Abstract
Tenomodulin (Tnmd) is a well-known gene marker for the tendon and ligament lineage, but its exact functions in these tissues still remain elusive. In this study, we investigated Tnmd loss of function in mouse tendon stem/progenitor cells (mTSPC) by implicating a previously established Tnmd knockout (KO) mouse model. mTSPC were isolated from control and Tnmd KO tail tendons and their stemness features, such as gene marker profile, multipotential, and self-renewal, were compared. Immunofluorescence and reverse transcriptase-polymerase chain reaction analyses for stem cell-, tenogenic-, osteogenic-, and chondrogenic-related genes confirmed their stemness and lineage specificity and demonstrated no profound differences between the two genotypes. Multipotential was not significantly affected since both cell types differentiated successfully into adipogenic, osteogenic, and chondrogenic lineages. In contrast, self-renewal assays validated that Tnmd KO TSPC exhibit significantly reduced proliferative potential, which was also reflected in lower Cyclin D1 levels. When analyzing possible cellular mechanisms behind the observed decreased self-renewability of Tnmd KO TSPC, we found that cellular senescence plays a major role, starting earlier and cumulating more in Tnmd KO compared with control TSPC. This was accompanied with augmented expression of the cell cycle inhibitor p53. Finally, the proliferative effect of Tnmd in TSPC was confirmed with transient transfection of Tnmd cDNA into Tnmd KO TSPC, which rescued their proliferative deficit. Taken together, we can report that loss of Tnmd affects significantly the self-renewal and senescence properties, but not the multipotential of TSPC.
Introduction
Tendons and ligaments are unique forms of connective tissues responsible for the integrity and mobility of the musculoskeletal system. The genetic profile of tendons and ligaments overlap in large with that of cartilage and bone, and presently, only few tissue-specific genes have been identified. One such gene is tenomodulin (Tnmd), which was discovered in 2001 by two different groups [1,2]. Initial northern blot data showed that Tnmd transcripts are expressed in muscle, diaphragm, and eye [1].
Additional northern blot, reverse transcriptase-polymerase chain reaction (RT-PCR), and in situ hybridization analyses revealed that the expression signal is prevalent in skeletal tendons and ligaments as well as in heart chordae tendineae cordis [1–5]. Hence, Tnmd was classified as a tendon gene marker.
Tnmd belongs to the family of type II transmembrane glycoproteins with a highly conserved cleavable C-terminal cysteine-rich domain. Immunohistochemical stainings in tendons and chordae tendineae have shown that the C-terminus is deposited in the extracellular matrix [3,4]. This domain has been proposed to be the functional part of the protein since Oshima et al. found out that the C-terminus of Tnmd is sufficient to inhibit vascular tube formation in vitro and suppression of tumor growth in vivo [6]. The extracellular part contains a BRICHOS domain, which was found in other genes associated with dementia, respiratory distress, and cancer; however, the exact role of this domain in Tnmd is still unclear [7,8].
Up to now, the binding partners of Tnmd and the signaling pathway, to which Tnmd belongs to, have not been identified, hence making the interpretation of the exact functions of Tnmd in tendons and ligaments very difficult. The first insight on the role of Tnmd in tendons and ligaments was derived from the establishment of a Tnmd knockout (KO) mouse line. Its characterization revealed no severe developmental phenotype, but reduced tendon cell density and proliferation in vivo. Furthermore, ultrastructural analyses of Tnmd-deficient Achilles tendons demonstrated that the collagen fibril size was pathologically increased, indicating impaired collagen fibrilogenesis and a status corresponding to premature matrix aging [3]. In addition, Komiyama et al. [9] recently showed that Tnmd is expressed in periodontal ligaments during murine tooth eruption. Together with the decreased cell adhesion of Tnmd KO mouse fibroblasts, it was proposed that Tnmd also acts on the maturation or maintenance of periodontal ligaments by positively regulating cell adhesion. In summary, the above studies suggested that Tnmd plays a regulating role on cell proliferation and adhesion in tendons and ligaments. Moreover when Tnmd is ablated, tendon tissues show signs of pathological aging.
A major breakthrough in the tendon field was the study by Bi et al. [10], which demonstrated that human and mouse tendons harbor a unique cell population, named tendon stem/progenitor cells (TSPC). These possess universal stem cell characteristics such as clonogenicity, self-renewal capacity, and multipotency [10]. Compared with bone marrow-derived mesenchymal stem cells (MSC), the TSPC expressed high levels of Scleraxis (Scx), a tendon-specific transcription factor, and Tnmd. Furthermore, the isolated cells showed the ability to regenerate tendon-like tissue after extended expansion in vitro and transplantation in vivo. This study also showed that TSPC reside within a unique niche, where two extracellular matrix proteins biglycan and fibromodulin control their function by modulating bone morphogenetic protein (BMP) signaling. The double KO model for these two proteoglycans is characterized by increased tendon cellularity together with decreased collagen fibril thickness, a phenotype somewhat opposite to that of Tnmd KO mice. Moreover, biglycan/fibromodulin-deficient TSPC had augmented clonogenicity and cell proliferation, but reduced type I collagen and Scx expression. Interestingly, Bi et al. were the first to show that there is a link between distorted TSPC functions and tendon pathology. TSPC within biglycan/fibromodulin-deficient tendon niche were far more responsive to BMP signaling leading to TSPC favoring the osteogenic lineage, which in turn resulted in the so called in-tendon ossification [10]. The above data suggested that the molecular environment provided by the niche is essential for the correct maintenance and differentiation of the stem/progenitor cells during development and repair.
Following the idea that altered TSPC properties can lead to tendon pathology, in this study we aimed to compare the basic cell functions of TSPC derived from control and Tnmd-KO tendons. We first focused on studying their gene transcription, multipotential, cell proliferation, and senescence, and then we performed rescue experiments by reintroducing Tnmd full length (FL) and C-terminus encoding cDNAs in Tnmd-deficient TSPC.
Materials and Methods
Mouse TSPC isolation and cell culture
Mouse TSPC (mTSPC) were isolated from Tnmd KO and control (wild type or heterozygous) tail tendons of 6-month-old mice. Husbandry, handling and sacrifice of Tnmd KO and control mice prior mTSPC isolation were strictly carried out according to the guidelines of the Bavarian authorities. To obtain a sufficient amount of mTSPC, a pool of 3–5 mouse tails per genotype were used. For this, tendon tissues were enzymatically treated overnight with collagenase type II (Worthington) in Dulbecco's modified Eagle's medium (DMEM)/Hams (1:1) (Biochrom) supplemented with 10% fetal bovine serum (FBS), l-ascorbic-acid-2-phosphate (both Sigma) and minimum essential medium (MEM)-Amino Acid (Biochrom). Then, the cell suspension was filtered, centrifuged at 500 g for 5 min, and resuspended in fresh culture media. mTSPC were grown at 37°C and 5% CO2 and kept up to 80% maximum confluence as the culture media was changed every third day. Cells in passages 1–5 were used for experiments.
Immunofluorescence
mTSPC were grown on 20 μg/mL collagen type I-coated slides and fixed with 4% paraformaldehyde. After permeabilization and blocking with 2% bovine serum albumin/phosphate buffered saline (PBS), cells were incubated overnight at 4°C with primary antibodies against CD146 (Millipore), CD105 (R&D Systems), CD90.2, CD73, and CD44 (all Novus Biologicals), Sca-1 (Abcam), Nestin (Proteintech), Nanog (R&D Systems), Scx (Abcam), and Tnmd [3]. Then, corresponding Alexa Fluor 488-conjugated secondary antibodies and nuclear dye 4′,6-diamidino-2-phenylindole (DAPI; both Life Technology) were applied at room temperature. Photomicrographs were taken on the Observer Z1 microscope equipped with the Axiocam MRm camera (Carl Zeiss). Immunofluorescence experiments were reproduced twice or thrice independently and representative images are shown.
