel lincRNAs name, ‘S’ and ‘AS’ indicates ‘sense’ and ‘antisense’ respectively. (c) Average expression values of previously annotated (white) and novel (brick red) lincRNAs in human lymphocyte subsets and lymphoid or non-lymphoid human tissues. (d) Validation of TH1 signature lincRNAs expression by RT-qPCR on primary CD4+ naïve, TH1 and Treg cells sorted from PBMC of healthy donors (average of three independent experiments ± SEM). (e) RT-qPCR analysis of TH1 signature lincRNAs expression in a time course of CD4+ naïve T cells differentiated in TH1 and TH2 polarizing conditions presented as relative quantity (RQ) relative to time zero (average of two independent experiments).
Finally, to confirm and extend our signature data, we assessed the expression of CD4+ TH1 lincRNAs by RT-qPCR in a new set of independent samples of primary human CD4+ naïve, regulatory T (Treg) and TH1 cells, as well as in naïve CD4+ T cells that were activated in vitro and induced to differentiate toward TH1 or TH2 cells. Specific subset expression was confirmed for 90% of the CD4+ TH1 signature lincRNAs (Fig. 3d). Moreover, 90% of CD4+ TH1 signature lincRNAs thatwere expressed in resting CD4+ TH1 cells purified ex vivo, were also highly expressed in naïve CD4+ T cells differentiated under TH1 polarizing conditions in vitro, whereas they were poorly expressed in naïve CD4+ T cells that were differentiated towards TH2 in vitro (Fig. 3e). As a corollary to these findings, we observed by RNA-seq that CD4+ naïve signature lincRNAs were mostly down-regulated during differentiation towards TH0 cells in vitro, when TH1, TH2 and TH17 signature lincRNAs were mostly up-regulated (Supplementary Fig. 3a). Taken together our data demonstrate that lincRNAs provide excellent signatures of human lymphocyte subsets, and suggest that human CD4+ T lymphocytes acquire most of their memory specific lincRNAs signatures during their activation-driven differentiation from naïve to memory cells.
Linc-MAF-4 downregulation skews CD4+ T cells towards TH2
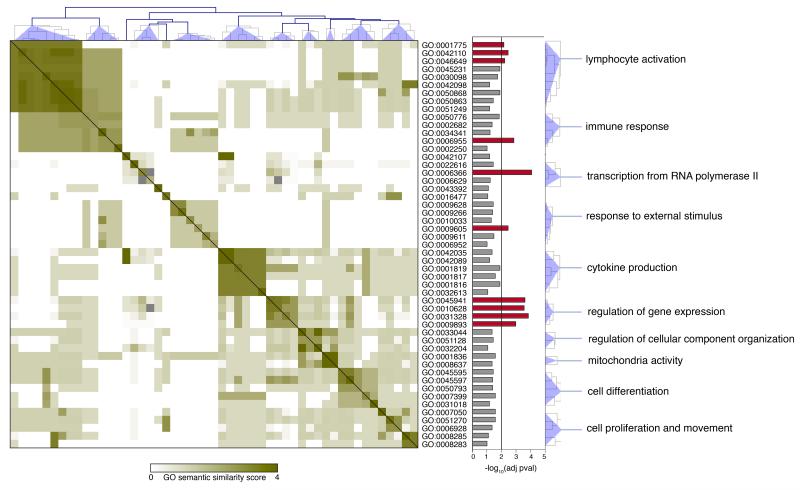
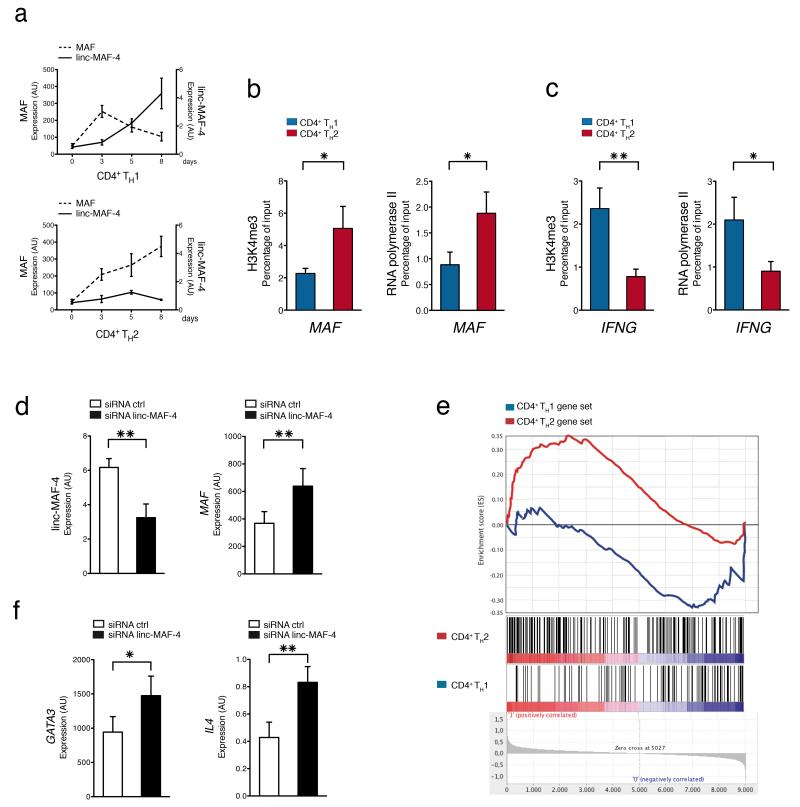
As lincRNAs have been reported to influence the expression of neighboring genes25, 26, 28, 39, we asked whether protein-coding genes proximal to lymphocytes signature lincRNAs were involved in key cell-functions. To this purpose we used the FatiGO tool from the Babelomics suite for functional enrichment analysis40 and found that protein-coding genes neighboring to signature lincRNAs were enriched for Gene Ontology terms strongly correlated with lymphocyte T cell activation (Fig. 4), pointing to a possible role of signature lincRNAs in important lymphocyte functions. To obtain proof of concept of this hypothesis, we chose to characterize in depth linc-MAF-4 (also referred to as linc-MAF-2 in LNCipedia database http://www.lncipedia.org41), a TH1 signature lincRNA, localized 139.5 kb upstream of the MAF gene. MAF encodes a transcription factor involved in TH2 differentiation42, which is also required for the efficient development of TH17 cells43 and controls IL4 transcription in CD4+ T follicular helper cells44. Our sequencing data showed that high expression of linc-MAF-4 correlated with a low amount of MAF transcript in CD4+ TH1 cells, conversely TH2 cells had low expression of linc-MAF-4 and abundant MAF transcripts. The anti-correlation of expression between lincRNAs and their neighboring genes is not a common feature of all lincRNAs12, 16, and it is probably restricted to a limited number of cis-acting lincRNAs. This observation was confirmed also in our dataset (data not shown). Moreover, no correlation was observed between the expression linc-MAF-4 and its proximal upstream protein coding genes: CDYL2 and DYNLRB2 (Supplementary Fig. 4a). A similar inverse relation between linc-MAF-4 and MAF was observed when naïve CD4+ T cells were differentiated in vitro towards TH1 or TH2 cells. In T lymphocytes differentiating towards TH1 cells, MAF transcript increased up to day 3 and then decreased thereafter (Fig. 5a). Conversely, linc-MAF-4 was poorly expressed for the first three days but then increased progressively. In CD4+ T lymphocytes differentiating towards TH2 cells, we found the opposite situation, both MAF transcript and protein abundance increased constantly up to day 8 while Iinc-MAF4 remained constantly low (Fig. 5a and Supplementary Fig. 4c), similarly to what was observed in CD4+ T lymphocytes differentiating towards TH17 cells (Supplementary Fig. 4d).
