Abstract
Human umbilical cord mesenchymal stromal cells (hUC-MSCs) of Wharton's jelly origin undergo adipogenic, osteogenic, and chondrogenic differentiation in vitro. Recent studies have consistently shown their therapeutic potential in various human disease models. However, the biological effects of major pregnancy complications on the cellular properties of hUC-MSCs remain to be studied. In this study, we compared the basic properties of hUC-MSCs obtained from gestational diabetes mellitus (GDM) patients (GDM-UC-MSCs) and normal pregnant women (N-UC-MSCs). Assessments of cumulative cell growth, MSC marker expression, cellular senescence, and mitochondrial function-related gene expression were performed using a cell count assay, senescence-associated β-galactosidase staining, quantitative real-time reverse transcription–polymerase chain reaction, immunoblotting, and cell-based mitochondrial functional assay system. When compared with N-UC-MSCs, GDM-UC-MSCs showed decreased cell growth and earlier cellular senescence with accumulation of p16 and p53, even though they expressed similar levels of CD105, CD90, and CD73 MSC marker proteins. GDM-UC-MSCs also displayed significantly lower osteogenic and adipogenic differentiation potentials than N-UC-MSCs. Furthermore, GDM-UC-MSCs exhibited a low mitochondrial activity and significantly reduced expression of the mitochondrial function regulatory genes ND2, ND9, COX1, PGC-1α, and TFAM. Here, we report intriguing and novel evidence that maternal metabolic derangement during gestation affects the biological properties of fetal cells, which may be a component of fetal programming. Our findings also underscore the importance of the critical assessment of the biological impact of maternal–fetal conditions in biological studies and clinical applications of hUC-MSCs.
Introduction
Various types of human stem cells have been isolated from a variety of sources, including embryos, bone marrow, fetal tissue, and cord blood. The stem cells obtained can be classified into embryonic stem cells, mesenchymal stromal cells (MSCs), and hematopoietic stem cells. These different types of stem cells have distinct biological characteristics and differentiation potentials. While embryonic stem cells are capable of differentiating into almost all tissues in the human body, the use of embryos and fetal tissue poses significant ethical problems [1]. Hematopoietic stem cells have a tendency to differentiate only into blood and blood-related cellular lineages. These practical limitations have led investigators to develop newer strategies in cell replacement therapies, and induced pluripotent stem cells are emerging as a promising cell source candidate in regenerative medicine, although many hurdles remain to be overcome, such as genomic instability and safe and effective delivery of reprogramming factors [2].
MSCs, which express defined surface markers, including CD90, CD73, and CD105, and lack expression of CD45, CD34, and HLA-DR, can differentiate to osteoblasts, adipocytes, and chondroblasts in vitro [3,4]. Even though their survival and differentiation potential into various cell lineages in vivo remain controversial, many studies have clearly shown the therapeutic efficacy of MSCs, mostly mediated by their paracrine effects. Although human MSCs have been traditionally isolated from adult tissues, such as the bone marrow and adipose tissue, recent studies show that human umbilical cord MSCs (hUC-MSCs) are a promising cell resource for therapeutic use [5–7]. UC-MSCs are isolated from Wharton's jelly of the umbilical cord and are distinguished from umbilical cord blood MSCs [8]. UC-MSCs are myofibroblast-like stromal cells with unique properties. They can be easily isolated in large quantities from postnatal cord tissues, which are usually discarded after birth. Moreover, UC-MSCs can be extensively expanded in culture, frozen/thawed, and also have immunomodulatory properties. Not surprisingly, a plethora of studies demonstrate the potential advantages of UC-MSCs in clinical applications for various diseases such as cancer, cardiovascular diseases, and osteoporosis [9–12].
Human pregnancy is inevitably associated with an intimate metabolic interaction between the mother and the fetus over a period of time. The maternal metabolic condition is closely linked to the intrauterine environment of the fetus. Fetal programming is the concept that maternal nutrition and medical risks during pregnancy such as gestational diabetes mellitus (GDM), preeclampsia, and obesity are reflected in the later life well-being and metabolism of the offspring [13]. Since fetal growth is primarily dependent on the functional integrity of the placenta, the placenta is physiologically dynamic to support proportional fetal growth, organ development, and differentiation, as well as to adapt to the mother's nutritional and metabolic states [14].
