SUMMARY
RNA polymerases are key multisubunit cellular enzymes. Microscopy studies indicated that RNA polymerase I assembles near its promoter. However, the mechanism by which RNA polymerase II is assembled from its 12 subunits remains unclear. We show here that RNA polymerase II subunits Rpb1 and Rpb3 accumulate in the cytoplasm when assembly is prevented and that nuclear import of Rpb1 requires the presence of all subunits. Using MS-based quantitative proteomics, we characterized assembly intermediates. These included a cytoplasmic complex containing subunits Rpb1 and Rpb8 associated with the HSP90 cochaperone hSpagh (RPAP3) and the R2TP/Prefoldin-like complex. Remarkably, HSP90 activity stabilized incompletely assembled Rpb1 in the cytoplasm. Our data indicate that RNA polymerase II is built in the cytoplasm and reveal quality-control mechanisms that link HSP90 to the nuclear import of fully assembled enzymes. hSpagh also bound the free RPA194 subunit of RNA polymerase I, suggesting a general role in assembling RNA polymerases.
INTRODUCTION
RNA polymerases play fundamental roles in the cell. RNA polymerases I and III synthesize the noncoding RNAs that form the translational apparatus, and their activity is intimately linked to the growth state of the cell (White, 2005). RNA polymerase II synthesizes capped noncoding RNAs as well as all mRNAs. This enzyme is at the heart of gene regulation and is subjected to many controls, including at the level of initiation, elongation, and termination (Fuda et al., 2009).
The activities, structures, and subunit composition of the three major RNA polymerases have been characterized in detail (Cramer et al., 2008). RNA polymerases I, II, and III are composed of 14, 12, and 17 subunits, respectively. The two largest subunits form the catalytic core of the enzyme, while the others are smaller and generally bind on their surface. The three RNA polymerases are related to each other, and this structural similarity is also highlighted by the fact that some subunits are shared by several polymerases, with a few being present in all three enzymes. Despite intensive studies on the structure and regulation of RNA polymerases, relatively little is known about the location and mechanism of their assembly. To date, this question has been principally addressed by live-cell microscopy techniques that used GFP-tagged subunits in mammalian cells. FRAP (fluorescence recovery after photobleaching) studies on RNA polymerase I have indicated that some subunits can either assemble or exchange directly at promoters (Dundr et al., 2002). Furthermore, the exchange/assembly rate depends on the phase of the cell cycle, indicating that assembly of RNA polymerase I in the nucleolus is a way to regulate gene expression (Gorski et al., 2008). Live-cell studies on RNA polymerase II promoters have also revealed a rapid exchange of basal and sequence-specific transcription factors at promoters. In yeast, the TATA binding factor TBP and the transcriptional activator Ace1p were shown to rapidly come on and off the chromatin, with a residency time in the range of seconds (Karpova et al., 2008; Sprouse et al., 2008). Moreover, the rapidly cycling Ace1p molecules were shown to be responsible for transcriptional activation. In mammalian cells, glucocorticoid and estradiol receptors were shown to have similarly high exchange rates with their target sites on DNA (McNally et al., 2000; Stenoien et al., 2001). More recently, the use of Drosophila salivary glands with polytene chromosomes and the generation of human cell lines carrying artificial arrays of reporter genes have allowed detailed studies of transcription in living cells (Janicki et al., 2004; Yao et al., 2006). In particular, FRAP studies of GFP-tagged subunits of RNA polymerase II were used to define the dynamics of their binding to promoters and the transcription kinetics of this key enzyme. In two studies that used HIV-1 reporters or natural Drosophila genes, an efficient initiation entry mode was found (Boireau et al., 2007; Yao et al., 2007). In contrast, a study that used a Tet-inducible promoter in human cells found that a large fraction of the polymerase exchanges rapidly at promoters, with only a few percent that go into productive elongation (Darzacq et al., 2007). Given the precedent example of RNA polymerase I assembly at its promoter, these data led to the suggestion that RNA polymerase II may also assemble at its promoter and that this could allow a gene-specific regulation of its assembly (Darzacq and Singer, 2008; Hager et al., 2009).
Recently, affinity purification of soluble human RNA polymerase II with TAP-tagged subunits identified a number of polymerase-associated factors of unknown function (Jeronimo et al., 2004, 2007). Four of these factors are homologous to the yeast R2TP complex (Boulon et al., 2008; Zhao et al., 2005), and further studies suggested that they indeed form a complex that contains the R2TP factors plus five prefoldin-like proteins (referred to as R2TP/Prefoldin-like) (Boulon et al., 2008; Cloutier et al., 2009; Sardiu et al., 2008). The yeast R2TP protein Tah1 was initially characterized as a cochaperone for HSP90 (Zhao et al., 2005) and, in agreement, we recently showed that the human Tah1 homolog hSpagh (also called RPAP3 or FLJ21908) is an HSP90 cofactor (Boulon et al., 2008). Furthermore, HSP90 and R2TP proteins, including hSpagh, play a key role during the assembly of snoRNPs (Boulon et al., 2008; Gonzales et al., 2005; King et al., 2001; Zhao et al., 2008), raising the possibility that the association of hSpagh with RNA polymerase II subunits could be involved in the assembly of this enzyme.
In this study, we used a combination of quantitative MS-based proteomics and fluorescence microscopy to characterize the mechanism of RNA polymerase II assembly in human cells and the role of hSpagh and Hsp90 in this process.