Figure 4. Gene Ontology semantic similarity matrix “protein coding” genes proximal to lincRNA signatures.
The semantic similarity scores for all GO term pairs were clustered using hierarchical clustering method. On the right of the matrix a bar plot of the adjusted p-values for each GO term is reported. Red bars represent GO terms that are significantly enriched in Gene Ontology analysis. Common ancestor is reported for each cluster.
Figure 5. Linc-MAF-4 contributes to TH1 cell differentiation.
(a) Expression of linc-MAF-4 and MAF assessed at different time points by RT-qPCR in activated CD4+ naïve T cells differentiated in TH1 or TH2 polarizing conditions (average of four technical replicates ± SEM). See also Supplementary Fig. 4 b,c. (b) ChIP-qPCR analysis of H3K4me3 and RNA polymerase II occupancy at MAF locus in CD4+ naïve T cells differentiated in TH1 or TH2 polarizing conditions at day 8 post-activation. Enrichment is a percentage of input (average of at least 5 independent experiments ± SEM). One-tailed t-test *P < 0.05. (c) As in (b) at IFNG locus as control (average of at least 10 independent experiments ± SEM). One-tailed t-test *P < 0.05; **P < 0.01. (d) Linc-MAF-4 and MAF expression determined by RT-qPCR in activated CD4+ naïve T cells (in the absence of polarizing cytokines) and transfected at the same time with linc-MAF-4 siRNA (black) or ctrl siRNA (white). Transcripts expression was detected 72 h post transfection (average of six independent experiments ± SEM). One-tailed t-test **P < 0.01; *P < 0.05. (e) Results of GSEA (Gene Set Enrichment Analysis) performed on gene expression data obtained from siRNA mediated knock-down of linc-MAF-4 in activated CD4 naïve T cells. Activation and transfection conditions were as in (d). The red and blue line represent the observed enrichment score profile of genes in the linc-MAF-4 / ctrl siRNA treated cells compared to the CD4 TH1 and TH2 reference gene sets respectively (average of four independent experiments). Nominal P < 0.05. (f) GATA3 and IL4 transcript expression determined by RT-qPCR in activated CD4+ naïve T cells transfected with linc-MAF-4 siRNA (black) or ctrl siRNA (white) (average of six independent experiments ± SEM). One-tailed t-test ** P < 0.01; * P < 0.05.
We further characterized MAF transcriptional regulation by looking at histone H3 lysine 4 tri-methylation (H3K4me3) abundance and RNA polymerase II occupancy at MAF promoter region in TH1 and TH2 cells. Consistent with a higher active transcription of MAF in CD4+ TH2 cells, we found that H3K4me3 content in TH2 cells was greater than in TH1 cells and that RNA polymerase II binding at MAF promoter was higher in TH2 than in TH1 cells (Fig. 5b). Intriguingly, linc-MAF-4 knock-down in activated CD4+ naïve T cells led to increased MAF expression (Fig. 5d and Supplementary Fig. 4e). All the above results indicate that modulation of MAF transcription in T cells depends on tuning of its promoter setting, and suggest a direct involvement of linc-MAF-4 in the regulation of MAF transcription.
We then assessed the overall impact of linc-MAF-4 knock-down on CD4+ T cell differentiation by performing transcriptome profiling and Gene Set Enrichment Analysis (GSEA). We defined as reference Gene-Sets the genes upregulated in CD4+ naïve T cells differentiated in vitro towards TH1 or TH2 types (Supplementary Table 1). We found that the CD4+ TH2 gene set was enriched for genes that were overexpressed in linc-MAF-4 knock-down cells, whereas the CD4+ TH1 gene set was depleted of these same genes (Fig. 5e). Concordant with these findings, the expression of GATA3 and IL4, two genes characteristic of TH2 cells, was increased after linc-MAF-4 knock-down (Fig. 5f and Supplementary Fig. 4f). Taken together these results demonstrate that linc-MAF-4 down regulation contributes to the skewing of CD4+ T cells differentiation towards TH2.