GDM is one of the common metabolic diseases during pregnancy, affecting up to about 14% of all pregnancies. It is becoming more prevalent due to the characteristics of modern living, such as an advanced maternal age at first pregnancy and changes in dietary habits [15]. Schwartz et al. [16] showed that umbilical cord insulin concentrations are strongly correlated with fetal growth and fetal insulin secretion, which support the Pedersen hypothesis that maternal hyperglycemia results in fetal hyperglycemia and macrosomia [17]. Although the exact mechanism by which GDM increases the risk of obesity and diabetes in offspring is poorly understood, recent studies indicated that placentas exposed to maternal GDM have modified patterns of DNA methylation in the leptin and adiponectin genes [18].
Collectively, these observations indicate that in utero exposure to GDM alters metabolic programming in newborns and their placenta, cord tissues, and cord blood. Thus, it is very plausible that maternal GDM can affect the cellular properties of UC-MSCs. However, there is a paucity of information regarding the relationship between metabolic derangement during pregnancy and alterations in the properties of UC-MSCs. In this study, we compared the cellular properties of UC-MSCs obtained from GDM patients (GDM-UC-MSCs) and normal pregnant women (N-UC-MSCs) to determine if GDM affects the biological features of UC-MSCs and if fetal cells show adaptive metabolic programming according to obstetrical conditions.
Materials and Methods
Ethics statement
This work was approved by the Institutional Review Board of Asan Medical Center, and all women provided written informed consent.
Isolation and expansion of UC-MSCs
The diagnosis of GDM was based on the International Association of Diabetes and Pregnancy Study Groups Consensus and Panel recommendations [19]. The umbilical cord samples were obtained at the time of cesarean delivery from GDM patients and healthy pregnant women. The cord was rinsed several times with sterile saline and sliced into 3-mm-thick pieces. The vessels and amnion were removed from cord segments. Wharton's jelly tissues were minced with scissors and digested for 3 h in Dulbecco's modified Eagle's medium (DMEM) (11995-075; Invitrogen-Gibco, Carlsbad, CA) containing 0.1% collagenase A (10103578001; Roche, Mannheim, Germany) at 37°C in a shaking incubator. To remove large pieces of tissue, the cells were filtered with a 100-μM mesh (BD Falcon, San Jose, CA) and pelleted by low-speed centrifugation at 200 g for 10 min. The isolated cells then were plated in DMEM supplemented with 10% fetal bovine serum (FBS), 50 μg/mL penicillin, and 100 μg/mL streptomycin (Invitrogen-Gibco) at 37°C in a humidified 5% CO2 incubator. The UC-MSC line, U150N6, was named according to the following system: U, umbilical cord; 150, serial preparation number; N, harvested from normal placenta (D from GDM-affected placenta); and 6, passage number. All of the placentas used in this study were subjected to histological examination to exclude cases with major placental lesions according to the criteria previously described [20].
Differentiation analysis
UC-MSCs were plated in 24-well plates at a density of 2×104 for adipogenic differentiation or 4×103 for osteogenic differentiation per well and allowed to attach overnight. Differentiation was induced using the Human Mesenchymal Stem Cell Functional Identification Kit (SC006; R&D Systems, Minneapolis, MN) according to the manufacturer's protocol in the α-minimum essential medium (11095-080; Invitrogen-Gibco). After 5 days of differentiation, total RNA was isolated using a Total RNA Mini Kit (FARBK001; Favorgen, Taiwan).
Cell growth assays
N-UC-MSCs and GDM-UC-MSCs were seeded on 12-well plates at 7,500 cells per well. After 3, 6, 9, and 12 days, the cells were counted using a hemocytometer after trypsinization. All experiments were performed using three replicates of each primary UC-MSC line.
Immunoblot analysis
Whole-cell lysates were prepared in the cell lysis buffer (9803; Cell Signaling Technology, Beverly, MA) containing protease inhibitor and phosphatase inhibitor cocktails (BP-477; Boston BioProducts, Worcester, MA), separated using 10% or 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes. Blots were probed with the following primary antibodies: CD105 (AF-1097; R&D Systems), CD90 (sc-9163; Santa Cruz, Santa Cruz, CA), CD73 (AP-2014; Abgent, San Diego, CA), p16 (sc-468; Santa Cruz), p53 (05-224; Millipore, Billerica, MA), phospho-p53 (Ser15) (9284; Cell Signaling Technology), and β-actin (cs-47778; Santa Cruz). TFAM antibodies were prepared in our laboratory [21]. Blots were developed using the SuperSignal West Pico Chemiluminescent Substrate (34080; Thermo Scientific, Rockford, IL).