Semiquantitative and quantitative PCR
Total RNA was extracted with the RNeasy Mini Kit (Qiagen). For cDNA synthesis, 1 μg total RNA and AMV First-Strand cDNA Synthesis Kit (Invitrogen) were used. Semiquantitative PCR was performed with Taq DNA Polymerase (Invitrogen) in MGResearch instrument (Bio-Rad). For primer sequences and PCR conditions (Table 1). For densitometric analysis, PCR bands were imaged on the Vilber Lourmat gel imager (Eberhardzell) and analyzed with the BioCapt software provided by the imager. Values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as fold change. Quantitative PCR was performed in a LightCycler 1.5 instrument equipped with the LightCycler 3.5 software (Roche). Primer kits for Scx and hypoxanthine-guanine phosphoribosyltransferase (HPRT) from Search-LC, and p16, p21, p53, and GAPDH from Qiagen were used in combination with the LightCycler Fast Start DNA Master SYBR Green Kit (Roche). Crossing points for each sample were determined by the second derivative maximum method and relative quantification was performed using the comparative ΔΔCt method. The relative gene expression was calculated as a ratio to HPRT or GAPDH. All PCR results have been reproduced minimum two independent times.
Table 1.
Semi-Quantitative Polymerase Chain Reaction Primers and Conditions
| Target gene | Primers | Annealing temperature (°C) | Cycle number | Product size (bp) | Reference |
|---|---|---|---|---|---|
| Stem cell-and lineage-related genes | |||||
| Aggrecan | F 5′-ttgccagggggagttgtattc-3′ R 5′-gacagttctcacgccaggtttg-3′ |
56 | 30 | 540 | Self-designed |
| aP2 | F 5′-ctactgctgctgctactctg-3′ R 5′-tggcttcacgacctgttc3′ |
53 | 33 | 185 | Self-designed |
| Biglycan | F 5′-gacaaccgtatccgcaaag-3′ R 5′-ccaggtgaagttcgttcag-3′ |
52 | 28 | 208 | Self-designed |
| Bone sialoprotein | F 5′-tgtctgctgaaacccgttc-3′ R 5′-ggggtctttaagtaccggc-3′ |
54 | 30 | 633 | Self-designed |
| CD31 | F 5′-ctgccagtccgaaaatggaac-3′ R 5′-cttcatccaccggggctatc-3′ |
54 | 33 | 218 | Self-designed |
| Collagen IIα1 | F 5′-ggtttggagagaccatgaac-3′ R 5′-tgggttcgcaatggattgtg-3′ |
55 | 30 | 463 | [36] |
| Collagen Iα2 | F 5′-gaacggtccacgattgcatg-3′ R 5′-ggcatgttgctaggcacgaag-3′ |
55 | 33 | 167 | Self-designed |
| Eya1 | F 5′-ttgtgggcaggcatcaag-3′ R 5′-agccgggataagacggatag-3′ |
53 | 28 | 238 | Self-designed |
| Fibromodulin | F 5′-catggcaaccagattacc-3′ R 5′-agatataaggccgtgagg-3′ |
52 | 28 | 212 | Self-designed |
| MyHC3 | F 5′-cttcacctctagccggatggt-3′ R 5′-aattgtcaggagccacgaaaat-3′ |
57 | 33 | 107 | Self-designed |
| MyoD1 | F 5′-ccactccgggacatagacttg-3′ R 5′-aaaagcgcaggtctggtgag-3′ |
57 | 33 | 109 | Self-designed |
| Nanog | F 5′-gaaatcccttccctcgccatc-3′ R 5′-ctcagtagcagacccttgtagc-3′ |
58 | 38 | 161 | Self-designed |
| Oct-4 | F 5′-tcaggttggactgggcctagt-3′ R 5′-ggaggttccctctgagttgctt-3′ |
57 | 38 | 100 | Self-designed |
| Osterix | F 5′-gaaaggaggcacaaagaag-3′ R 5′-caccaaggagtaggtgtgtt-3′ |
54 | 30 | 348 | [34] |
| PPARγ | F 5′-ctccgtgatggaagaccactc-3′ R 5′-agactcggaactcaatggc-3′ |
52 | 33 | 275 | Self-designed |
| Runx2 | F 5′-atgcgtattcctgtagatccg-3′ R 5′-ttggggaggatttgtgaagac-3′ |
56 | 30 | 606 | Self-designed |
| Six1 | F 5′-accagttctcgcctcacaatc-3′ R 5′-ttttcggtgttctccctttcc-3′ |
57 | 28 | 358 | [35] |
| Sox9 | F 5′-tggcagaccagtacccgcatct-3′ R 5′-tctttcttgtgctgcacgcgc-3′ |
55 | 30 | 136 | [36] |
| VEGFα | F 5′-tcactgtgagccttgttcaga-3′ R 5′-tttacctcaagctgcctcgcc-3′ |
49 | 33 | 117 | Self-designed |
| Cell cycle | |||||
| Cyclin D1 | F 5′-ctggccatgaactacctgga-3′ R 5′-atccgcctctggcattttgg-3′ |
56 | 28 | 279 | [37] |
| Tenomodulin and FLAG | |||||
| Tnmd (Fig. 1) | F 5′-aactccacctcagcagtagtcc-3′ R 5′-tttcttggatacctcgggccagaa-3′ |
51 | 35 | 249 | Self-designed |
| Tnmd (Fig. 5) | F 5′-gactacaaagacgatgacgacaag-3′ R 5′-cctcgacggcagtaaatacaac-3′ |
51 | 30 | FL: 836 Cterm: 248 |
Self-designed |
| Housekeeping gene | |||||
| GAPDH | F 5′-caactacatggtttacatgttc-3′ R 5′-gccagtggactccacgac-3′ |
50 | 30 | 181 | [38] |
Primer sequences used for quantitative polymerase chain reaction are not included in the table, because they are the property of the manufacturers of the Primer Kits (Search-LC for Scleraxis, and Qiagen for p16, p21, and p53).
FL, PCR product of Tnmd full length cDNA; Cterm, PCR product of Tnmd C-terminus cDNA.
Adipogenic, osteogenic, and chondrogenic differentiation assays
Three-lineage differentiation protocols were carried out as described in Alberton et al. [11] with slight modifications. In brief, for adipogenic differentiation, 8×103 cells/cm2 mTSPC were seeded in triplicates in six-well plates, and were cultivated in an induction media for 5 days [DMEM-high glucose with 10% FBS, 1 μM dexamethasone, 0.2 mM indomethacin, 0.1 mg/mL insulin, and 1 mM 3-isobutyl-1-methylxanthine and (all Sigma)] followed by 2 days in preservation media [DMEM-high glucose medium supplemented with 10% FBS, 0.1 mg/mL insulin (all Sigma)]. The process was repeated for 21 days. The adipogenic differentiation was estimated by staining and digital signal quantification of Oil Red O (Sigma). Using automatic color pixel quantification tool in the Adobe Photoshop CS5 software, the Oil Red O-positive areas were estimated and calculated in percentage to the image total pixel size. For osteogenic differentiation 4×103 cells/cm2 were seeded in triplicates in six-well plates. Then, the osteogenic stimulation media [DMEM-high glucose medium (PAA) supplemented with 10% FBS, 10 mM β-glycerophosphate, 50 μM l-ascorbic acid 2-phosphate, 100 nM dexamethasone (all Sigma)] was applied every 3 days for a period of 21 days. The extent of osteogenic differentiation was determined with Alizarin Red staining and quantification by using Osteogenic Quantification Kit (Millipore), as recommended by the manufacturer.
For chondrogenic differentiation, TSPC were preconditioned during monolayer expansion in hypoxia incubator (Sanyo) for 4 days. Next, 4.5×105 cells/pellet were spun down in V-bottom 96-well plates and stimulated for 28 days with differentiation media composed of DMEM-high glucose, 10 μM dexamethasone, 1 nM sodium pyruvate, 0.195 mM l-ascorbic acid, and 1% insulin transferrin selenium (all Sigma) supplemented with 10 ng/mL transforming growth factor (TGF)β1, and 100 ng/mL BMP2 (both R&D Systems). The extent of chondrogenic differentiation was evaluated with Safranin Orange staining for cartilage glycosaminoglycans. Using the polygonal tool of the Image Pro Plus software (Media Cybernetics), the Safranin Orange-positive area was measured and results were shown as percentage of the total pellet area. In the adipogenic and osteogenic differentiation protocols, unstimulated cells maintained in DMEM-high glucose with 10% FBS were used as controls, whereas in the chondrogenic differentiation protocol, the control pellets were cultured in differentiation media without growth factors. Photomicrographs were taken with the AxioCam ICc3 color camera mounted on Axiovert 40 CFL microscope (Carl Zeiss). All differentiation assays were reproduced twice independently (consisting of triplicates), and in each adipogenic and chondrogenic experiments four images were digitally analyzed.