Epigenetic regulation of MAF transcription by linc-MAF-4
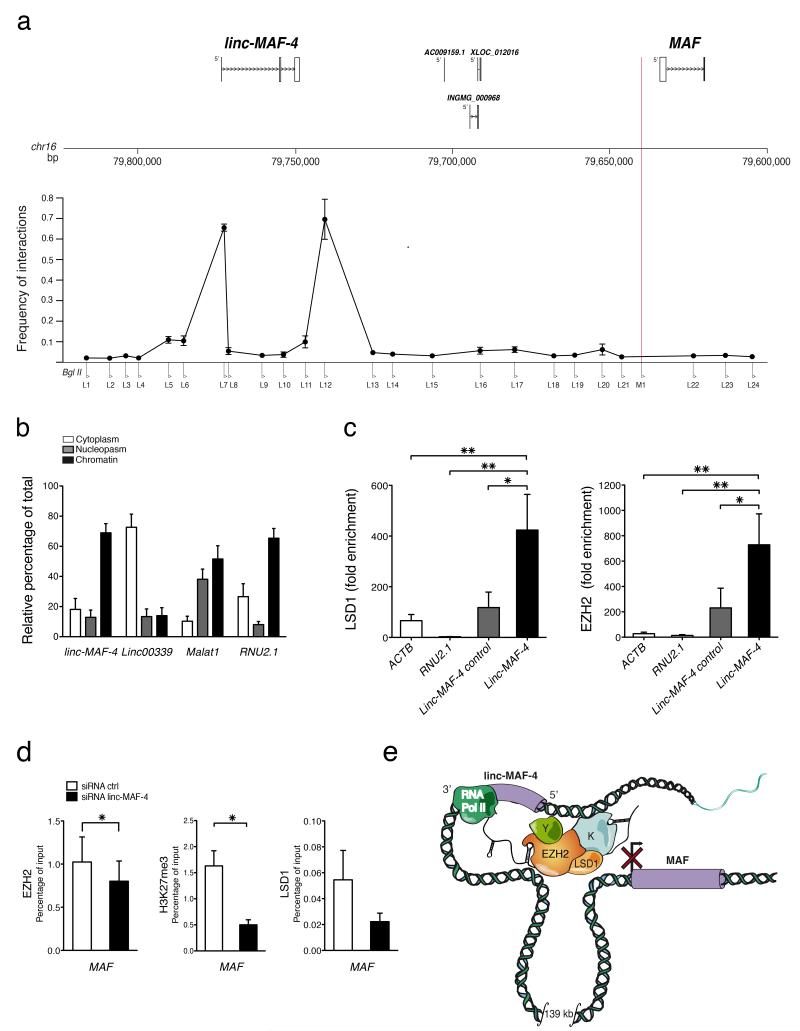
Since linc-MAF-4 gene maps in relative proximity (139.5 kb) to MAF gene we asked whether linc-MAF-4 can down-regulate MAF transcription, and, we investigated whether their genomic regions could physically interact. Chromosome conformation capture (3C) analysis was exploited to determine relative crosslinking frequencies among regions of interest. We tested the conformation of the linc-MAF-4 – MAF genomic region in differentiated CD4+ TH1 cells. A common reverse primer mapping within the MAF promoter region, was used in combination with a set of primers spanning the locus, and interactions were analyzed by PCR. Specific interactions between MAF promoter and 5′ and 3′ end regions of linc-MAF-4 were detected (Fig. 6a and Supplementary Fig. 5a,b), indicating the existence of an in cis chromatin looping conformation that brings linc-MAF-4 in close proximity to MAF promoter. Interestingly, the subcellular fractionation of in vitro differentiated CD4+ TH1 lymphocytes revealed a strong enrichment of linc-MAF-4 in the chromatin fraction (Fig. 6b). Because other chromatin-associated lincRNAs regulate neighboring genes by recruiting specific chromatin remodelers, we tested in RNA immunoprecipitation (RIP) assays the interaction of linc-MAF-4 with different chromatin modifiers, including activators and repressors (data not shown), and found a specific enrichment of linc-MAF-4 in the immunoprecipitates of two repressors, EZH2 and LSD1 (Fig. 6c and Supplementary Fig. 5c). In agreement with these findings, we found that linc-MAF-4 knock-down in activated CD4+ naïve T cells reduced both EZH2 and LSD1 abundance and correlated with the reduction of EZH2 enzymatic activity at MAF promoter as demonstrated by the H3K27me3 reduction at this locus (Fig. 6d). Remarkably, H3K27me3 content was not reduced at either the MYOD1 promoter region (a known target of EZH2) or at a region within the chromatin loop between linc-MAF-4 and MAF marked by H3K27me3 (Supplementary Fig. 5d). Altogether, these results demonstrate that there is a long distance interaction between linc-MAF-4 and MAF genomic regions, through which linc-MAF-4 could act as a scaffold to recruit both EZH2 and LSD1 and modulate the enzymatic activity of EZH2 on MAF promoter, thus regulating its transcription (Fig. 6e).
Figure 6. Epigenetic characterization of linc-MAF4/MAF genomic locus.