RESULTS
Unassembled RNA Polymerase II Subunits Accumulate in the Cytoplasm
To study assembly of RNA polymerase II, we took advantage of α-amanitin. This drug binds the large Rpb1 subunit of human RNA polymerase II with high affinity. In vitro, this reduces the elongation rate more than 100-fold and makes the polymerase prone to essentially irreversible stalling (Rudd and Luse, 1996). In vivo, this results in transcriptional arrest and concomitant destruction of subunit Rpb1. The other subunits remain intact, suggesting that α-amanitin may result in enzyme disassembly (Nguyen et al., 1996). First, the localization of RNA polymerase II subunit Rpb3 was analyzed by fluorescence microscopy using a U2OS cell line stably expressing a GFP-Rpb3 fusion protein known to incorporate into functional RNA polymerase (Boireau et al., 2007 and see below). The GFP-Rpb3 fusion protein normally accumulates in the nucleus, with only a faint cytoplasmic staining (Figure 1A). However, this changed following α-amanitin addition, with GFP-Rpb3 now accumulating in the cytoplasm. To test whether this resulted either from a failure to import newly synthesized Rpb3 or instead from export of the nuclear subunit, cells were treated with both α-amanitin and leptomycin B (LMB), a specific inhibitor of the exportin CRM1 (Fornerod et al., 1997). This resulted in the retention of GFP-Rpb3 in the nucleus (Figure 1A), indicating that an active export mechanism removes GFP-Rpb3 from nuclei when Rpb1 is degraded.
Figure 1. Disruption of RNA Polymerase II Assembly Induces the Accumulation of Subunits Rpb1 and Rpb3 in the Cytoplasm.
(A) Cytoplasmic accumulation of subunits Rpb1 and Rpb3 following treatment with LMB or α-amanitin. U2OS cells expressing a GFP-Rpb3 fusion were untreated (NT) or treated for 15 hr with α-amanitin (α-am, 10 μg/ml) and/or leptomycin B (LMB, 15 nM) and processed for immunofluorescence against Rpb1. Each field is 83.75 × 83.75 μm. DAPI stains nuclei. Similar results were obtained after 3 and 6 hr of treatments, although at these time points α-amanitin affects only a fraction of cells (not shown).
(B) Depletion of subunit Rpb2 leads to the accumulation of newly synthesized Rpb1 in the cytoplasm. U2OS cells were transfected for 48 hr with siRNA against luciferase (siFFL) or Rpb2 (siRpb2) and treated or not with α-amanitin for 15 hr (10 μg/ml). Cells were processed for immunofluorescence against Rpb1. Each field is 83.75 × 117.25 μm.
(C) Inhibition of RNA polymerase II transcription by DRB has no effect on the localization of Rpb1. U2OS cells were treated for 15 hr with DRB (100 μM), and the localization of Rpb1 was determined by immunofluorescence (panels α-Rpb1). FISH against polyA + RNAs was performed in parallel to assess the inhibition of transcription (panel PolyA+). Each field is 67 × 67 μm.
(D) Depletion of RNA polymerase II subunits leads to the accumulation of Rpb1 in the cytoplasm. U2OS cells were transfected for 48 hr with siRNA against luciferase (siFFL) or the indicated subunit. Cells were processed for immunofluorescence with an antibody against Rpb1. Each field is 83.75 × 117.25 μm and contains few nondepleted cells that show wild-type nuclear Rpb1 levels.
Surprisingly, LMB prevented nuclear import of Rpb1 (Figure 1A) and made this subunit insensitive to α-amanitin (Figures 1A and S1). Because α-amanitin specifically promotes degradation of the elongating form of RNA polymerase II (Nguyen et al., 1996), the cytoplasmic accumulation of Rpb1 most likely represents newly synthesized subunits that were never engaged in transcription elongation. These data suggest that factors required for Rpb1 nuclear import may be retained in the nucleus upon LMB treatment. Since Rpb3 remains nuclear in these conditions, an intriguing possibility would be that Rpb1 needs to be incorporated in fully assembled enzymes before being imported into the nucleus.
To test this hypothesis, RNA polymerase II assembly was prevented by inhibiting de novo synthesis of individual subunits with siRNAs. Depletion of Rpb2, the second largest subunit, prevented nuclear import of Rpb1 (Figure 1B). The pool of Rpb1 accumulated in the cytoplasm was resistant to α-amanitin, indicating that it corresponds to newly synthesized proteins that were never engaged in elongation. Furthermore, analysis with a panel of antibodies that recognize various phosphoisoforms of the CTD heptad-repeat indicated that the cytoplasmic Rpb1 is mostly unphosphorylated (see Supplemental Experimental Procedures and Figure S1). This effect on Rpb1 caused by siRNA-mediated depletion of Rpb2 was not an indirect consequence of transcription inhibition. Indeed, the transcription inhibitors DRB and actinomycin D, which do not trigger subunit degradation (Nguyen et al., 1996), did not induce a cytoplasmic accumulation of Rpb1 (Figures 1C and S1). These results were extended by examining siRNA-mediated removal of other RNA polymerase II subunits. Remarkably, depletion of any subunit, including Rpb3, resulted in a similar accumulation of Rpb1 in the cytoplasm (Figures 1D and S1, and see below for quantitative measurements of Rpb1 localization). Together with the fact that degradation of elongating Rpb1 by α-amanitin triggered nuclear export of Rpb3, these data suggested that assembly of RNA polymerase II occurs in the cytoplasm and is an obligatory step prior to nuclear import.