Population doubling analysis
Long-term cell growth was evaluated by calculation of cumulative population doubling (PD) and population doubling time (PDT). The following formulas were used:
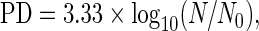 |
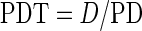 |
In PD, N is the cell number at the end of the experiment and N0 at the beginning. Growth curves were built by calculating cumulative PD, which is a sum of individual PDs. For PDT, D is the number of hours of the experimental period N0 to N.
Colony unit forming assay
Clonogenic potential of the mTSPC was performed as described in Alberton et al. [11]. Shortly, 26 cells/cm2 were seeded in 10 cm Petri dishes. After 14 days of cultivation, formed colonies were visualized with 0.5% crystal violet/methanol staining and imaged. Colony unit forming (CFU) efficiency was calculated as follows: CFU=(number of colonies/number of plated cells)×100. CFU assay was reproduced twice in triplicates.
WST-1 assay
The WST-1 Proliferation Kit (Roche) was used according to the manufacturer's recommendations. mTSPC at a density of 15×103 cells/cm2 were plated in triplicates in 96-well dishes. After 48 h of cultivation, cells were incubated with the WST-1 reagent for 4 h. Optical density was measured using the microtiter-plate reader (Thermo Scientific) at a wavelength of 420 nm with reference filter at 620 nm. The WST-1 experiments were carried out thrice independently.
β-Galactosidase assay
Cellular senescence was visualized by the Senescent Histochemical Staining Kit (Sigma). mTSPC (15×102 cells/cm2) were plated in triplicates and incubated for 3 days, fixed with fixation buffer, and incubated with staining solution overnight at 37°C. Then, the staining mixture was replaced with 70% glycerol and images were taken with the AxioCam ICc3 color camera mounted on the Axiovert 40 CFL microscope (Carl Zeiss). Percentage of senescent cells was calculated as the number of blue cells versus the total cell count from 10 images per genotype. Senescence assays were carried out twice independently in passages 3–5.
Detection of p16, p21, and p53 on tendon tissue sections
Paraffin Achilles tendon sections (7 μm thick) from 6-month-old mice were collected with Microtome (Hyrax M55; Carl Zeiss) and treated with 0.2% hyaluronidase/PBS for 30 min. After washing, the sections were blocked with the Mouse-on-Mouse kit (Vector Laboratories) and then incubated with primary mouse antibodies against p16 (Santa Cruz Biotechnology), p21 (BD Biosciences), and p53 (Abcam) overnight at 4°C. Secondary Alexa Flour 546 antibody (Life Technology) was applied for 1 h at room temperature, followed by DAPI staining for 5 min. Fluorescent images were taken with the Axiocam MRm camera on the Observer Z1 microscope (Carl Zeiss). Two independent experiments were performed.
Transfection of mTSPC
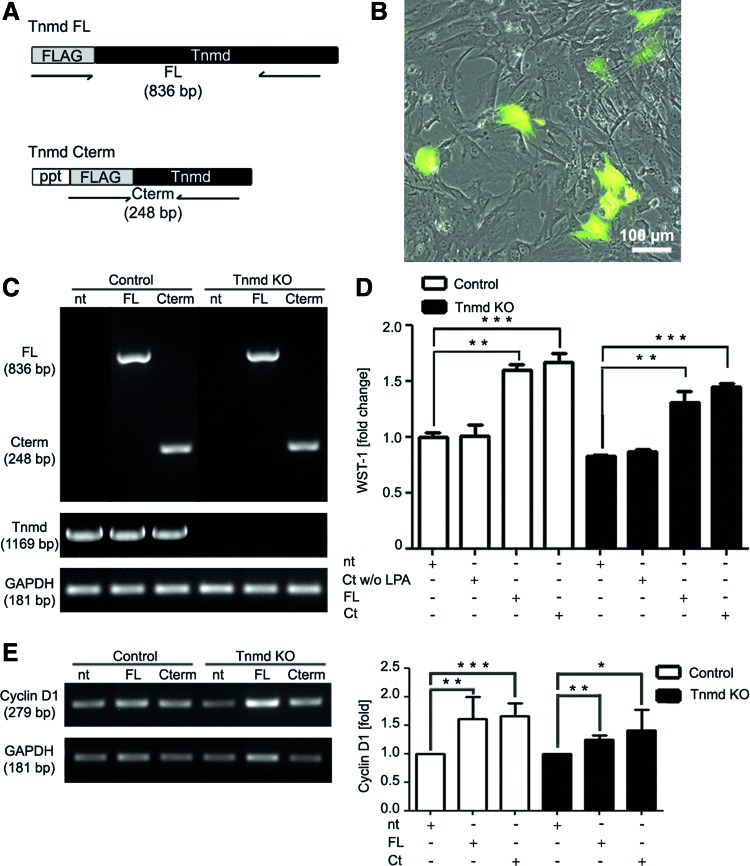
mTSPC were transiently transfected with three plasmids carrying cDNA encoding: (1) FL Tnmd, (2) C-terminal domain of Tnmd (Cterm) and (3) enhanced green fluorescent protein (EGFP) (positive control). Both of the Tnmd plasmids were kindly provided by Prof. Chisa Shukunami, Kyoto University, Japan. The transfection protocol was previously described in Alberton et al. [11]. In brief, 2×104 cells/cm2 mTSPC were transfected with 2.5 μg plasmid DNA using the Opti-MEM media and the Lipofectamine 2000 Kit (Life Technology). After 6 h, transfection media was replaced with complete growth media and cells were maintained in culture for another 2 days. The transfection efficiency was monitored by the GFP expression. Posttransfection, cells were used for the RNA isolation or WST-1 assays. Negative controls used were: nontransfected mTSPC and cells treated with the Cterm plasmid without Lipofectamine. Transfection experiments were reproduced two independent times.
Statistics
Quantitative data were analyzed with the GraphPad Prism 5 software (GraphPad). Bar charts show mean values and standard deviations. All quantitative data were acquired from two or three independent experiments, each performed in triplicates. Unpaired t-test was used and a P-value of 0.05% was considered statistically significant and it was indicated in the figures as follows *P<0.05, **P<0.005, ***P<0.0005.