(a) Schematic representation of the genomic region analyzed by 3C. Position relative to linc-MAF-4 and MAF of three lincRNAs present in the region is shown in the upper part of the panel. The M1 primer at the 5′ end of MAF (red line) was used as bait. Primers (L1-L24) spanning the region between linc-MAF-4 and MAF were tested for interaction. Relative frequency of interaction between MAF and linc-MAF-4 5 ′ (L7) and 3 ′ (L12) ends is shown in CD4+ naïve T cells differentiated in TH1 polarizing conditions (day 8) (average of three independent experiments ± SEM). (b) Relative abundance of linc-MAF-4 transcript in cytoplasm, nucleus and chromatin in CD4+ naïve T cells differentiated in TH1 polarizing conditions (day 8). Linc-00339, MALAT1 and RNU2.1 were used respectively as cytoplasmic, nuclear and chromatin-associated controls (average of three independent experiments ± SEM). (c) RIP assay for LSD1 and EZH2 in CD4+ naïve T cells differentiated in TH1 polarizing conditions (day 8). Fold enrichment is relative to mock. ACTB, RNU2.1 and a region upstream the TSS of linc-MAF-4 were chosen as controls (average of six independent experiments ± SEM). The statistical significance was determined with ANOVA and Dunnet post-hoc test: *P < 0.05; **P < 0.01. (d) ChIP-qPCR analysis of EZH2, H3K27me3 and LSD1 occupancy at MAF locus in activated CD4+ naïve T cells transfected with linc-MAF-4 siRNA (black) or ctrl siRNA (white) (average of at least three independent experiments ± SEM). One-tailed t-test * P < 0.05. (e) Model for linc-MAF-4-mediated MAF repression in TH1 lymphocytes. When linc-MAF-4 is expressed, it recruits chromatin remodelers (i.e. LSD1 and EZH2) at MAF 5′-end, taking advantage of a DNA loop that brings linc-MAF-4 5′ and 3′ end in close proximity to MAF 5′ end.
Discussion
Mammalian genomes encode more long non-coding RNAs than previously thought16, 45 and the number of lincRNAs playing a role in cellular processes steadily grows. As there are relatively few examples of functional long non-coding RNAs in the immune system24-28, with the present study we depict a comprehensive landscape of lincRNAs expression in thirteen subsets of human primary lymphocytes. Moreover, we identified a lincRNA (linc-MAF-4) that appear to play a key role in CD4+ T helper cell differentiation.
LincRNAs have been reported to have high tissue specificity12 and our study of lincRNAs expression in highly pure primary human lymphocyte provides an added value because it allows the identification of lincRNAs whose expression is restricted to a given lymphocyte cell subset. Interestingly, we found that lincRNAs define the cellular identity better than protein coding genes, including those that encode surface receptor coding genes that are generally considered the most precise markers of lymphocytes subsets. Due to their specificity of expression, human lymphocytes lincRNAs that are not yet annotated in public resources would have not been identified without performing de novo transcriptome reconstruction. Indeed by exploiting three different de novo strategies we identified 563 novel lincRNAs and increased by 11.8% the number of lincRNAs expressed in human lymphocytes. As our conservative analysis was limited to thirteen cellular subsets, one may wonder how many novel lincRNAs could be identified by transcriptome analysis of all of the several hundreds human cell types.
We compared our data with previous analyses of lincRNAs expression in mouse immune system28 exploiting the LNCipedia database (http://www.lncipedia.org) 41. We found that 51% of the human lincRNA signatures are conserved in mouse, which is similar to the overall conservation between human and mouse lincRNAs (60%). However further studies will be necessary to asses that also their function is conserved.
Based on our findings, signature lincRNAs might be exploited to discriminate and differentiate at the molecular level those cell subsets that cannot be distinguished easily based on cell surface markers because of their cellular heterogeneity, such as CD4+ Treg cells. However, as lincRNAs expression in a tissue is averaged across all the cell types composing that tissue, a transcriptome analysis of unfractionated tissue-derived cells may underestimate the expression of cell specific lincRNAs. In fact, the great majority of our lymphocyte lincRNA signatures cannot be detected in RNAs extracted from total lymphoid tissues (peripheral blood and lymph nodes), although these same tissues contain cells from all of the lymphocytes subsets we assessed.
The lincRNAs role in differentiation has been described in different cell types17, 20, 23, 46, 47. In the mouse immune system it has been found that lincRNAs expression changes during naïve to memory CD8+ T cell differentiation24 and during naïve CD4+ T cells differentiation into distinct helper T cell lineages28. We show in human primary lymphocytes that activation-induced differentiation of CD4+ naïve T cells was associated with increased expression of lincRNAs belonging to the CD4+ TH1 signature, suggesting that upregulation of TH1 lincRNAs is part of the cell differentiation transcriptional program. Indeed, linc-MAF-4, one of the TH1 signature lincRNA, was poorly expressed in TH2 cells and its experimental downregulation skewed differentiating T helper cells toward a TH2 transcription profile. We found that linc-MAF-4 regulated transcription by exploiting a chromatin loop that brings its genomic region close to the promoter of MAF gene. We propose that the chromatin organization of this region allows linc-MAF-4 transcript to recruit both EZH2 and LSD1 and modulate the enzymatic activity of EZH2 negatively regulating MAF transcription with a mechanism of action similar to that shown for the lincRNAs HOTAIR48 and MEG349. We therefore provide a mechanistic proof of concept that lincRNAs can be important regulators of CD4+ T cell differentiation. Given the number of specific lincRNAs expressed in the different lymphocytes subsets, it can be postulated that many other lincRNAs might contribute to cell differentiation and to the definition of cell identity in human lymphocytes. These findings and the high cell specificity of lincRNAs suggest lincRNAs as novel and highly specific molecular targets for the development of new therapies for diseases (such as autoimmunity, allergy and cancer) in which altered CD4+ T cell functions play a pathogenic role.