GFP-Tagged Subunits Rpb1 and Rpb3 Have Similar Dynamics at the Transcription Site of an HIV-1 Reporter Gene
Assembly of RNA polymerase I in the nucleolus was demonstrated by FRAP using GFP-tagged subunits (Dundr et al., 2002). Indeed, RNA polymerase I subunits have biphasic recovery curves in the nucleolus, with an initial rapid recovery of about 1 min that is specific for each subunit and that represents exchange of their unassembled form. To further substantiate that RNA polymerase II is assembled in the cytoplasm, rather than at promoters, we determined the dynamics of different polymerase subunits at the transcription site of an HIV-1 reporter gene that was tagged with MS2 sites for visualization in living cells (Fusco et al., 2003; Boireau et al., 2007) (Figure 2). To this end, we used the Exo1 cell line, which has integrated many copies of the reporter gene and was shown to accumulate high levels of RNA polymerase II at its transcription site (Boireau et al., 2007). We previously observed that in this system, GFP-tagged subunit Rpb3 was initiating transcription efficiently and did not show a rapid exchange phase during the first minute of the recovery (Boireau et al., 2007). However, this did not preclude a different behavior for the other subunits and, in particular, for subunit Rpb1 (Darzacq et al., 2007). To analyze the dynamics of this subunit, we transfected the Exo1 cell line with a GFP-tagged Rpb1 construct that carried an α-amanitin resistance mutation and selected α-amanitin-resistant cell lines to replace the endogenous polymerase by the GFP-tagged one. Remarkably, the analysis of GFP-Rpb1 dynamics by FRAP revealed a recovery rate at the HIV-1 transcription site that was not significantly different from the GFP-Rpb3 subunit, with an absence of an initial rapid exchange phase. This was consistent with the idea that RNA polymerase II was recruited to the HIV-1 promoter as a preformed complex rather than as free subunits.
Figure 2. GFP-Rpb1 and GFP-Rpb3 Associate with an HIV-1 Promoter with Similar Dynamics.
(A) Schematic of the reporter construct used in the assay. The HIV-1 reporter plasmid was inserted in multiple tandem copies in the chromatin of U2OS cells.
(B) Recovery of GFP-Rpb1 at the transcription site of the HIV-1 reporter. MS2-Cherry accumulates at the reporter transcription site (top left panel), together with GFP-Rpb1 (panel “prebleach”). Images of GFP-Rpb1 at the indicated time points show the regular and slow recovery of the protein at the transcription site.
(C) Comparison of the recovery rate of GFP-Rpb3 and GFP-Rpb1 at the transcription site of the HIV-1 reporter gene. The recovery curves show very similar profiles for the two proteins. Time is in seconds.
Characterization of RNA Polymerase II Assembly Intermediates by SILAC Quantitative Proteomics Using GFP-Rpb3 as a Bait
Next, a quantitative MS-based proteomics approach was used to characterize the changes in RNA polymerase II complexes in either the absence or presence of α-amanitin, i.e., when RNA polymerase II enzyme is either mostly nuclear and active or mostly cytoplasmic and inactive, respectively (see Figure 1). A triple isotope SILAC labeling scheme was employed (Figure 3A) (Trinkle-Mulcahy et al., 2006). Control U2OS cells were cultivated in medium containing the normal, “light” amino acids, while cells stably expressing GFP-Rpb3 were grown either with “medium” (no treatment) or with “heavy” (α-amanitin treatment) isotope-labeled amino acids. Soluble cell extract was prepared from each of the light, medium, and heavy cell cultures; GFP-Rpb3 and associated partners were affinity purified, and tryptic peptides were analyzed by MS. Intensity ratios for the three isotopic forms of each protein were determined using Max-Quant (Cox et al., 2009) and analyzed within Peptracker (Y.A. and A.I.L., unpublished data). Data were plotted in 2D logarithmic graphs, with the x axis representing enrichment of GFP-Rpb3-associated proteins in comparison with the control IP (medium/light ratio) and the y axis representing enrichment of GFP-Rpb3-associated proteins in α-amanitin-treated versus untreated cells (heavy/medium ratio) (Figure 3A). Contaminant proteins are clustered around the origin, while proteins specifically copurified with GFP-Rpb3 in untreated cells appear to the right of the graph. Proteins whose specific copurification with GFP-Rpb3 decreased upon treatment with α-amanitin appear below GFP-Rpb3, while those present above correspond to proteins whose specific copurification with GFP-Rpb3 increased after treatment. Of the 12 known RNA polymerase II subunits, 11 were specifically copurified with GFP-Rpb3, with a high number of peptides identified and quantified (see Table S1). Five of these—subunits Rpb4, Rpb5, Rpb7, Rpb8, and Rpb9—clearly dissociated from GFP-Rpb3 in cells treated with α-amanitin, showing that RNA polymerase II complex is, at least in part, disassembled after Rpb1 degradation. In agreement, structural analyses have shown that except for Rpb9, four subunits cited above interact with Rpb3 via subunit Rpb1 (Figure 3B) (Armache et al., 2003; Bushnell and Kornberg, 2003). Several additional factors were found to associate with subunit Rpb3 with high specificity, i.e., RPAP1, RPAP2, GrinL1A, GPN1, GPN2, and GPN3, and the SILAC data show that these factors preferentially associate with Rpb3 subunits that are not assembled in a complete RNA polymerase II enzyme (Figures 3A and 3B and Table S1). Systematic yeast two-hybrid assays also revealed an extensive set of protein-protein interactions (Figure 3B and Table S2). Together, these data indicate the existence of a cytoplasmic RNA polymerase II complex formed by subunits Rpb2, Rpb3, Rpb10, Rpb11, and Rpb12 and identify additional factors associated with this complex.
Figure 3. Characterization of Partially Assembled RNA Polymerase II Subcomplexes by Quantitative SILAC Proteomic Analysis.