Results
Control and Tnmd-KO mTSPC possess stem cell- and tendon-related gene expression
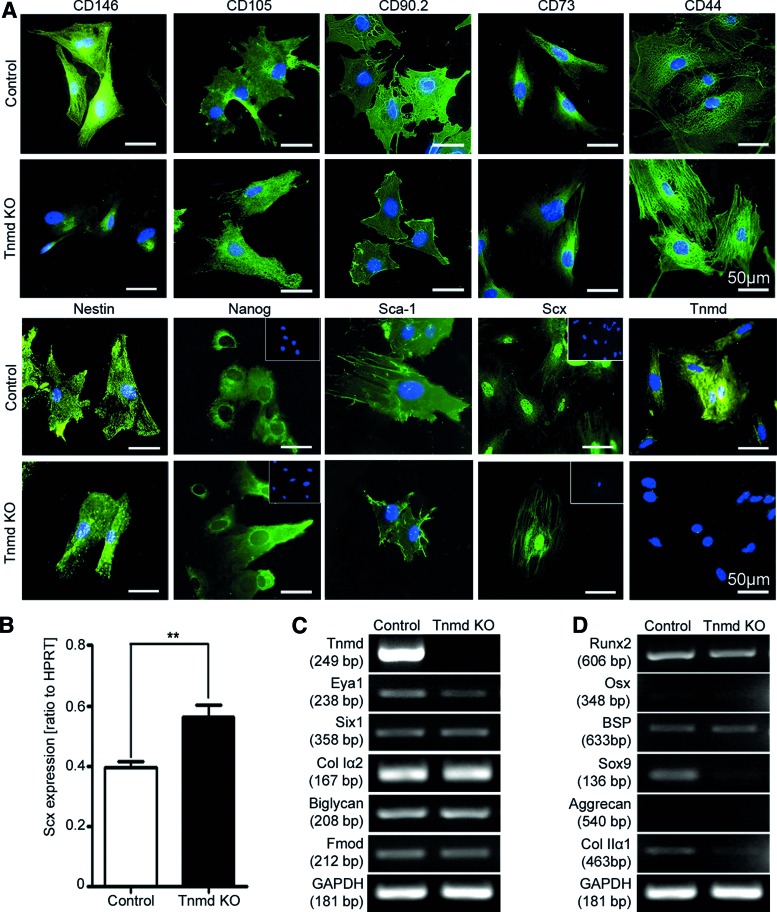
To compare the stem cell profile between control and Tnmd KO mTSPC, we first examined the expression of well-known stem cell markers CD146, CD105, CD90.2, CD73, CD44, Nestin, Nanog, and Sca-1 [12–17]. The immunocytochemical results clearly demonstrated that the eight tested markers were expressed in both mTSPC types (Fig. 1A). To verify the tenogenic origin of the mTSPC we performed immunocytochemical stainings for Tnmd and the tendon-specific transcription factor Scx (Fig. 1A). The Scx antibody staining revealed that both control and Tnmd KO mTSPC have the typical nuclear expression. In comparison, Tnmd staining was observed only in control mTSPC whereas Tnmd KO cells showed no Tnmd expression confirming that in these cells Tnmd is deleted. Next, we screened our cells for the expression of characteristic genes for tenogenic, osteogenic, and chondrogenic lineages. For this, we performed quantitative PCR for Scx and semiquantitative PCR analyses for the tendon-related—Tnmd, transcription factors Eya1 and Six1, collagen type I (Col Iα2), and proteoglycans, biglycan, and fibromodulin; bone-related—transcription factors Runx2 and Osterix, and matrix protein bone sialoprotein; cartilage-related—transcription factor Sox9, and matrix proteins aggrecan and collagen type II (Col IIα1). Interestingly, in Tnmd KO mTSPC we observed significantly higher levels of Scx expression in comparison to the controls (Fig. 1B). The semiquantitative PCR analysis for Tnmd validated the lack of Tnmd transcripts and confirmed the protein data. Both mTSPC types demonstrated comparable expression of Eya1, Six1, Col Iα2, biglycan, and fibromodulin (Fig. 1C). With regard to the osteogenic lineage markers, we detected low mRNA levels of Runx2 and bone sialoprotein, but not of Osterix gene (Fig. 1D). Among the chondrogenic lineage markers, we observed weak expression of Sox9 and collagen type 2 only in control mTSPC (Fig. 1D), whereas aggrecan expression was not detected in any of the samples.
FIG. 1.
Expression profile of mTSPC. (A) Immunocytochemical detection of stem cell markers CD146, CD105, CD90.2, CD73, CD44, Nestin, Nanog, and Sca-1 and tenogenic markers Scx and Tnmd. Insets show nuclear DAP1 stainings. (B) Quantitative PCR for Scx. **P<0.005. Semiquantitative PCR for tendon- (C), bone- and cartilage-related genes (D). Staining and PCRs were performed with mTSPC in passage 3, and were repeated minimum two independent times. mTSPC, mouse tendon stem/progenitor cells; PCR, polymerase chain reaction; KO, knockout; Scx, Scleraxis; Tnmd, tenomodulin; HPRT, hypoxanthine-guanine phosphoribosyltransferase; Col, collagen; Fmod, fibromodulin; Osx, osterix; BSP, bone sialoprotein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Color images available online at www.liebertpub.com/scd
To further verify the mTSPC purity we performed additional semiquantitative PCR analyses for the following markers: Oct-4 and Nanog (stem cells); myogenic differentiation 1 (MyoD1) and myosin heavy chain 3 (MyHC3, muscle); peroxisome proliferator-activated receptor gamma (PPARγ) and adipose protein 2 (aP2, adipose), and vascular endothelial growth factor alpha (VEGFα) and CD31 (vessels) (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd). Our results demonstrated basal expression levels of the transcription factors Nanog, PPARγ, and aP2, which were comparable between both cell types, and no expression of muscle- and vessel-related markers.
Taken together, our results confirmed the TSCP character of the obtained cells and furthermore, demonstrated that control and Tnmd KO cells have very similar expression of stem cell- and lineage-characteristic genes.
Multipotential is permissive in Tnmd KO mTSPC
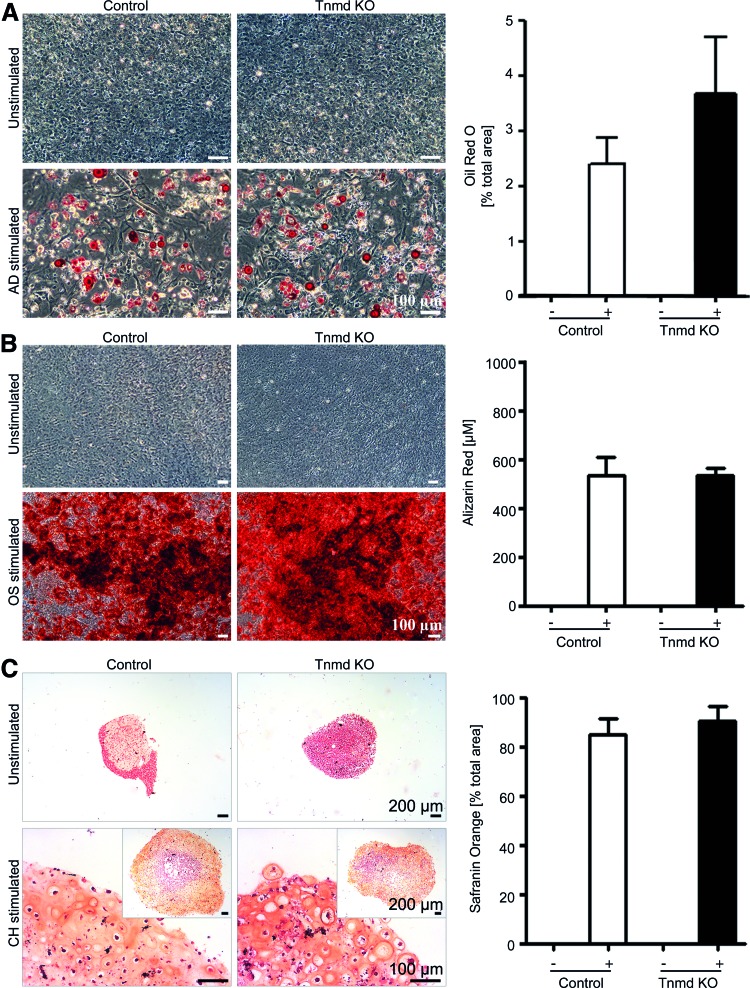
To rule out whether control and Tnmd KO mTSPC preserved their plasticity in vitro and to investigate if Tnmd deficiency could influence mTSPC differentiation toward other mesenchymal lineages, we subjected the mTSPC to three typical differentiation protocols, namely adipogenic, osteogenic, and chondrogenic. By performing the Oil Red O, Alizarin Red, and Safranin Orange stainings, we observed that both mTSPC types were able to form lipid vacuoles (Fig. 2A), to mineralize their extracellular matrix (Fig. 2B), and to deposit abundant amount of glycosaminoglycans (Fig. 2C). The quantitative analyses of the accumulated Oil Red O, Alizarin Red, and Safranin Orange dyes showed no significant differences between the control and Tnmd KO mTSPC. These results suggested that loss of Tnmd has no profound effect on mTSPC multipotential.
FIG. 2.