ONLINE METHODS
Purification of primary immunological cell subsets
Blood buffy coat cells of healthy donors were obtained from Fondazione I.R.C.C.S. Ca’Granda Ospedale Maggiore Policlinico in Milan and peripheral blood mononuclear cells were isolated by Ficoll-hypaque density gradient centrifugation. The ethical committee of Fondazione I.R.C.C.S. Ca’Granda Ospedale Maggiore Policlinico approved the use of PBMCs from healthy donors for research purposes, and informed consent was obtained from subjects. Human blood primary lymphocyte subsets were purified >95% by cell sorting using different combinations of surface markers (Table 1). For in vitro differentiation experiments resting naïve CD4+ T cells were purified >95% by negative selection with magnetic beads with the isolation kit for human CD4+ Naïve T cells of Miltenyi and stimulated with Dynabeads Human T-Activator CD3/CD28 (Life Technologies). IL-2 was added at 20 IU/ml (R&D Systems 202-IL). TH1 polarization was initiated with 10 ng/ml IL12 (R&D Systems 219-IL) and TH2 neutralizing antibody anti-IL-4 (2 μg/ml) (R&D Systems MAB3007). TH2 polarization was induced by activation with Phytohemagglutinin, PHA (4 μg/mL Sigma L2769) in the presence of IL-4 (10 ng/ml) (R&D Systems 204-IL), and neutralizing antibodies to IFN-γ (2 μg/ml) (R&D Systems MAB 285) and anti-IL-12 (2 μg/ml) (R&D Systems MAB219). For GATA-3 and c-Maf intracellular staining, cells were harvested and then fixed for 30 min in Fixation/permeabilization Buffer (eBioscience) at 4 °C. Cells were stained with antibodies anti-GATA-3 (eBioscience clone TWAJ) and anti-c-Maf (eBioscience clone sym0F1) in washing buffer for 30 min at 4 °C. Cells were then washed two times, resuspended in FACS washing buffer and analyzed by flow cytometry.
RNA isolation and RNA sequencing
Total RNA was isolated using mirVana Isolation Kit. Libraries for Illumina sequencing were constructed from 100 ng of total RNA with the Illumina TruSeq RNA Sample Preparation Kit v2 (Set A). The generated libraries were loaded on to the cBot (Illumina) for clustering on a HiSeq Flow Cell v3. The flow cell was then sequenced using a HiScanSQ (Illumina). A paired-end (2×101) run was performed using the SBS Kit v3 (Illumina). Real-time analysis and base calling was performed using the HiSeq Control Software Version 1.5 (Illumina).
RNA-seq
RNA-seq data representative of 13 lymphocyte populations were collected for transcriptome reconstruction. Five biological replicates were analyzed for all populations except for CD8+ TCM and B CD5+ (four samples). The whole dataset was aligned to GRCh37 (Genome Reference Consortium Human Build 37) with TopHat v.1.4.1 32 for a total of over 1.7 billions mapped paired-end reads (30 million reads per sample on average). These data were also mapped with the aligner STAR v.2.2.0 31. RNA-seq datasets of 16 human tissues belonging to the Illumina Human BodyMap 2.0 project (ArrayExpress accession no. E-MTAB-513) were mapped following the same criteria.
Reference annotation
An initial custom reference annotation of unique, non-redundant transcripts was built by integrating the Ensembl database (version 67 from May 2012) with the lincRNAs identified by another group13 using Cuffcompare v.2.1.1 33. The annotated human lincRNAs were extracted from Ensembl using BioMart v.67 and subset by gene biotype ‘lincRNA’ (5,804 genes). Other classes of genes were integrated in the annotation: the list of protein coding genes (21,976 genes), the receptors genes collection defined in BioMart under GO term GO:000487 (2,043 genes with receptor activity function) and the class of genes involved in metabolic processes corresponding to GO term GO:0008152 (7,756 genes). Hence, the complete reference annotation consisted of 195,392 transcripts that referred to 62,641 genes, 11,170 of which are non-redundant lincRNA genes.
De novo genome-based transcripts reconstruction
A comprehensive catalogue of lincRNAs specifically expressed in human lymphocyte subsets was generated using a de novo genome-based transcripts reconstruction procedure with three different approaches. Two aligners were used: TopHat v.1.4.1 and STAR v. 2.2.0. The de novo transcriptome assembly was performed on the aligned sequences (samples of the same population were concatenated into one “population alignment”) generated by STAR and TopHat using Cufflinks v. 2.1.1 with reference annotation to guide the assembly (-g option) coupled with multi-read (-u option) and fragment bias correction (-b option) to improve the accuracy of transcripts abundance estimates. With this method, about 30,000–50,000 new transcripts were identified in each lymphocyte population. The third approach employed the genome-guided Trinity software (http://pasa.sourceforge.net/#A_ComprehensiveTranscriptome), which generates novel transcripts performing a local assembly on previously mapped reads from specific location. The Trinity50 default aligner was substituted with STAR. Each candidate transcript was then processed using the PASA pipeline, which reconstructs the complete transcript and gene structures, resolving incongruences derived from transcript misalignments and alternatively splices events, refining the reference annotation when there are enough evidence and proposing new transcripts and genes in case no previous annotation can explain the new data.
Novel lincRNA genes identification
Annotated transcripts and new isoforms of known genes were discarded, retaining only novel genes and their isoforms located in intergenic position. In order to filter out artifactual transcripts due to transcriptional noise or low polymerase fidelity, only multi-exonic transcripts longer than 200 bases were retained. Then, the HMMER3 algorithm35 was run for each transcript to identify occurrences of any protein family domain documented in the Pfam database (release 26; used both PfamA and PfamB). All six possible frames were considered for the analysis, and the matching transcripts were excluded from the final catalogue.
The coding potential for all the remaining transcripts was then evaluated using PhyloCSF (phylogenetic codon substitution frequency)36 (PhyloCSF was run on a multiple sequence alignment of 29 mammalian genomes (in MAF format) (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/) to obtain the best scoring ORF greater than 29 amino acids across all three reading frames. To efficiently access the multialignment files (MAF) the bio-maf (https://github.com/csw/bioruby-maf) Ruby biogem51 was employed. This library provides indexed and sequential access to MAF data, as well as performing fast manipulations on it and writing modified MAF files. Transcripts with at least one open reading frame with a PhyloCSF score greater than 100 were excluded from the final catalogue. The PhyloCSF score threshold of 100 was determined previously described13 to optimize specificity and sensitivity when classifying coding and non coding transcripts annotated in RefSeq (RefSeq coding and RefSeq lincRNAs). PhyloCSF score =100 corresponds to a false negative rate of 6% for coding genes (i.e., 6% of coding genes are classified as non-coding) and a false positive rate of ~10% (i.e., 9.5% of noncoding transcripts are classified as coding).