(A) Comparison of GFP-Rpb3 complexes in the presence or absence of α-amanitin. Left panel: design of the triple-encoding SILAC GFP-Rpb3 pull-down experiment (see text). Right graph: SILAC results visualized on a 2D logarithmic graph for all proteins identified. SILAC ratios have been normalized so that IP contaminants are clustered at the origin of the graph. On the x axis, log2(M/L ratio) correlates with the enrichment in GFP-Rpb3 IP versus CTL IP. On the y axis, log2(H/M ratio) correlates with the enrichment in α-amanitin versus nontreated cells. The bait, Rpb3, is spotted in red, and a red line separates the proteins whose interaction with Rpb3 is increased after α-amanitin treatment (above the line) or decreased (below). Variations lower than 2-fold are not considered significant. Only proteins that are significantly enriched in GFP-Rpb3 IP (log[M/L] > 2) are labeled (spotted in orange), plus all R2TP/Prefoldin-like components (spotted in green) and all polymerase subunits (spotted in purple). Proteins that are identified or quantified with less than two peptides are labeled in gray and italic. SILAC ratio values of labeled proteins are listed in Table S1.
(B) Model of changes in RNA polymerase II complex and associated factors in untreated cells versus α-amanitin-treated cells, according to SILAC results. The bait is circled in red and the color code is similar to the graph, except that RNA polymerase II subunits that show a decreased association with GFP-Rpb3 upon α-amanitin treatment are displayed in white. Gray lines represent two-hybrid interactions.
Large Subunits of RNA Polymerase I and II Associate with the HSP90 Cochaperone hSpagh when Enzyme Assembly Is Prevented
Next, a similar SILAC-based proteomics strategy was used to identify factors bound to newly synthesized Rpb1 in cells treated with α-amanitin and LMB (Figure 4A). In this case, an antibody specific for the endogenous Rpb1 subunit was used for immunoaffinity purification (antibody PB-7C2), rather than a GFP fusion protein. Treatment of cells with α-amanitin and LMB prevented the association of most subunits with Rpb1. While Rpb8 was not much affected, all the other subunits were present in Rpb1 complexes in lower amount, from ~3-fold to ~6-fold (Figure 4A and Table S3). Several additional factors copurified specifically with Rpb1, including some that were previously described (Jeronimo et al., 2007). Remarkably, association of Rpb1 with RPAP2, GPN1, GPN3, GrinL1A, and the R2TP/Prefoldin-like components hSpagh, PFDN2, and UXT was largely unaffected by treatment with α-amanitin and LMB, indicating that they all bind Rpb1 prior to its assembly into a complete enzyme. Systematic yeast two-hybrid tests further revealed that UXT interacts with Rpb1 (Table S4) and could thus link it to the R2TP/Prefoldin-like complex (Figure 4D).
Figure 4. Unassembled Rpb1 Is Associated with the R2TP/Prefoldin-like Complex.
(A) Comparison of Rpb1 complexes when RNA polymerase II assembly is normal or inhibited by α-amanitin and LMB. The experimental design is indicated on the left. It is similar to Figure 2, except that cells are treated simultaneously with α-amanitin and LMB (for 15 hr) and that an antibody against endogenous Rpb1 is used. Legend as in Figure 2.
(B) Transcriptional shutdown does not result in polymerase disassembly (legend as in A). The experimental design is indicated on the left. Legend is as in Figure 2, except that cells were untreated or treated with α-amanitin + LMB (15 hr) or with actinomycin D (7 hr).
(C) Unassembled Rpb1 is specifically associated with the R2TP/Prefoldin-like complex. SILAC ratios are from the experiment depicted in (B) and correspond to cells treated with α-amanitin + LMB versus those treated with actinomycin D. The ratios are normalized to that of Rpb1.
(D) Model of the complex containing unassembled Rpb1 (legend as in Figure 3).
Analysis of protein half-lives using pulsed isotope labeling and quantitative MS showed no correlation with loss or enrichment in the immunoprecipitates upon α-amanitin treatment (F.M. Boisvert and A.I.L., unpublished data), indicating that changes in complex composition were not due to loss of proteins induced by transcriptional shutdown (data not shown). To further rule out this possibility, we performed a similar SILAC experiment in which we analyzed the status of polymerase assembly in cells treated with actinomycin D. The time of actinomycin D treatment (7 hr) was chosen to match the time of effective transcriptional arrest during α-amanitin treatment (Figure S2). For this purpose, a triple-labeling SILAC experiment was performed, where we purified Rpb1 complexes from (1) untreated cells (light amino acids), (2) cells treated with actinomycin D (medium amino acids), and (3) cells treated with α-amanitin + LMB (heavy amino acids). The data were displayed in a 2D graph, with SILAC ratios of actinomycin D versus untreated cells on the x axis and ratios of α-amanitin + LMB versus untreated cells on the y axis. Again, we observed that α-amanitin + LMB induced RNA polymerase II disassembly and association of the Rpb1/Rpb8 dimer with the full R2TP/Prefoldin-like complex (Figure 4B). In contrast, polymerase disassembly was not observed upon treatment with actinomycin D. Although an increased association was observed with the R2TP/Prefoldin-like complex, it was clearly less than in cells treated with α-amanitin + LMB (Figures 4B and 4C). Thus, unassembled Rpb1 is specifically associated with the R2TP/Prefoldin-like complex.
hSpagh is a central component of the R2TP/Prefoldin-like complex that binds HSP90 (Boulon et al., 2008). To determine possible functions of hSpagh and HSP90 in the biogenesis of RNA polymerase II, we first confirmed the binding of HSP90 and hSpagh to Rpb1 by western blotting. Coimmunoprecipitation (coIP) experiments of endogenous proteins in S100 extracts revealed that Rpb1 interacted with hSpagh and HSP90, that hSpagh was associated with HSP90, and that these interactions did not depend on HSP90 ATPase activity (Figure S3). Next, we generated a stable U2OS clone expressing a GFP-tagged version of hSpagh and immunoaffinity-purified GFP-hSpagh. This confirmed that Rpb1 was bound to GFP-hSpagh, even after treating cells with α-amanitin and LMB (Figure 5A). We then depleted the Rpb2 subunit with siRNA. Remarkably, interaction of Rpb1 with GFP-hSpagh increased upon knockdown of Rpb2 (Figure 5B), confirming that hSpagh preferentially binds unassembled Rpb1. Interestingly, hSpagh has also been reported to bind the large subunit of RNA polymerase I, RPA194 (Jeronimo et al., 2007). To test whether this also occurs preferentially on the unassembled form of this subunit, cells were treated with siRNA against the second largest subunit of RNA polymerase I, RPA135, and GFP-hSpagh immunoprecipitates were probed for the presence of RPA194. Indeed, an increased association of RPA194 with hSpagh was seen upon knockdown of RPA135 (Figure 5C), suggesting that hSpagh preferentially associates with free RPA194.