Three-lineage differentiation of control and Tnmd KO mTSPC. (A) On the left, representative pictures of adipogenic differentiation. Accumulated lipid vacuoles were detected by Oil Red O staining after 21 days stimulation (lipid vacuoles in red). On the right, quantitative analyses of the Oil Red O-positive areas. (B) On the left, representative pictures of osteogenic differentiation. Deposited calcified matrix was visualized by Alizarin Red staining at day 21 (calcified matrix in red). On the right, quantification of the Alizarin Red accumulation. (C) On the left, representative images of chondrogenic differentiation. Cartilage glycosaminoglycans were visualized by Safranin Orange staining at day 28 (glycosaminoglycans in orange, cell nuclei in dark red). On the right, quantification analyses of positively stained areas. In (A–C), unstimulated mTSPC were used as negative controls. Differentiation protocols were carried out in triplicates with cells in passage 1 and were reproduced twice independently. AD, adipogenic; OS, osteogenic; CH, chondrogenic; −, unstimulated; +, stimulated. Color images available online at www.liebertpub.com/scd
Tnmd KO mTSPC have impaired capacity to self-renew
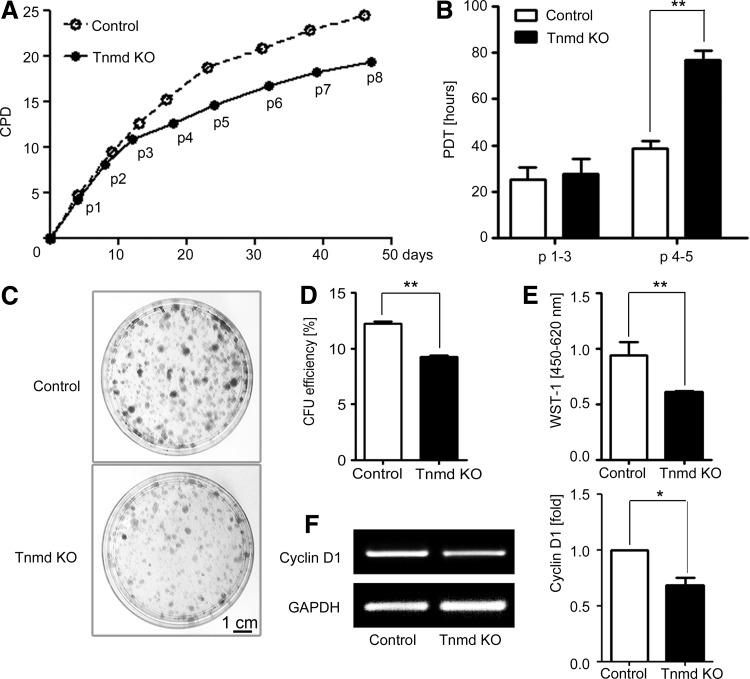
To compare the self-renewability of control and Tnmd KO mTSPC, we first performed a long-term proliferation study by estimating their PDs for a period of 50 days. We found that at the beginning of the growth curve, both mTSPC types divided similarly (Fig. 3A). However, already at passage 3 the Tnmd KO cells demonstrated a reduced proliferation rate, and the differences between the two cell populations further increased with in vitro cultivation. These changes were even more apparent when we calculated the PDTs of both mTSPC types at two different time points from their growth curves (Fig. 3B). At passage 3, control and Tnmd KO cells had PDT of 25.4±5.1 and 27.6±6.5 h, respectively. At passage 5, the PDT of control cells was 38.7±2.9 h, whereas the PDT of Tnmd KO cells was significantly increased to 76.9±3.9 h. Next, by performing the CFU assays, we detected that Tnmd KO cells have limited clonogenic potential (Fig. 3C), as their CFU efficiency was only 9.2±0.2% in contrast to 12.2±0.2% of the control cells (Fig. 3D). Short-term proliferation analysis with the WST-1 reagent further confirmed that Tnmd KO mTSPC have a significantly lower proliferative activity than control mTSPC (Fig. 3E). Finally, we examined the expression of Cyclin D1 gene, which drives G1/S phase transition of the cell cycle, and found that Tnmd KO mTSPC have significantly decreased with∼25% Cyclin D1 mRNA levels (Fig. 3F). In summary, by implicating different experimental methods we clearly demonstrated that Tnmd is an important regulator of mTSPC proliferation because its loss results in significantly impaired self-renewal of mTSPC.
FIG. 3.
Investigation of mTPSC self-renewal capacity. (A) Long-term proliferation was assessed by estimating cumulative population doubling for a period of 50 days (corresponding to eight consecutive passages). (B) Calculation of population doubling time of control and Tnmd KO mTSPCs from early passages (p1–3) and middle passages (4–5). (C) Colony-forming unit assay. Formed colonies were visualized with crystal violet staining after 14 days. (D) CFU efficiency was estimated by counting colony numbers. CFU assays were performed in triplicates and reproduced twice independently. (E) The WST-1 proliferation assay (average of three independent experiments). (F) Semiquantitative PCR for Cyclin D1 (left panel) and densitometric quantification of PCR bands (right panel). PCRs were reproduced three times. *P<0.05, **P<0.005. CPD, cumulative population doubling, PDT, population doubling time; p, passage; CFU, colony forming unit.
Cellular senescence is augmented in Tnmd KO mTSPC
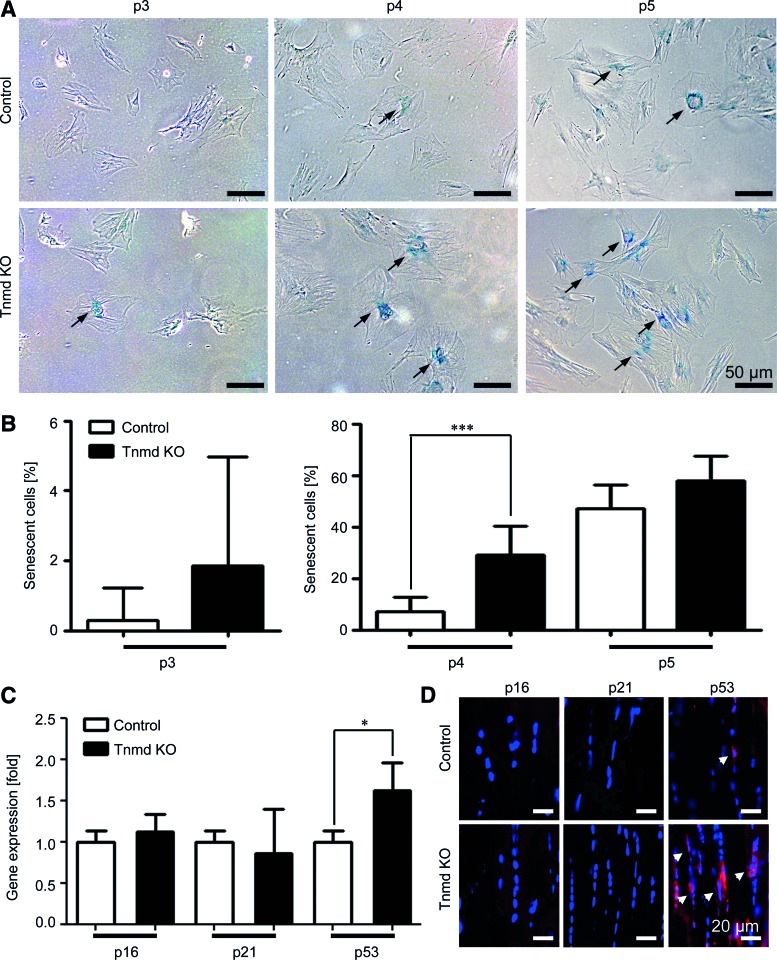
Following our major observation of the reduced self-renewability of Tnmd KO mTSPC, we investigated the possibility that these cells might undergo earlier senescence or activate apoptosis. For this, we first performed histochemical stainings for β-galactosidase activity, an enzyme that is highly active in senescent cells. We analyzed control and Tnmd KO cells from three different passages 3, 4, and 5, and found that already at passage 3 there are senescent cells present in the Tnmd KO mTSPC population (Fig. 4A). Quantification of the percentage of senescent cells revealed that at passage 3 control mTSPC had almost no senescent cells, whereas 2% of Tnmd KO cells were senescent (Fig. 4B). At passage 4, we observed more senescent cells in both cell types, yet the numbers were significantly higher in Tnmd KO (7.04±1.1% and 28.9±3.6% for control and Tnmd KO cells, correspondingly). At passage 5, the number of senescent cells increased in control cells (47.0±4.2%), nevertheless it remained lower than in Tnmd KO cell, which at this point had 58.0±4.3% β-galactosidase-positive cells. Next, we investigated the mRNA levels of three well-known cell cycle inhibitors and senescence gene markers, namely p16, p21, and p53. Quantitative PCR analysis demonstrated a slight elevation of p16 and significant increase of p53 expression in Tnmd KO TSPC, whereas p21 levels were unchanged (Fig. 4C). To confirm this finding on protein level, we performed immunofluorescent detection of cells expressing p16, p21, or p53 in collected Achilles tendon sections. Our results showed a higher prevalence of p53-positive cells in Tnmd KO tendons in comparison to the controls, which further validated the PCR data (Fig. 4D).
FIG. 4.