Senescence-associated β-galactosidase staining assay
Cells were seeded in six-well plates at a density of 3×105 cells per well and allowed to attach overnight. The cells were stained using a beta-Galactosidase Staining Kit (K802-250; BioVision, Milpitas, CA). Briefly, cells were fixed for 15 min at room temperature in 3% formaldehyde. After washing with phosphate-buffered saline (PBS), the cells were incubated with a staining solution mix overnight at 37°C. The percentage of positive cells was analyzed with a microscope with a digital charged-coupled device capturing and image analysis system (Olympus BX51/QImaging Evolution MP 5.5/UIC MetaMorph).
Quantitative real-time reverse transcription–polymerase chain reaction
Total RNA (1 μg) was reverse transcribed using Superscript III Reverse Transcriptase (18080-051; Invitrogen-Gibco). Quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR) was performed with Power SYBR® Green PCR Master Mix (4367659; Applied Biosystems, Foster City, CA) on a MyiQ Single Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using the following primer sets: CD105, 5′-GCCAGCATTG TCTCACTTCA TG-3′ and 5′-GCAACAAGCT CTTTCTTTAG TACCA-3′; CD90, TCAGGAAATG GCTTTTCCCA and TCCTCAATGA GATGCCATAA GCT; CD73, CGCAACAATG GCACAATTAC and CAGGTTTTCG GGAAAGATCA; PPARγ, GCAGTGGGGA TGTCTCATAA TGC and CAGGGGGGTG ATGTGTTTGA A; alkaline phosphatase (ALP), GGACATGCAG TACGAGCTGA and TGTCTTCCGA GGAGGTCAAG; osteocalcin (OC), CACTCCTCGC CCTATTGGC and CCCTCCTGCT TGGACACAAA G; collagen type 1 alpha 1 (Col1α1), TAGGGTCTAG ACATGTTCAG CTTTGT and CCGTTCTGTA CGCAGGTGAT T; p16, GCCTTTTCAC TGTGTTGGAG and TGCCATTTGC TAGCAGTGTG [22]; p21, ACTTCGCCTG GGAGCGTGTG and CGTTTGGAGT GGTAGAAATC TGTCAT; p27, CGGCTAACTC TGAGGACACG C and ATTTGGGGAA CCGTCTGAAA CATT [23]; and GAPDH, GAAGGTGAAG GTCGGAGTC and GAAGATGGTG ATGGGATTTC. The thermal cycling conditions consisted of an initial denaturation step at 95°C for 5 min, then 50 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by a final extension step at 72°C for 10 min. The expression levels of the above genes were normalized to that of GAPDH.
Semiquantitative RT-PCR
Total RNA was isolated and cDNA was synthesized as described for qRT-PCR. The semiquantitative RT-PCR was performed at cycles at which the PCR products did not reach saturation levels to measure the expression levels of mitochondria-related genes, as described previously [21]. Primer sets and amplification cycles are listed in Table 1. The RT-PCR products were analyzed on a 1.2% agarose gel or analyzed by qRT-PCR normalized to a control gene, GAPDH.
Table 1.