Figure 5. hSpagh Preferentially Associates with Unassembled Forms of the Large Subunits of RNA Polymerases I and II.
(A and B) hSpagh preferentially binds incompletely assembled subunit Rpb1. U2OS cells stably expressing GFP-hSpagh were subjected to GFP-TRAP immunoprecipitation. Inputs and pellets were analyzed by western blots with the indicated antibodies. Cells were untreated (NT) or treated with α-amanitin and LMB (α-am LMB) (A). Cells were treated with siRNA against Rpb2 (GFP-hSpagh si-Rpb2) or luciferase as control (GFP-hSpagh) (B). The input lanes were loaded with one-tenth of the amount used for the immunoprecipitation.
(C) hSpagh associates with unassembled RNA polymerase I subunit RPA194. Cells were treated with siRNA against RPA135 (GFP-hSpagh siRPA135) or luciferase (GFP-hSpagh), subjected to GFP-TRAP purification, and analyzed by western blotting with the indicated antibody. The input lanes were loaded with one-tenth the amount of extracts used in the purification.
(D) Quantitative SILAC proteomic analyses of GFP-hSpagh complexes in cells treated or not with α-amanitin. Left panel shows the experimental design. U2OS cells stably expressing GFP-hSpagh were treated or not with α-amanitin and subjected to SILAC proteomic analysis. Right panel shows SILAC results visualized on a 2D logarithmic graph. x axis: log(M/L ratio) correlates with the enrichment of proteins in GFP-hSpagh IP versus control IP (untreated cells). y axis: log(H/M ratio) correlates with the enrichment of proteins in GFP-hSpagh IP in treated versus untreated cells.
(E) Diagram displaying GFP-hSpagh complexes according to SILAC results. Legend is as in Figure 2, except that the thick bars represent direct physical interactions previously demonstrated (Boulon et al., 2008).
To further characterize the RNA polymerase II complexes associated with hSpagh, we performed SILAC-based proteomic analyses using GFP-hSpagh as bait, and we compared protein complexes from control and α-amanitin-treated cells (Figure 5D and Tables S5 and S6). Components of the R2TP/Prefoldin-like complex were readily identified copurifying with GFP-hSpagh, but only four RNA polymerase II subunits were found, i.e., Rpb1, Rpb2, Rpb8, and Rpb5. Furthermore, α-amanitin-induced destruction of Rpb1 strongly decreased the levels of Rpb2 and Rpb8 copurified with GFP-hSpagh, while levels of Rpb5 remained unchanged. Previous SILAC immunoprecipitation showed that Rpb1 dissociates from Rpb5 but remains associated with hSpagh upon treatment with LMB and α-amanitin (Figure 4A). Altogether, these data suggest that hSpagh associates independently with Rpb5- and Rpb1-containing subcomplexes (Figure 5E).
Interaction of hSpagh with Unassembled Subunit Rpb1 Occurs in the Cytoplasm
Unassembled subunit Rpb1 accumulates in the cytoplasm, suggesting that its interaction with hSpagh may take place in this compartment. To test this possibility, we set up a subcellular corecruitment assay (Figure 6) (Darzacq et al., 2006). In these experiments, hSpagh was artificially targeted to distinct subcellular compartments: either to cytoplasmic P-bodies, by fusion to a fragment of p54/RCK (Minshall et al., 2009), or to nucleoplasmic LacO arrays, by fusion to Laci (Darzacq et al., 2006). Subsequent recruitment of interacting partners to either P-bodies or LacO arrays was then tested to assess their binding to hSpagh in the cytoplasm or the nucleus, respectively. First, corecruitment of GFP-HSP90 was tested. In cells that expressed either p54- or Laci-importin Kpna3 fusion protein as a negative control, GFP-HSP90 localized diffusely and did not concentrate in P-bodies or at the LacO array (Figure 6A). In contrast, in cells that expressed hSpagh fused to either p54 or Laci, GFP-HSP90 accumulated in P-bodies or at the LacO array, respectively (Figure 6B). Next, the localization of Rpb1 was analyzed by immunofluorescence. We found that it was weakly detectable in P-bodies containing p54-hSpagh. However, it accumulated there strongly upon depletion of Rpb2, and this was specific, because no recruitment was seen in cells expressing p54-Kpna3 in the same conditions (Figure 6C). Interestingly, p54 fusion with truncated versions of hSpagh revealed that different domains are responsible for HSP90 and Rpb1 binding (Figure S4). Moreover, Rpb1 could not be detected at the LacO array bound by Laci-hSpagh. This indicated that interaction of Rpb1 with hSpagh predominantly occurs in the cytoplasm and suggested that the nuclear pool of subunit Rpb1 is mostly present in a completely assembled enzyme that doesn’t interact with hSpagh.
Figure 6. hSpagh Interacts with HSP90 and Unassembled Subunit Rpb1 in the Cytoplasm.
(A) hSpagh interacts with HSP90 in the nucleus and the cytoplasm. U2OS cells were transfected with the indicated constructs and processed for epifluorescence microscopy. The p54 and Laci constructs were also fused to a red fluorescent protein. Each field is 38 × 38 μm. Insets: zoom on the boxed area (4.3 × 4.3 μm).