Cellular senescence analyses. (A) Representative images of mTSPC stained for active β-galactosidase (arrows) at three different passages (p3–5). (B) Quantification of senescent cell numbers. For each passage, β-galactosidase staining was repeated two independent times in triplicates. (C) Quantitative PCR for cell cycle inhibitors and senescent markers p16, p21, and p53. PCRs were performed three independent times. (D) Representative images of p16, p21, and p53 immunofluorescent detection in vivo. Stainings were reproduced twice using Achilles tendon sections from 6-month-old mice. *P<0.05, ***P<0.0005. Color images available online at www.liebertpub.com/scd
At the end, by using TUNEL and JC-1 assays, we analyzed both cell types for apoptosis in passages 3–5. TUNEL method is based on terminal transferase dUTP nick end labeling, in which the nuclei of apoptotic cells are labeled in fluorescent green. JC-1 is 5,50,6,60-tetrachloro-1,10,3,30-tetraethylbenzimidazolylcarbo-cyanine iodide dye and in viable cells, it aggregates in the intact mitochondria and produces red fluorescent signal. In apoptotic cells, which have a mitochondrial leakage, JC-1 resides in the cytoplasm in a monomeric form and stains the cell cytoplasm green (experimental protocols are given in Supplementary Data). Both assays clearly showed no sign of apoptosis neither in control nor in Tnmd KO mTSPC (Supplementary Fig. S2). Thus, we concluded that the reduced self-renewal potential of Tnmd KO mTSPC is due to their early entry into cellular senescence, which we confirmed by demonstrating that this cell population contains higher number of senescent cells along with upregulated p16 and p53.
Transfection of full or truncated Tnmd cDNA rescues the proliferation deficit of Tnmd KO mTSPC
To clarify that indeed the lack of Tnmd expression in mTSPC is the sole reason for their reduced proliferative capacity, we transfected transiently control and Tnmd KO mTSPC with plasmids encoding either FL or C-terminal domain (Cterm) of the Tnmd gene (Fig. 5A). To control transfection efficiency and to estimate the optimal time of gene expression in our primary mTSPC, we also used an EGFP-expressing plasmid (Fig. 5B). Posttransfection, one part of the cells were used for mRNA isolation and semiquantitative PCR, which confirmed the successful expression of FL or Cterm transgenes in mTSPC (Fig. 5C). The other part of cells was implicated for a short-term proliferation analysis with WST-1 assay (Fig. 5D). Hence, we determined that when Tnmd KO cells were transfected with both types of Tnmd plasmids, their proliferation rate increased significantly. Interestingly, re-expressing just Tnmd C-terminal domain was sufficient to rescue the proliferative deficit of Tnmd KO mTSPC. Furthermore, we observed a significantly enhanced proliferation also in transfected control mTSPC suggesting an additive effect of the introduced FL or Cterm transgenes to the endogenous Tnmd expression. To further confirm the observation drawn by the WST-1 assay, we performed the semiquantitative PCR analysis for Cyclin D1, where a significant upregulation correlated with the transfection of Tnmd FL or Cterm plasmids in both cell types (Fig. 5E). Taken together, our results showed that the ectopic expression of full or truncated Tnmd cDNA rescued the impaired self-renewal of Tnmd KO mTSPC; hence confirming the positive role of Tnmd in regulating the proliferation of mTSPC.
FIG. 5.
Rescue of Tnmd expression in Tnmd KO mTSPC. (A) Schematic representation of the Tnmd full length and Tnmd C-terminal plasmids used for transfection. Transfections were carried out two independent times. (B) A representative picture of EGFP-transfected mTSPC, which was used to monitor transfection efficiency. (C) Upper panel: PCR analysis for the expression of the transfected Tnmd FL and Tnmd Cterm cDNA performed with primer sets indicated in (A). Lower panel: PCR for the expression of endogenous Tnmd. PCRs were performed twice independently. (D) The WST-1 proliferation assay with control and Tnmd KO mTSPC after transfection. Fold change was calculated to WST-1 value of nontransfected control mTSPC. The WST-1 experiments were reproduced twice in triplicates. (E) Semiquantitative PCR for Cyclin D1 (right panel) and densitometrical quantification of the PCR product (left panel). PCR were repeated twice independently. *P<0.05, **P<0.005, ***P<0.0005. FL, full length; Cterm, C-terminal; ppt, pre-pro-trypsin secretion signal; nt, nontransfected; LPA, Lipofectamine; Ct w/LPA, C-terminal without Lipofectamine; Ct, C-terminal. Color images available online at www.liebertpub.com/scd
Discussion
Understanding the molecular nature and function of tendons is crucial for the development of successful therapeutic strategies for tendon repair. To date, one of the main problems in tendon biology is the shortage of tendon-specific genes distinguishing tendon stem, progenitor, and mature cells from other mesenchymal cell types. Therefore, enriching our knowledge on the tenogenic markers and their role in tendon tissue and cell niche is essential for understanding tendon development, maintenance, and healing. In view of the above, we focused our investigation on Tnmd, which is one of the few known tendon marker genes. Our data on the Tnmd KO mouse suggested that Tnmd plays an in vivo regulatory function in tendon cell density and proliferation as well as in collagen fibril maturation [3]. However the mechanisms behind the observed phenotype remain unknown. In 2007, Bi et al. [10] also reported that tendons enclose their own TSPC and furthermore, suggested that distorted TSPC functions relate to abnormalities in tendon tissues. Therefore, by implementing TSPC isolated from Tnmd KOs and control mice, in this study we analyzed if the loss of Tnmd leads to significant changes in mTSPC functions in vitro. Hence providing an explanation on the observed in vivo phenotype as well as novel information on a potential role of Tnmd in TSPC.
First, to validate the stem/progenitor character of the obtained mTSPC, we analyzed the expression of several adult stem cell- and mesenchyme-related genes. Immunocytochemical stainings confirmed the presence of the MSC markers CD146, CD105, CD90.2, CD73, CD44, Nestin, and Sca-1 in control and Tnmd KO mTSPC [10,14,16]. Analysis of the early stem cell markers Oct-4 and Nanog revealed no expression of Oct-4, but weak expression of Nanog on mRNA and protein levels. RT-PCR profiling of genes related to tenogenic, osteogenic, and chondrogenic lineages showed strong expression of the transcription factors Scx, Eya1, and Six1, and the matrix proteins Col Iα1, biglycan, and fibromodulin, all related to tendon cell lineage. Regarding the mesenchymal lineages, we detected a basal expression of the osteogenic Runx2, chondrogenic Sox9, and adipogenic PPARγ and aP2 transcription factors. Interestingly, in vivo data by Sugimoto et al. [18] have shown the existence of Scx and Sox9 double-positive progenitors during limb development and proposed that they are a unique multipotent cell population that can commit into tenocytes, ligamentocytes, and chondrocytes. In addition, this study suggested that a fine interplay between Sox9 and Scx expression levels determines the commitment of the early progenitors toward one or the other lineage. In our mTSPC we detected also a very weak PCR band for bone sialoprotein and collagen type II, which we attributed to the minor presence of osteoblast and chondrocyte-like cells possibly deriving from the tendon enthesis side, which contain such cells. However, our additional PCR data, including muscle (MyoD1 and MyHC3) and vessel (VEGFα and CD31)-related markers clearly excluded contamination of muscle and blood cells. In summary, the obtained mTSPC in this study demonstrated a phenotype that is comparable to the mTSPC described in the Bi et al. [10]. The cells expressed basic levels of a variety of mesenchyme-related transcription factors, but demonstrated enriched tendon-related gene marker expression. Interestingly, our gene expression screening suggested that the deficiency of Tnmd has an effect only on the expression of Scx, Sox9, and Col IIα1, whereas the expression levels of the other markers were comparable between control and Tnmd KO mTSPC. Two studies by Brent et al. [19] and Shukunami et al. [20], showed that Tnmd expression domains coincide with those of Scx. An overexpression of Scx in cultured chicken tenocytes triggered the upregulation of Tnmd [20], whereas in Scx KO mice, the Tnmd expression was profoundly diminished [21]. The above finding led to the suggestion that Tnmd is a direct target of Scx. However, to clarify whether the observed changes of Scx, Sox9, and Col IIα1 basal expression in Tnmd KO TSPC represents a feedback response to the loss of Tnmd, will need further investigation. In summary, our results on the expression of mesenchymal stem- and lineage-specific genes clearly validated the TSPC character of our cells and are in line with the characterization data of other human and mTSPC published by Bi et al. [10], Tempfer et al. [16], Zhou et al. [22], Mienaltowski et al. [23], and Kohler et al. [14].