Primer Sets Used for Semiquantitative Reverse Transcription–Polymerase Chain Reaction
| Category | Genes | Sense primer | Antisense primer | Product size (bp) | Annealing temp. (°C) | Number of cycles |
|---|---|---|---|---|---|---|
| NADH dehydrogenase | ND2 | CCTCAATTACCCACATAGGA | TTGAGTAGTAGGAATGCGGT | 115 | 49 | 21 |
| ATP synthase | AS6 | AACAACCGACTAATCACCAC | GTAAATGAGTGAGGCAGGAG | 164 | 60 | 21 |
| AS8 | ACTATTCCTCATCACCCAAC | GGCAATGAATGAAGCGAACA | 138 | 60 | 21 | |
| Complex I | NDUFB9 | CAGCATCCACAGCCATACAT | GCAGCTGCTTAACCTCTCGT | 199 | 60 | 28 |
| Mitochondrial biogenesis-related genes | PGC1α | TCAGTCCTCACTGGTGGACA | TGCTTCGTCGTCAAAAACAG | 531 | 56 | 28 |
| TFAM | GGCACAGGAAACCAGTTAGG | CAGAACACCGTGGCTTCTAC | 366 | 55 | 28 | |
| Control | 18S rRNA | GAGCGAAAGCATTTGCCAAG | GGCATCGTTTATGGTCGGAA | 102 | 60 | 26 |
Cell viability and mitochondrial activity assays
The cell viability and mitochondrial activity of UC-MSCs were analyzed using 96-well plate assays as described previously [21,24]. Quantitative measurement of cell number, calcein viability, mitochondrial membrane potential (ΔΨm) (with tetramethylrhodamine, ethyl ester [TMRE]), reactive oxygene species (ROS) with 2′, 7′-dichlorofluorescin diacetate (DCF-DA) and MitoSOX, and intracellular ATP content was performed. Mitochondrial superoxide levels were measured after incubating cells with 5 μM MitoSOX (Molecular Probes, Eugene, OR) for 10 min at 37°C, protected from light. Cells were washed with PBS and counterstained with 2 μg/mL Hoechst for 10 min, and fluorescent intensities were determined at 510/580 nm. MitoSOX intensity was normalized to Hoechst staining.
Bioenergetic profile measurement using the XF-24 analyzer
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured in adherent cells using a Seahorse XF-24 Analyzer (Seahorse Bioscience, Billerica, MA) following the manufacturer's protocol with minor modifications [25]. UC-MSCs (1.0×104 cells/well) were seeded in XF-24 microplates in 250 μL DMEM containing 10% FBS and incubated at 37°C/5% CO2 for 24 h. Assays were initiated by replacing the medium in each well with 590 μL assay medium (DMEM without sodium bicarbonate) prewarmed to 37°C. After gentle mixing for 10 min in the XF-24 Analyzer, the OCR (basal OCR) and ECAR baselines were measured simultaneously for 3 min. Oligomycin (75 μL of a 10 μg/mL stock), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP; 83 μL of a 3 μM stock), and rotenone (93 μL of a 1 μM stock) were consecutively injected into each well to reach the desired final working concentration for quantifying the ATP turnover rate (basal OCR–oligomycin-OCR) and total respiratory capacity (FCCP-OCR–rotenone-OCR). The OCR and ECAR were calculated from 3-min measurement cycles. Results were normalized to the cell number.
Statistics
Data represent the mean±standard deviation (n=3 or 4). Comparisons between groups were analyzed using two-tailed Student's t tests. Statistical significance is represented in the figures by an asterisk (*) indicating P<0.05.
Results
GDM-UC-MSCs display decreased cell growth
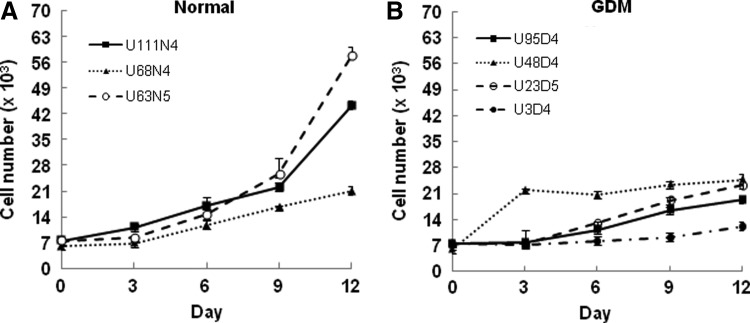
To assess whether GDM-UC-MSCs showed any changes in cell growth, we initially cultured seven UC-MSC lines obtained from three normal and four GDM-affected placentas under standard conditions in the presence of 10% serum. Growth curve analyses of N-UC-MSCs and GDM-UC-MSCs showed that the proliferation rate of early-passage GDM-UC-MSCs was substantially lower compared with N-UC-MSCs (Fig. 1A, B). Twelve days after plating, the four GDM-UC-MSC lines grew to a mean of 1.9×104 cells per plate versus a mean of 4.1×104 cells per plate for the N-UC-MSCs, indicating that GDM-UC-MSCs have a greatly reduced population doubling rate.
FIG. 1.