(B) hSpagh interacts with subunit Rpb1 in the cytoplasm. U2OS cells were transfected with the indicated construct and siRNAs and then processed for immunofluorescence against Rpb1. Legend is as in (A).
(C) hSpagh fails to recruit Rpb1 subunit at LacO array in the nucleoplasm. Legend is as in (A) and (B).
HSP90 and hSpagh Are Required to Stabilize Unassembled Cytoplasmic Subunit Rpb1
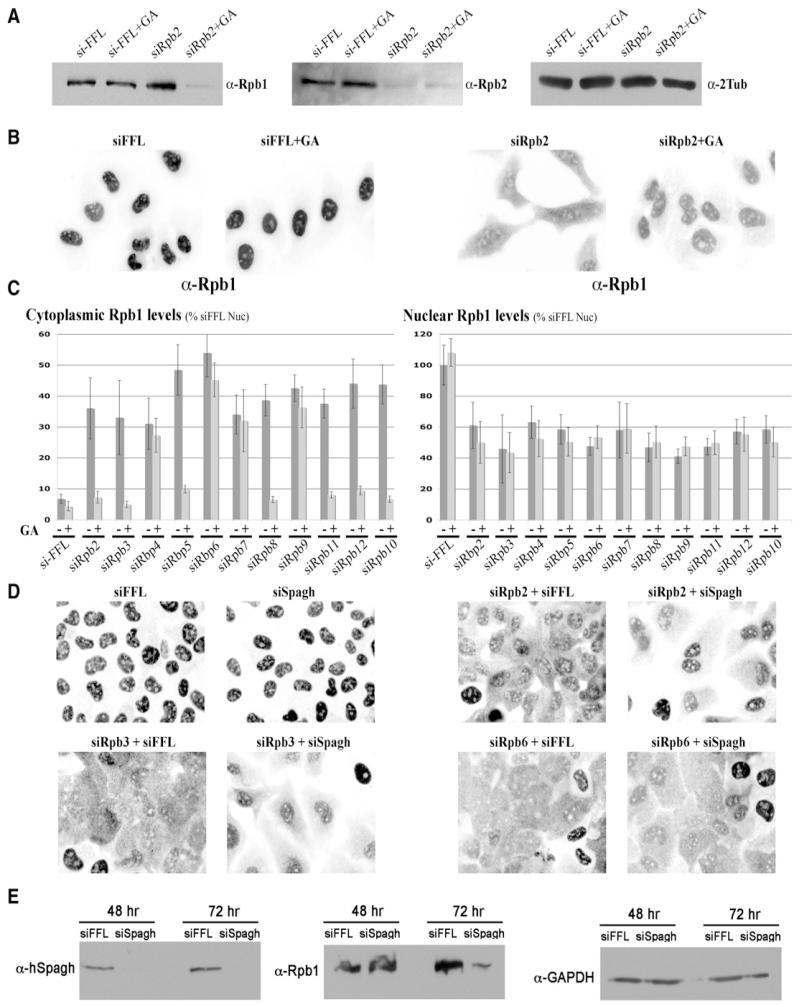
hSpagh is an HSP90 cofactor, and we thus analyzed the effects of inhibiting HSP90 activity with geldanamycin (GA). Treatment with GA usually results in the degradation of client proteins (Whitesell et al., 1994), but it had little or no effect on the accumulation of Rpb1 following an overnight treatment (Figures 7A and 7B). Because hSpagh is more specifically associated with the unassembled Rpb1 subunit, we tested the effect of GA when combined with polymerase assembly inhibition. Remarkably, GA destabilized unassembled Rpb1 that accumulated upon either α-amanitin + LMB treatment or siRNA-mediated Rpb2 depletion (Figures 7A and S1). In addition, quantification of fluorescence microscopy images revealed that GA destabilized specifically the cytoplasmic form of Rpb1 (Figure 7B). This also occurred upon siRNA-mediated depletion of every subunit except Rpb4, Rpb6, Rpb7, and Rpb9 (Figure 7C). We therefore propose that HSP90 is required to stabilize Rpb1 through most of the assembly pathway and, in particular, for the joining of Rpb1 with Rpb8, Rpb5, and with the Rpb2-Rpb3-Rpb10-Rpb11-Rpb12 subcomplex. To then test the role of hSpagh in this process, it was depleted with siRNA. We observed that removal of hSpagh mimicked the effect of HSP90 inhibition: it destabilized the cytoplasmic Rpb1 that accumulated upon depletion of either Rpb2 or Rpb3, but not Rpb6 (Figures 7D and S5). Interestingly, long-term depletion of hSpagh, for 3 days, resulted in the loss of Rpb1, consistent with hSpagh having a role in the assembly of RNA polymerase II (Figure 7E).
Figure 7. hSpagh and HSP90 Activity Stabilize Unassembled Cytoplasmic Rpb1 Subunit.
(A) Inhibition of HSP90 activity with GA selectively destabilizes the unassembled Rpb1 subunit. U2OS cells were treated for 43 hr with control siRNAs (siFFL) or with siRNA against Rpb2 (siRpb2) and incubated with GA in the last 15 hr (GA, 2 μM) when indicated. Extracts were prepared in HNTG and analyzed by western blotting with antibodies against Rpb1, Rpb2, and tubulin.
(B) Inhibition of HSP90 activity with GA selectively destabilizes cytoplasmic Rpb1 subunit. U2OS cells were treated as above and subjected to immunofluorescence against Rpb1. Each field is 83.75 × 117.25 μm.
(C) All stages of polymerase II assembly are not equally sensitive to GA. The amount of subunit Rpb1 in the cytoplasm and nucleus was quantified as follows. Subunit Rpb1 was labeled by immunofluorescence, and images of cells treated as in (B) were taken with identical microscopic settings. Background was removed, and Rpb1 signals in the nucleus and cytoplasm were normalized to the nuclear signal of Rpb1 in untreated cells. Numbers are averages of more than 80 cells; bars are the standard deviations.