De novo data integration
Duplicates among the transcripts identified with the same de novo method were resolved using Cuffcompare v2.1.1. In the same way, the resulting three datasets were further merged to generate a non-redundant atlas of lincRNAs in human lymphocytes and only genes identified by at least 2 out of 3 software were considered. A unique name was given to each newly identified lincRNA gene composed by the prefix “linc-” followed by the Ensembl gene name of the nearest protein coding gene (irrespective of the strand). The additional designation “up” or “down” defines the location of the lincRNA with respect to the sense of transcription of the nearest protein coding gene. In addition, either “sense” or “antisense” was added to describe the concordance of transcription between the lincRNA and its nearest coding gene. A numerical counter only of newly identified lincRNAs related to the same protein coding gene is added as suffix (such as ‘linc-geneX-(up|down)-(sense|antisense)_#n’). This final non-redundant catalogue of newly identified lincRNAs includes 4,666 new transcripts referring to 3,005 new genes.
LincRNA signatures definition
A differential expression analysis among the thirteen cell subsets profiled was performed using Cuffdiff v.2.1.1. This analysis was run using --multi-read-correction (-u option) and upper quartile normalization (--library-norm-method quartile) to improve robustness of differential expression calls for less abundant genes and transcripts. Only genes expressed over 0.21 FPKM29 were considered in the downstream analysis to filter out genes that are merely by-products of leaky gene expression, sequencing errors, and/or off-target read mapping. After adding a pseudo-count of 1 to the raw FPKM for each gene, applying log2 transformation and Z-score normalization, K-means clustering with Euclidean metric was performed on lincRNAs expression values using MultiExperiment Viewer v.4.6 tool. The same procedure was then applied to the expression values of protein coding, metabolic and receptors genes. The Silhouette function52 was used to select an appropriate K (number of clusters). A K ranging from 13 to 60 was tested, and the value associated with the highest Silhouette score for each class of genes was selected. The number of clusters that maximizes the Silhouette score is 15 for lincRNA (Supplementary Fig. 2a), 24 for protein coding genes and 23 and 36 for receptors and metabolic genes respectively. The centroid-expression profile of each cluster was then evaluated in order to associate each cluster to a single cellular population (Fig. 2).
In order to select specifically expressed lincRNA genes, K-means results were subsequently intersected with the JS score, a cell-specificity measure based on Jensen-Shannon divergence and only the genes assigned to the same cellular population by both techniques were retained for further analysis. The estimation procedure for the JS score was adapted by building a reference model composed of 13 cell subsets. For the selected lincRNAs, the intrapopulation consistency among different samples was subsequently evaluated to minimize the biological variability: only genes expressed in at least 3/5 (or 3/4 replicates for CD8+ CM and CD5+ B) of the profiled samples whose maximal expression value was >2.5 fold compared to all other lymphocyte subsets were considered. Finally, non-parametric Kruskal-Wallis test was applied to select only lincRNA genes with a significant difference across the medians of the different lymphocyte populations: a P-value lower than 0.05 was considered and the lincRNA genes that meet these selection criteria were selected as signature genes.
Gene Ontology Enrichment Analysis
A Gene Ontology (GO) enrichment analysis was performed for biological process terms associated with protein coding genes that are proximal to lincRNA signatures at genomic level. For each lincRNA signature, the proximal protein-coding gene was selected regardless of the sense of transcription. FatiGO tool of Babelomics suite (version 4.3.0) was used to identify the enriched GO terms of the 158 protein coding genes (input list). All protein coding genes that are expressed in lymphocyte subsets (19,246 genes) (except the genes proximal to a lincRNA signature gene [input list]) defined the background list. Only GO terms with adjusted P value lower than 0.01 were considered (10 GO terms). Moreover, we performed a gene ontology semantic similarity analysis on the 51 GO terms with adjusted P value lower than 0.1 resulting from previous analysis using G-SESAME tool. This analysis provides as a result a symmetric matrix where each value represents a similarity score between GO term pairs. Then, we carried out a hierarchical clustering based on semantic similarity matrix to group together all GO terms with common GO parent.
Naïve CD4+ T cells siRNA transfection
Activated CD4+ naïve T Cells, were transfected with 300 nM FITC-labeled- linc-MAF-4 siRNA or FITC-labeled-AllStars negative control (Qiagen) with Lipofectamine 2000 (Life Technologies) according to the manufacturer protocol. FITC-positive cells were sorted and lysed 72 h post transfection. siRNAs sequences are provided in Supplementary Table 2.
Gene Expression Analysis
Gene expression analysis of transfected activated CD4+ naive cells was performed with Illumina Direct Hybridization Assays according to the standard protocol (Illumina). Total RNA was isolated, quality controlled and quantified as described above; for each sample 500 ng of total RNA were reverse transcribed according to the Illumina TotalPrep RNA Amplification kit (AMIL1791 - LifeTechnologies) and cRNA was generated by in vitro transcription (14 h). Hybridization was performed according to the standard Illumina protocol on Illumina HumanHT-12 v4 Expression BeadChip arrays (BD-103-0204 - Illumina). Scanning was performed on an Illumina HiScanSQ System and data were processed with Genome Studio; arrays were quantile normalized, with no background subtraction, and average signals were calculated on gene-level data for genes whose detection P value was lower than 0.001 in at least one of the cohorts considered.