Next, we performed three lineage differentiation experiments to investigate if Tnmd plays a role in mTSPC commitment. Bi et al. [10] have shown that extracellular proteins can affect the TSPC differentiation capacity since upon loss of byglican and fibromodulin, mTSPC from the double mutant mice were exhibiting an increased osteogenic potential due to altered BMP signaling. With regard to Tnmd deficiency in mTSPC, we observed by specific stainings for adipocytes, osteoblasts, and chondrocytes that both cell types were able to form lipid vacuoles, mineralize their extracellular matrix, and deposit cartilage glycosaminoglycans, suggesting no profound differentiation phenotype due to the lack of Tnmd. Still, similarly to Zhou et al. [22], who have demonstrated that TSPC from aged rats have enhanced adipogenic differentiation, we detected that Tnmd KO mTSPC exhibit slight inclination to differentiate into adipocytes in comparison to control mTSPC. Interestingly, Tolppanen et al. [24,25] and Saiki et al. [26] have suggested a possible role of Tnmd in obesity, therefore, to further clarify if lower levels of Tnmd correlate with increased adipogenesis of mTSPC, follow-up studies using mTSPC from Tnmd KO mice in different developmental ages or fed on high calorie diet will be relevant. Altogether, we concluded that mTSPC multipotential is permissive and not profoundly affected by the loss of Tnmd expression.
Distinctive hallmarks of stem cells are their clonogenic and self-renewal abilities, which together with asymmetric division determine stem cells as the major cell type responsible for tissue homeostasis and regeneration. Thus, our next aim was to evaluate whether Tnmd deficiency will result in some changes in mTSPC colony formation and proliferation in vitro. Long-term analyses of the control and Tnmd KO mTSPC growth curves revealed decreased self-renewability of the Tnmd KO cell pool. Tnmd KO cells displayed inferior proliferative capacity and significantly increased PDT already after passage 3 and had significantly lower number of colonies. Based on the quantitative results from the WST-1 assays, we found that also short-term proliferation was negatively influenced by the loss of Tnmd. The above functional analyses were additionally confirmed on a molecular level by RT-PCR for the well-known proliferative marker—Cyclin D1, which demonstrated reduced mRNA levels in Tnmd KO mTSPC. Finally, we performed in parallel Tnmd rescue and overexpression experiments in Tnmd KO and control cells, respectively. The obtained results clearly showed that, first, the Tnmd transfection indeed results in rescue of the proliferation deficit of Tnmd KO mTSPC and also it can further enhance the proliferation of control cells, and second, that the effect of Tnmd C-terminus is identical to that of the FL Tnmd cDNA. Based on the above observation we convincingly prove that Tnmd is a positive regulator of mTSPC self-renewal and that this action is mediated by the C-terminal part of the molecule. Our findings corroborate with the previous studies on the antiangiogenic effect of Tnmd and its homologous gene chondromodulin-I in vascular tube formation and tumor growth, which have evidently proposed that the C-terminus is the functional domain of these proteins (reviewed in Shukunami et al. [27], Shukunami and Hiraki [28]). It has been also reported that the C-terminal domain of chodromodulin-I can strongly induce the proliferation of chondrocytes in vitro, which is similar to our novel result on the positive effect of Tnmd on TSPC self-renewal [3,29]. Interestingly, the data by Shukunami's group and ours demonstrate that Tnmd and chondromodulin-I can exert diverse proliferative effects depending on the cell type. They can powerfully suppress the proliferation of endothelial cells, whereas they can stimulate the proliferation of tenocytes and chondrocytes. Up to now, the signaling pathways in which Tnmd and chondromodulin-I participate are not known mostly due to their novel protein domain structure as well as the lack of knowledge about the potential binding partners of these proteins. Therefore, we can only speculate that Tnmd might be a cofactor regulating the function of growth factors or growth factor receptors and hence, depending on the availability, act as stimulator or inhibitor of proliferation. However, to clarify the molecular mode of action of Tnmd, future research focusing on identifying Tnmd-binding proteins is of upmost importance.
Finally, we investigated for possible reasons behind the reduced proliferative capacity of Tnmd KO mTSPC, as we tested if these cells are entering earlier into senescence or if they might have higher apoptotic index. Based on two different apoptotic assays, we excluded apoptosis as a major cause, which is also in line with a lack of increased apoptotic cell numbers in Tnmd-deficient tendon tissues reported in Docheva et al. [3]. In contrast, when we studied senescence by β-galactosidase stainings and quantitative PCR analysis for cell cycle inhibitory gene, we found higher number of senescent cells in Tnmd KO cell population, which was accompanied by slightly upregulated levels of p16 and significantly increased p53 mRNAs. Similar changes in TSPC functions, such as reduced self-renewal and increased senescence, were detected during human tendon aging and degeneration [14]. Analyzing human TSPC, obtained from Achilles tendons of young and aged patients, it was observed that aged TSPC exhibit profound self-renewal and clonogenic deficits as well as premature entry into senescence and significant upregulation of p16 expression. Other studies with rat TSPC [22] and mouse bone marrow MSC [30] have also shown that there is a reduction in stem cell frequency in aged tissues as well as when isolated in vitro the aged stem cells proliferated significantly slower than their counterparts obtained from young animals. Shibata et al. [31], Stolzing et al. [32], and Kasper et al. [33] have reported augmented senescence features of MSC from aged bone marrow aspirates. Hence, we suggest that the altered self-renewal and senescence abilities of Tnmd-deficient mTSPC result in reduced cell proliferation and density detected in Tnmd KO tendons in vivo. In our study, we could not compare the TSPC availability in control and Tnmd-KO mice in vivo, because up to date, the exact location of TSPC in tendon tissues has not been mapped due to the lack of unique markers discriminating pure stem cells from progenitors and mature tenocytes.
Taken together, in this study, we report novel data on the role of Tnmd in mTSPC proliferation and senescence in vitro. Loss of Tnmd resulted in reduced mTSPC clonogenicity and self-renewal together with higher incidence of senescence, but it had no profound effect on TSPC multipotentiality. By rescuing the proliferation deficit of Tnmd KO cells with transfection of exogenous Tnmd cDNA, we could clearly demonstrate the positive effect of Tnmd on the self-renewal of the tendon stem cell niche. We believe that our findings on Tnmd role in TSPC will provide a new insight into the molecular mechanisms regulating TSPC and will be helpful in understanding tendon development and repair processes where these cells are majorly involved.
Supplementary Material
Acknowledgments
D.D. acknowledges the German Research Foundation (DFG grant. DD 1414/3-1). P.A. and D.D. acknowledge the support of the Bavarian Research Foundation (Project: DOK-100-08). C.P. was supported by the German Research Foundation (DFG grant: PO 1718/1-1). M.S. current address is at Amgen GmbH, Munich.
Author Disclosure Statement
The authors have nothing to disclose.