Umbilical cord mesenchymal stromal cells (UC-MSCs) derived from gestational diabetes mellitus (GDM) patients exhibit retarded growth proliferation. The growth of 7.5×103 UC-MSCs isolated from patients with normal pregnancies (A) and GDM (B) was monitored over a period of 12 days. GDM-UC-MSCs consistently showed decreased proliferation compared with normal pregnant women (N-UC-MSCs). Points represent the mean values from three independent experiments; bars denote standard deviation (SD).
GDM-UC-MSCs undergo premature senescence
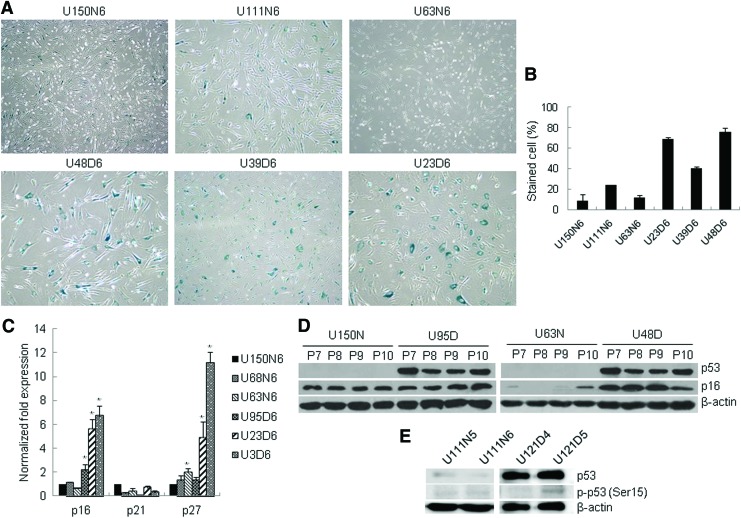
Since GDM-UC-MSCs showed markedly decreased cell growth, we next investigated whether GDM induces replicative senescence in UC-MSCs. Senescence is characterized by a large, flatter cell morphology and senescence-associated β-galactosidase (SA-β-gal) expression [26]. GDM-UC-MSCs exhibited increased SA-β-gal staining, which was detected cytochemically as blue perinuclear staining (Fig. 2A, B). The SA-β-gal-stained cells were larger and rounder with a flatter cytoplasmic appearance than the N-UC-MSC controls in the relatively early passages.
FIG. 2.
GDM-UC-MSCs enter a premature senescence at early passages. (A) Representative photomicrographs of senescence-associated β-galactosidase (SA-β-gal) staining in UC-MSCs at passage 6. (B) Quantification of the percentage of SA-β-gal-positive cells shown in (A). (C) The relative expressions of the cyclin-dependent kinase inhibitor genes p16, p21, and p27 were analyzed using quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR). Data are presented as levels of p16, p21, and p27 transcripts relative to the control gene GAPDH. The mean of three experiments is shown in each column; bar denotes SD (*P<0.05 determined by t test). (D) Induction of the senescence marker proteins p16 and p53 was examined by western blotting in N-UC-MSCs (U150N and U63N) and GDM-UC-MSCs (U95D and U48D) with increasing passage number from P7 to P10. The p53 protein was only detected in GDM UC-MSCs. (E) Whole-cell lysates were prepared at the indicated passage and analyzed by western blotting for phosphorylated p53 (Ser15). β-Actin served as a loading control. Color images available online at www.liebertpub.com/scd
Derepression of cyclin-dependent kinase inhibitors (CDKIs) can lead to the induction of cellular senescence in MSCs. CDKN2A (p16INK4a), CDKN1A (p21CIP1), and p27KIP1 inhibit CDK4/6, leading to senescence, and their gradual increases are observed with aging in most mammalian tissues [27–29]. Therefore, we next investigated whether the aging phenotype of GDM-UC-MSCs was mediated by CDKI p16, p21, and p27 induction. As expected, p16 and p27 transcripts were markedly elevated in GDM-UC-MSCs compared with those in N-UC-MSCs (Fig. 2C). In addition, the p53 protein was only detected in GDM-UC-MSCs with relatively increased levels of p16 protein at passage 7–10 (Fig. 2D). Early experiments [30,31] suggested that p53 becomes phosphorylated in senescence. We next examined phosphorylated p53 (Ser15) expression. Phosphorylated p53 was weakly detected in GDM-UC-MSCs at passage 5, but not in N-UC-MSCs at passages 5 or 6 (Fig. 2E). These results support the viewpoint that proliferative arrest of GDM-UC-MSCs is driven by premature senescence involving the CDKI proteins.