(D) hSpagh stabilizes unassembled cytoplasmic Rpb1. U2OS cells were treated with the indicated siRNA for 43 hr and processed for immunofluorescence against Rpb1. Each field is 83.75 × 117.25 μm.
(E) Destabilization of Rbp1 after long-term depletion of hSpagh. U2OS cells were treated with siRNAs against hSpagh for 48 or 72 hr and analyzed by western blotting with the indicated antibodies.
DISCUSSION
In this study, we have identified a mechanism for the formation of RNA polymerase II from its subunits, which suggests that the assembly pathway occurs predominantly, if not entirely, within the cytoplasm. Analysis of the SILAC quantitative MS data permits rigorous and comprehensive evaluation of the dynamic behavior of RNA polymerase II complexes, while fluorescence microscopy assays determine their subcellular localization. This identified putative assembly intermediates containing a subset of RNA polymerase II subunits and showed that these complexes occur predominantly in the cytoplasm. Altogether, we propose a model whereby nuclear entry of RNA polymerase II is restricted to fully assembled enzymes. Previous studies on RNA polymerase I postulated the existence of an assembly compartment in the nucleolus (Dundr et al., 2002). For RNA polymerase II, our results indicate that this assembly compartment is the cytoplasm. This model does not exclude the possibility of additional nuclear reassembly mechanisms for disassembled polymerases. However, recycling of RNA polymerase II subunits may also be predominantly a cytoplasmic process, rather than occurring at polymerase II promoters. In agreement, FRAP experiments using GFP-tagged subunits Rpb1 and Rpb3 show virtually identical recovery rates at an HIV-1 promoter, with an absence of a rapid recovery phase during the first minute of the experiment. This indicates that RNA polymerase II is recruited as a preformed complex, allowing for an efficient initiation entry mode. Our data support the idea that assembly of RNA polymerase II is regulated at the level of the entire cell, rather than in a gene-specific manner, at individual promoters.
RNA polymerase II is a central enzyme required for the transcription of essentially all pre-mRNAs in eukaryotic cells. The other main macromolecular machines involved in eukaryotic gene expression, i.e., ribosomes and spliceosomes, are also composed of multisubunit complexes that are largely assembled in compartments distinct from their final functional location. Thus, ribosomes are mostly assembled within nucleoli but function in mRNA translation in the cytoplasm (Boisvert et al., 2007), while snRNPs mature in the cytoplasm but function to remove introns exclusively in the nucleus (Matera et al., 2007). Separating sites of assembly and function might be beneficial in preventing aberrant and potentially disruptive interactions between partially or incorrectly assembled complexes and their substrates.
Our proteomics data identified factors that associate specifically with the putative RNA polymerase II assembly intermediates. Further studies will be required to characterize the precise role of these different factors in RNA polymerase II biogenesis. Interestingly, RPAP2 might be the key import factor of RNA polymerase II, as it binds the fully assembled enzyme (Jeronimo et al., 2007); it is required for its import and it shuttles in an LMB-dependent manner (data not shown). In this study, we deciphered the function of hSpagh, which interacts with unassembled Rpb1, the largest RNA polymerase II subunit, and which is a cofactor for HSP90. We show that HSP90 stabilizes free Rpb1 and that Rpb1 and HSP90 bind separable domains on hSpagh (Figure S4). This suggests that hSpagh delivers unassembled Rpb1 to HSP90, thereby linking this important chaperone to the assembly mechanism of RNA polymerase II. HSP90 is unusual in that it functions at late stages of protein folding, with clients in a near-native state, whereas most chaperones function at earlier stages to facilitate the folding and maturation of nascent proteins (Wandinger et al., 2008). HSP90 thus appears ideally suited to facilitate assembly of protein complexes. Recent systematic studies on HSP90 have suggested a large number of potential client proteins, with diverse functions, but few in vivo roles have been clearly demonstrated (McClellan et al., 2007; Zhao et al., 2005). Here, we provide evidence that HSP90 and its cochaperone hSpagh coordinate the assembly of RNA polymerase II in the cytoplasm. The precise mode of action of HSP90 will require detailed structural data, but possible roles could be to maintain Rpb1 in a conformation compatible with assembly, to prevent association with incorrect partners, and/or to provide a quality-control mechanism to ensure that the enzyme is properly assembled. Our data also suggest that HSP90 may buffer a transient imbalance in free subunit stoichiometry, which could, for instance, arise upon spontaneous, stochastic fluctuations in gene expression (Raser and O’Shea, 2005).
Purification of tagged hSpagh identified not only Rpb1, but also the large subunits of RNA polymerases I and III (Jeronimo et al., 2007 and this study). By western blot analysis, binding of subunit RPA194 to hSpagh was barely detectable under normal conditions but increased markedly upon depletion of subunit RPA135 (Figure 5C). This indicates that hSpagh preferentially associates with unassembled RPA194, suggesting that HSP90 and hSpagh may be involved in the assembly of all three RNA polymerases. HSP90 and hSpagh are also involved in the production of snoRNPs (Boulon et al., 2008), and they could thus assemble the machineries that produce tRNAs, mRNAs, and ribosomes. These molecules form the translational apparatus, whose activity directly controls production of the cellular mass (White, 2005). We therefore propose that the function of HSP90 in snoRNP and RNA polymerase assembly may contribute to its established role in cell proliferation and oncogenic transformation (Calderwood et al., 2006).