GSEA (Gene Set Enrichment Analysis)
GSEA is a statistical methodology used to evaluate whether a given gene set is significantly enriched in a list of gene markers ranked by their correlation with a phenotype of interest. To evaluate this degree of ‘enrichment’, the software calculates an enrichment score (ES) by moving down the ranked list, i.e., increasing the value of the sum if the marker is included in the gene set and decreasing this value if the marker is not in the gene set. The value of the increase depends on the gene-phenotype correlation. GSEA was performed comparing gene expression data obtained from activated CD4+ naïve T cells transfected with linc-MAF-4 siRNAs vs. control siRNAs. The experimentally generated dataset from the in vitro differentiated cells (in TH1 or TH2 polarizing conditions respectively) derived from CD4+ naïve T cells of the same donors where linc-MAF-4 down-regulation was performed, were used to construct reference gene sets for TH1 and a TH2 cells. RNA for gene expression analysis of TH1 and TH2 differentiating cells was collected 72 h after activation (i.e., the same time-point of RNA collection in the linc-MAF-4 downregulation experiments) but a fraction of cells was further differentiated up to day 8 to assess IFN-γ and IL-13 production by TH1 and TH2 cells. The TH1 and TH2 datasets were ranked as log2 ratios of the expression values for each gene in the two conditions (TH1/TH2), and the most upregulated/downregulated genes (having log2 ratios ranging from |3| to |0.6|) were assigned to the TH1 and TH2 reference sets respectively.
Genes from the TH1 gene list which were downregulated in a TH1 vs. control-siRNA comparison and genes from the TH2 gene list that were downregulated in a TH2 vs. control-siRNA comparison were filtered out, obtaining a TH1-specific gene set (74 genes) and a TH2-specific gene set (141 genes) (Supplementary Table 1). GSEA was then performed on the linc-MAF-4 specific siRNA vs. control siRNA dataset. The metric used for the analysis is the log2 Ratio of Classes, with 1,000 gene set permutations for significance testing.
RT-qPCR Analysis
For reverse transcription, equal amounts of DNA-free RNA (500 ng) were reverse-transcribed with SuperScript III (LifeTechnologies) following the suggested conditions. Diluted cDNA was then used as input for RT-qPCR to assess MAF (Hs00193519_m1), IL4 (Hs00174122_m1), GATA3 (Hs01651755_m1), TBX21 (Hs00203436_m1), RORC (Hs01076119_m1), IL17 (Hs00174383_m1), Linc00339 (Hs04331223_m1), MALAT1 (Hs01910177_s1), RNU2.1 (Hs03023892_g1) and GAPDH (Hs02758991_g1) gene expression with Inventoried TaqMan Gene Expression assays (LifeTechnologies) were used. For assessment of linc-MAF-4 and validation of CD4+ TH1 signature lincRNAs specific primers were designed and 2.5 μg of CD4+ TH1, Treg or naive cells RNA were used for reverse transcription with SuperScript III (LifeTechnologies). RT-qPCR was performed on diluted cDNA with PowerSyberGreen (LifeTechnologies) and specificity of the amplified products was monitored by performing melting curves at the end of each amplification reaction. The primers used in qPCR are listed in Supplementary Table 2.
Cell fractionation
In vitro differentiated TH1 cells were resuspended in RLN1 buffer (50 mM Tris-HCl pH 8, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) supplemented with SUPERase•In (Ambion) for 10 min on ice. After a centrifugation at 300g for 2 min, the supernatant was collected as the cytoplasmic fraction. The pellet was resuspended in RLN2 buffer (50 mM Tris-HCl pH 8, 500 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40) supplemented with RNase inhibitors for 10 min on ice. Chromatin was pelletted at maximum speed for 3 min. The supernatant represents the nuclear fraction. All the fractions were resuspended in TRIzol (Ambion) to 1 ml and RNA was extracted following the standard protocol.
RNA immunoprecipitation (RIP)
In vitro differentiated TH1 cells were UV-crosslinked at 400 mJ/cm2 in ice-cold D-PBS and then pelleted at 1350g for 5 min. The pellet was resuspended in ice-cold lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% NP-40) supplemented with 0.5 mM β-mercaptoethanol, Protease Inhibitor Cocktail Tablets cOmplete, EDTA-free (Roche) and SUPERase•In (Ambion) and left rocking at 4 °C until lysis was complete. Debris was centrifuged at 13000g for 10 min. The lysate was precleared with Dynabeads® Protein G (Novex®) for 30 min at 4 °C and then incubated for 2 h with 7 μg of antibodies specific for EZH2 (Active Motif - 39875); LSD1 (Abcam – ab17721), or HA (Santa Cruz - sc7392) as mock control. The lysate was coupled with Dynabeads® Protein G (Novex®) for 1 h at 4 °C. Immunoprecipitates were washed for five times with lysis buffer. RNA was then extracted following mirVana miRNA Isolation Kit (Ambion) protocol. The RNA transcript abundance of Linc-MAF-4 or of the negative controls β-actin, RNU2.1 and a region upstream the TSS of linc-MAF-4 (linc-MAF-4 control) was assessed by RT-qPCR.