References
- 1.Brandau O, Meindl A, Fassler R. and Aszodi A. (2001). A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Dev Dyn 221:72–80 [DOI] [PubMed] [Google Scholar]
- 2.Shukunami C, Oshima Y. and Hiraki Y. (2001). Molecular cloning of tenomodulin, a novel chondromodulin-I related gene. Biochem Biophys Res Commun 280:1323–1327 [DOI] [PubMed] [Google Scholar]
- 3.Docheva D, Hunziker EB, Fassler R. and Brandau O. (2005). Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol Cell Biol 25:699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura N, Shukunami C, Hakuno D, Yoshioka M, Miura S, Docheva D, Kimura T, Okada Y, Matsumura G, et al. (2008). Local tenomodulin absence, angiogenesis, and matrix metalloproteinase activation are associated with the rupture of the chordae tendineae cordis. Circulation 118:1737–1747 [DOI] [PubMed] [Google Scholar]
- 5.Takimoto A, Oro M, Hiraki Y. and Shukunami C. (2012). Direct conversion of tenocytes into chondrocytes by Sox9. Exp Cell Res 318:1492–1507 [DOI] [PubMed] [Google Scholar]
- 6.Oshima Y, Sato K, Tashiro F, Miyazaki J, Nishida K, Hiraki Y, Tano Y. and Shukunami C. (2004). Anti-angiogenic action of the C-terminal domain of tenomodulin that shares homology with chondromodulin-I. J Cell Sci 117:2731–2744 [DOI] [PubMed] [Google Scholar]
- 7.Sanchez-Pulido L, Devos D. and Valencia A. (2002). BRICHOS: a conserved domain in proteins associated with dementia, respiratory distress and cancer. Trends Biochem Sci 27:329–332 [DOI] [PubMed] [Google Scholar]
- 8.Willander H, Hermansson E, Johansson J. and Presto J. (2011). BRICHOS domain associated with lung fibrosis, dementia and cancer–a chaperone that prevents amyloid fibril formation?. FEBS J 278:3893–3904 [DOI] [PubMed] [Google Scholar]
- 9.Komiyama Y, Ohba S, Shimohata N, Nakajima K, Hojo H, Yano F, Takato T, Docheva D, Shukunami C, Hiraki Y. and Chung UI. (2013). Tenomodulin expression in the periodontal ligament enhances cellular adhesion. PLoS One 8:e60203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi Y, Ehirchiou D, Kilts TM, Inkson CA, Embree MC, Sonoyama W, Li L, Leet AI, Seo BM, et al. (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat Med 13:1219–1227 [DOI] [PubMed] [Google Scholar]
- 11.Alberton P, Popov C, Pragert M, Kohler J, Shukunami C, Schieker M. and Docheva D. (2012). Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev 21:846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann JM, Riethmuller G. and Johnson JP. (1989). MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc Natl Acad Sci U S A 86:9891–9895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baksh D, Yao R. and Tuan RS. (2007). Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cells 25:1384–1392 [DOI] [PubMed] [Google Scholar]
- 14.Kohler J, Popov C, Klotz B, Alberton P, Prall WC, Haasters F, Muller-Deubert S, Ebert R, Klein-Hitpass L, et al. (2013). Uncovering the cellular and molecular changes in tendon stem/progenitor cells attributed to tendon aging and degeneration. Aging Cell 12:988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lendahl U. and McKay RD. (1990). The use of cell lines in neurobiology. Trends Neurosci 13:132–137 [DOI] [PubMed] [Google Scholar]
- 16.Tempfer H, Wagner A, Gehwolf R, Lehner C, Tauber M, Resch H. and Bauer HC. (2009). Perivascular cells of the supraspinatus tendon express both tendon- and stem cell-related markers. Histochem Cell Biol 131:733–741 [DOI] [PubMed] [Google Scholar]
- 17.Spring FA, Dalchau R, Daniels GL, Mallinson G, Judson PA, Parsons SF, Fabre JW. and Anstee DJ. (1988). The Ina and Inb blood group antigens are located on a glycoprotein of 80,000 MW (the CDw44 glycoprotein) whose expression is influenced by the In(Lu) gene. Immunology 64:37–43 [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto Y, Takimoto A, Akiyama H, Kist R, Scherer G, Nakamura T, Hiraki Y. and Shukunami C. (2013). Scx+/Sox9+progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 140:2280–2288 [DOI] [PubMed] [Google Scholar]
- 19.Brent AE, Braun T. and Tabin CJ. (2005). Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development 132:515–528 [DOI] [PubMed] [Google Scholar]
- 20.Shukunami C, Takimoto A, Oro M. and Hiraki Y. (2006). Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol 298:234–247 [DOI] [PubMed] [Google Scholar]
- 21.Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ. and Schweitzer R. (2007). Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134:2697–2708 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Akinbiyi T, Xu L, Ramcharan M, Leong DJ, Ros SJ, Colvin AC, Schaffler MB, Majeska RJ, Flatow EL. and Sun HB. (2010). Tendon-derived stem/progenitor cell aging: defective self-renewal and altered fate. Aging Cell 9:911–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mienaltowski MJ, Adams SM. and Birk DE. (2013). Regional differences in stem cell/progenitor cell populations from the mouse achilles tendon. Tissue Eng Part A 19:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolppanen AM, Pulkkinen L, Kolehmainen M, Schwab U, Lindstrom J, Tuomilehto J, Uusitupa Mand the Finnish Diabetes Prevention Study Group. (2007). Tenomodulin is associated with obesity and diabetes risk: the Finnish diabetes prevention study. Obesity (Silver Spring) 15:1082–1088 [DOI] [PubMed] [Google Scholar]
- 25.Tolppanen AM, Kolehmainen M, Pulkkinen L. and Uusitupa M. (2010). Tenomodulin gene and obesity-related phenotypes. Ann Med 42:265–275 [DOI] [PubMed] [Google Scholar]
- 26.Saiki A, Olsson M, Jernas M, Gummesson A, McTernan PG, Andersson J, Jacobson P, Sjoholm K, Olsson B, et al. (2009). Tenomodulin is highly expressed in adipose tissue, increased in obesity, and down-regulated during diet-induced weight loss. J Clin Endocrinol Metab 94:3987–3994 [DOI] [PubMed] [Google Scholar]
- 27.Shukunami C, Oshima Y. and Hiraki Y. (2005). Chondromodulin-I and tenomodulin: a new class of tissue-specific angiogenesis inhibitors found in hypovascular connective tissues. Biochem Biophys Res Commun 333:299–307 [DOI] [PubMed] [Google Scholar]
- 28.Shukunami C. and Hiraki Y. (2007). Chondromodulin-I and tenomodulin: the negative control of angiogenesis in connective tissue. Curr Pharm Des 13:2101–2112 [DOI] [PubMed] [Google Scholar]
- 29.Inoue H, Kondo J, Koike T, Shukunami C. and Hiraki Y. (1997). Identification of an autocrine chondrocyte colony-stimulating factor: chondromodulin-I stimulates the colony formation of growth plate chondrocytes in agarose culture. Biochem Biophys Res Commun 241:395–400 [DOI] [PubMed] [Google Scholar]
- 30.Katsara O, Mahaira LG, Iliopoulou EG, Moustaki A, Antsaklis A, Loutradis D, Stefanidis K, Baxevanis CN, Papamichail M. and Perez SA. (2011). Effects of donor age, gender, and in vitro cellular aging on the phenotypic, functional, and molecular characteristics of mouse bone marrow-derived mesenchymal stem cells. Stem Cells Dev 20:1549–1561 [DOI] [PubMed] [Google Scholar]
- 31.Shibata KR, Aoyama T, Shima Y, Fukiage K, Otsuka S, Furu M, Kohno Y, Ito K, Fujibayashi S, et al. (2007). Expression of the p16INK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem Cells 25:2371–2382 [DOI] [PubMed] [Google Scholar]
- 32.Stolzing A, Jones E, McGonagle D. and Scutt A. (2008). Age-related changes in human bone marrow-derived mesenchymal stem cells: consequences for cell therapies. Mech Ageing Dev 129:163–173 [DOI] [PubMed] [Google Scholar]
- 33.Kasper G, Mao L, Geissler S, Draycheva A, Trippens J, Kuhnisch J, Tschirschmann M, Kaspar K, Perka C, Duda GN. and Klose J. (2009). Insights into mesenchymal stem cell aging: involvement of antioxidant defense and actin cytoskeleton. Stem Cells 27:1288–1297 [DOI] [PubMed] [Google Scholar]
- 34.Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R. and Yoneda T. (2008). BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem 283:29119–29125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salingcarnboriboon R, Yoshitake H, Tsuji K, Obinata M, Amagasa T, Nifuji A. and Noda M. (2003). Establishment of tendon-derived cell lines exhibiting pluripotent mesenchymal stem cell-like property. Exp Cell Res 287:289–300 [DOI] [PubMed] [Google Scholar]
- 36.Gouttenoire J, Bougault C, Aubert-Foucher E, Perrier E, Ronziere MC, Sandell L, Lundgren-Akerlund E. and Mallein-Gerin F. (2010). BMP-2 and TGF-beta1 differentially control expression of type II procollagen and alpha 10 and alpha 11 integrins in mouse chondrocytes. Eur J Cell Biol 89:307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y. and Takeuchi T. (2009). A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem 284:733–739 [DOI] [PubMed] [Google Scholar]
- 38.Bocker W, Docheva D, Prall WC, Egea V, Pappou E, Rossmann O, Popov C, Mutschler W, Ries C. and Schieker M. (2008). IKK-2 is required for TNF-alpha-induced invasion and proliferation of human mesenchymal stem cells. J Mol Med (Berl) 86:1183–1192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.