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Antibodies
U2OS cells were cultivated in DMEM plus antibiotics and 10% FBS. Cells were transfected with Ca-P coprecipitates or with lipofectamine and plus reagent (Invitrogen). Drugs were used at the following concentrations: actinomycin D, 5 μg/ml; DRB, 100 μM; α-amanitin, 10 μg/ml; GA, 2 μM; LMB, 15 nM, except for the SILAC experiments, where it was reduced to 8 nM. The sequence of siRNAs is given in the Supplemental Information.
L30-GFP-Rpb3 was described previously (Boireau et al., 2007). The other vectors were made using the Gateway technology (Invitrogen), with donor vectors from the human ORFeome (Lamesch et al., 2007) or generated by PCR. Detailed maps are available upon request. hSpagh TPR1 and TPR2 domains encompassed nucleotides 390–690 and 850–1160, respectively.
Antibodies against Rpb1 recognized the CTD heptads-repeat (both phosphorylated and unphosphorylated forms [Figure S1]) and were mouse monoclonal from Euromedex (PB-7C2) (Souffelweyersheim, France). Antibodies against Rpb5 and RPA194 were mouse monoclonals from Euromedex (PB-4H7) and Santa Cruz (C-1), respectively. Antibody against Rpb2 was a goat polyclonal from Santa Cruz (S20). GFP-TRAP beads were previously described and contained a recombinant llama monoclonal and monochain antibody against GFP (Chromotek; Planegg-Martinsried, Germany) (Rothbauer et al., 2008). Tubulin was detected using a rat antibody from Serotec (MCA78G) (Colmar, France) and GAPDH using an antibody from Santa Cruz (FL-335).
Two-Hybrid Assays
Two-hybrid assays were performed as previously described (Boulon et al., 2008).
Cellular Imaging
For analyses in fixed cells, cells were grown on glass coverslips, treated as indicated, and fixed in 4% formaldehyde, 1× PBS for 30 min at RT. After permeabilization in 0.5% Triton for 10 min at RT and labeling with the appropriate antibodies, slides were mounted in Vectashield. For quantitative imaging of Rpb1 localization, images were captured with a 40× objective using identical settings. Average gray levels in the cytoplasm and nucleus were measured for a large number of cells (between 80 and 200). Background levels were taken in an area devoid of cells and subtracted to cytoplasmic and nuclear values. Signals from individual cells were then averaged and normalized to the nuclear signal of untreated cells.
For FRAP analyses in live cells, U2OS Exo1 cells were transfected with GFP-Rpb3 and MS2-Cherry vectors, transcription sites were bleached, and fluorescence recovery was recorded as previously described (Boireau et al., 2007). For studies of GFP-Rpb1, U2OS Exo1 cells were transfected with a GFP-tagged α-amanitin-resistant form of subunit Rpb1 (Sugaya et al., 2000), and clones were selected in 5 μg/ml α-amanitin. Individual clones were checked for expression of the HIV-1 reporter gene and GFP-Rpb1, and suitable cells were then transfected with an MS2-Cherry vector and analyzed by FRAP.
Immunoprecipitation
Cells were rinsed in PBS and lysed in HNTG for 20 min at 4°C. Cellular debris was removed by centrifugation (10 min at 20,000 g), and extracts were incubated with appropriate beads (2 hr at 4°C). The beads were washed four times in PBS/Triton 0.1% and resuspended in Laemmli buffer. Bound proteins were analyzed by western blotting. HNTG contained 20 mM HEPES (pH 7.9), 150 mM NaCl, 1% Triton, 10% glycerol, 1 mM MgCl2, 1 mM EGTA, and protease inhibitors (Roche).
Proteomic Analyses
The SILAC procedures are described in detail in the Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Kress for the gift of DsRed2-p54/RCK and the idea of using it to target fusion proteins to P-bodies. We thank M. Vidal for the gift of the human ORFeome 3.1 and U. Rothbauer and H. Leonhardt for the gift of GFP-TRAP beads. We thank R. Bordonné for critical reading of the manuscript. A.I.L. is a Wellcome Trust Principal Research Fellow. S. Boulon is a long-term fellow of the Human Frontier Science Program (HFSP), and D.M. had a fellowship from ANRS. This work was supported by grants from SIDACTION; ANRS; La Ligue Contre le Cancer; ARC; ANR; Eurasnet; PROSPECTS (PROteomics SPECification in Time and Space), which stems from the European Commission’s FP7 (GA HEALTH-F4-2008-201648); and RASOR (Radical Solutions for Researching the proteome).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, Supplemental References, five figures, and six tables and can be found with this article online at doi:10.1016/j.molcel.2010.08.023.
REFERENCES
- Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Bäcker V, Kornblihtt A, Marcello A, Bertrand E. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jády BE, Rothé B, Pescia C, Robert MC, Kiss T, et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Cloutier P, Al-Khoury R, Lavallée-Adam M, Faubert D, Jiang H, Poitras C, Bouchard A, Forget D, Blanchette M, Coulombe B. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods. 2009;48:381–386. doi: 10.1016/j.ymeth.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Singer RH. The dynamic range of transcription. Mol. Cell. 2008;30:545–546. doi: 10.1016/j.molcel.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales FA, Zanchin NI, Luz JS, Oliveira CC. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 2005;346:437–455. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol. Cell. 2008;30:486–497. doi: 10.1016/j.molcel.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell. Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kédinger C, Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Rudd MD, Luse DS. Amanitin greatly reduces the rate of transcription by RNA polymerase II ternary complexes but fails to inhibit some transcript cleavage modes. J. Biol. Chem. 1996;271:21549–21558. doi: 10.1074/jbc.271.35.21549. [DOI] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. USA. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT. Regulation of TATA-binding protein dynamics in living yeast cells. Proc. Natl. Acad. Sci. USA. 2008;105:13304–13308. doi: 10.1073/pnas.0801901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, Mancini MA. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Sugaya K, Vigneron M, Cook PR. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 2000;113:2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, Lamond AI. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J. Biol. Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.