Chromatin Immunoprecipitation analysis (ChIP)
In vitro differentiated TH1 and TH2 cells were crosslinked in their medium with 1/10 of fresh formaldehyde solution (50 mM HEPES-KOH pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 11% formaldehyde) for 12 min. Subsequently they were treated with 1/10 of 1.25 M glycine for 5 min and centrifuged at 1350g for 5 min at 4 °C. Cells were lysed in LB1 (50 mM HEPES-KOH pH 7.5, 10 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 and 0.25% Triton X-100) supplemented with Protease Inhibitor Cocktail Tablets cOmplete, EDTA-free (Roche) and Phenylmethanesulfonyl fluoride (Sigma) at 4 °C. Nuclei were pelleted at 1350g for 5 min at 4 °C and washed in LB2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) supplemented protease inhibitors. Nuclei were again pelleted at 1350g for 5 min at 4 °C and resuspended with a syringe in 200 μl LB3 (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylscarcosine) supplemented with protease inhibitors. Cell debris were pelleted at 20000g for 10 min at 4 °C and a ChIP was set up in LB3 supplemented with 1% Triton X-100, protease inhibitors and antibodies against H3K4me3 (Millipore - 07-473), H3K27me3 (Millipore – 07-449), RNA polymerase II STD repeat YSPTSPS (Abcam – ab5408), LSD1 (Abcam – ab17721), EZH2 (Active Motif - 39875) or no antibody (as negative control) o/n at 4 °C. The day after Dynabeads® Protein G (Novex®) were added at left at 4 °C rocking for 2 h. Then the beads were washed twice with Low salt wash buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% SDS, 2 mM EDTA, 1% Triton X-100) and with High salt wash buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.1% SDS, 2 mM EDTA, 1% Triton X-100). Histones IPs were also washed with a LiCl solution (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 1% NP-40, 1 mM EDTA). All samples were finally washed with 50 mM NaCl in 1× TE. Elution was performed overnight at 65 °C in 50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS. Samples were treated with 0.02 μg/μl RNase A (Sigma) for 2 h at 37 °C and with 0.04 μg/μl proteinase K (Sigma) for 2 h at 55 °C. DNA was purified with phenol/chloroform extraction.
Chromosome Conformation Capture (3C)
For 3C analysis cells were crosslinked and digested as described for ChIP53. Nuclei were resuspended in 500 μl of 1.2× NEB3 buffer (New England BioLabs) with 0.3% SDS and incubated at 37 °C for 1 h and then with 2% Triton X-100 for another 1 h. Digestion was performed with 800 U of BglII (New England BioLabs)overnight at 37 °C shaking. Digestion was checked loading digested and undigested controls on a 0.6% agarose gel. Then the sample was incubated with 1.6% SDS for 25 min at 65 °C and with 1.15× ligation buffer (New England BioLabs) and 1% Triton X-100 for 1 h at 37 °C. Ligation was performed with 1000 U of T4 DNA ligase (New England BioLabs) for 8 h at 16 °C and at 22°C for 30 min. DNA was purified with phenol-chloroform extraction after RNase A (Sigma) and Proteinase K (Sigma) digestion. As controls, BACs corresponding to the region of interested were digested with 100 U BglII in NEB3 buffer in 50 μl o/n at 37 °C. Then fragments were ligated with 400 U T4 DNA ligase overnight at 22°C in 40 μl. PCR products amplified with GoTaq Flexi (Promega) for BACs and samples were run on 2.5% agarose gels and quantified with ImageJ software. Primers are listed in Supplementary Table 3.
Statistical analysis
Unless indicated otherwise in the figure legend(s), a one-tailed, paired t-test was performed on experimental data with Prism (GraphPad Software). For multiple comparisons between human lymphocytes subsets a non-parametric Kruskal-Wallis test was used. ANOVA and Dunnet post-hoc test was applied for statistical analysis of RNA immunoprecipitation experiments in Fig.6c. Number of experimental replicates is indicated in the legend of each figure.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank C. Cheroni for support in statistical analysis; M. Moro and M.C. Crosti for technical assistance with cell sorting; S. Biffo D. Gabellini (Dulbecco Telethon Institute at San Raffaele Scientific Institute Milan, Italy), P. Della Bona (San Raffaele Scientific Institute, Milan, Italy) and A. Lanzavecchia (Institute for Research in Biomedicine, Bellinzona, Switzerland) for discussions and critical revision of the manuscript; B.J. Haas and A. Dobin (Broad Institute, Cambridge, MA USA) for helping the integration of genome guided Trinity with STAR aligner. The INGM Bioinformatics facility for support. Google Summer of Code Project for supporting Clayton Wheeler in the development of https://github.com/csw/bioruby-maf.
This study was supported by: the Flagship CNR-MIUR grant “EPIGEN”, CARIPLO grant n° 2013-0955, AIRC grant n° IG2013-ID14596, ERC Advanced Grant n° 269022 to S.A, ERC Consolidator Grant n° 617978 to M.P, and by an unrestricted grant of the “Fondazione Romeo ed Enrica Invernizzi”.
Footnotes
ACCESSION CODES
ArrayExpress accession: E-MTAB-2319
REFERENCES
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–89. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–55. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 3.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–31. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 6.Pagani M, et al. Role of microRNAs and long-non-coding RNAs in CD4(+) Tcell differentiation. Immunol Rev. 2013;253:82–96. doi: 10.1111/imr.12055. [DOI] [PubMed] [Google Scholar]
- 7.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–73. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koralov SB, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 9.O’Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi RL, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 12.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrdlickova B, et al. Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity. Genome Med. 2014;6:88. doi: 10.1186/s13073-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 16.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JH, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang KC, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182:7738–48. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 25.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189:2084–8. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez JA, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu G, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–8. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hart T, Komori HK, LaMere S, Podshivalova K, Salomon DR. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics. 2013;14:778. doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–6. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–22. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–82. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–4. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volders PJ, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–51. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho IC, Lo D, Glimcher LH. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and - independent mechanisms. J Exp Med. 1998;188:1859–66. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Nurieva RI, Dong C. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunol Rev. 2013;252:139–45. doi: 10.1111/imr.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato K, et al. Marked induction of c-Maf protein during Th17 cell differentiation and its implication in memory Th cell development. J Biol Chem. 2011;286:14963–71. doi: 10.1074/jbc.M111.218867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cabianca DS, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–31. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaneko S, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonnal RJ, et al. Biogem: an effective tool-based approach for scaling up open source software development in bioinformatics. Bioinformatics. 2012;28:1035–7. doi: 10.1093/bioinformatics/bts080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rousseeuw PJ, Leroy AM, John Wiley & Sons . Wiley series in probability and mathematical statistics Applied probability and statistics. Wiley; New York: 1987. [Google Scholar]
- 53.Bodega B, et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.