GDM-UC-MSCs and N-UC-MSCs express comparable amounts of stem cell marker proteins
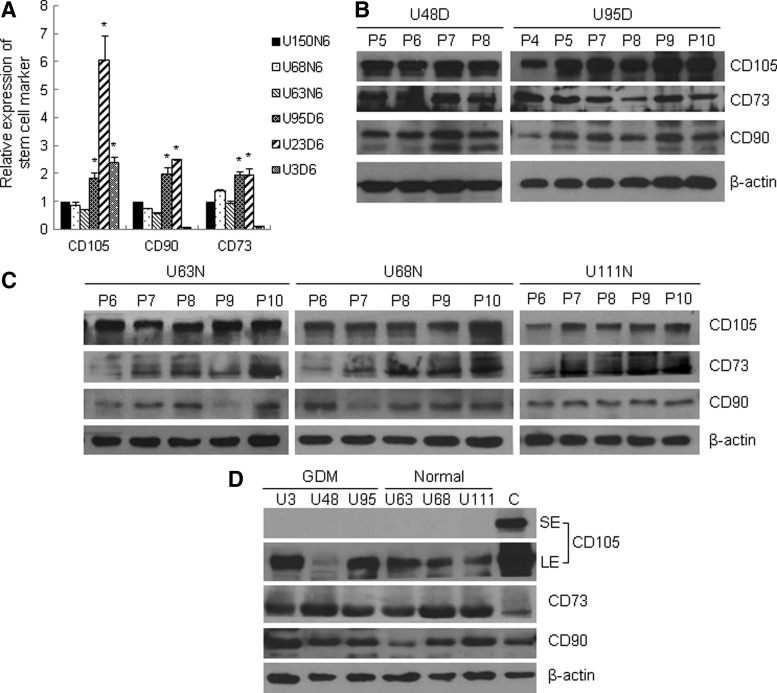
UC-MSCs express a defined set of the surface markers CD90, CD73, and CD105. To examine whether GDM-UC-MSCs express these stem cell marker proteins, total RNA was extracted and subjected to qRT-PCR. The results showed that the levels of stem cell markers in GDM-UC-MSCs were similar to those in N-UC-MSCs, except for CD105 expression in a GDM-UC-MSC line, U23D6 (Fig. 3A). To compare the levels of these marker proteins, immunoblot analyses were performed with five different UC-MSCs at their early passages. The levels of CD90, CD73, and CD105 proteins were similar in all five UC-MSC lines (Fig. 3B, C). Weiss et al. [9] reported a decrease in CD105 during continuous culture. However, in our culture conditions, the expression levels of CD90, CD103, and CD 105 were not much different across passages 4–10 in both groups of UC-MSCs.
FIG. 3.
GDM-UC-MSCs and N-UC-MSCs express similar levels of stem cell marker mRNA and proteins. (A) The relative transcript levels of the stem cell markers CD105, CD90, and CD70 in N-UC-MSCs (U150N6, U68N6, and U63N6) and GDM-UC-MSCs (U95D6, U23D6, and U3D6) at passage 6 were analyzed by quantitative RT-PCR. Expression values were normalized to that of GAPDH. The mean value obtained from three independent experiments is shown in each column; bar denotes SD (*P<0.05 determined by t test). (B, C) Total cell extracts of GDM-UC-MSCs (B) and N-UC-MSCs (C) were analyzed by western blotting using antibodies against CD105, CD73, CD90, and β-actin (loading control). (D) Total cellular proteins of UC-MSCs in umbilical cords and cultured N-UC-MSCs as a control were analyzed by immunoblotting as described above. SE, short-term exposure; LE, long-term exposure.
To further analyze whether these MSC markers are induced under FBS-supplemented culture conditions, the UC-MSCs from Wharton's jelly were subjected to immunoblot analysis without culture steps. CD90, CD73, and CD105 expression was readily detected in both normal and GDM-affected UC-MSCs (Fig. 3D). Taken together, these results suggest that MSC markers are not induced during in vitro culture steps in UC-MSCs and that GDM does not change their expression levels.
Lineage-specific differentiation is impaired in GDM-UC-MSCs
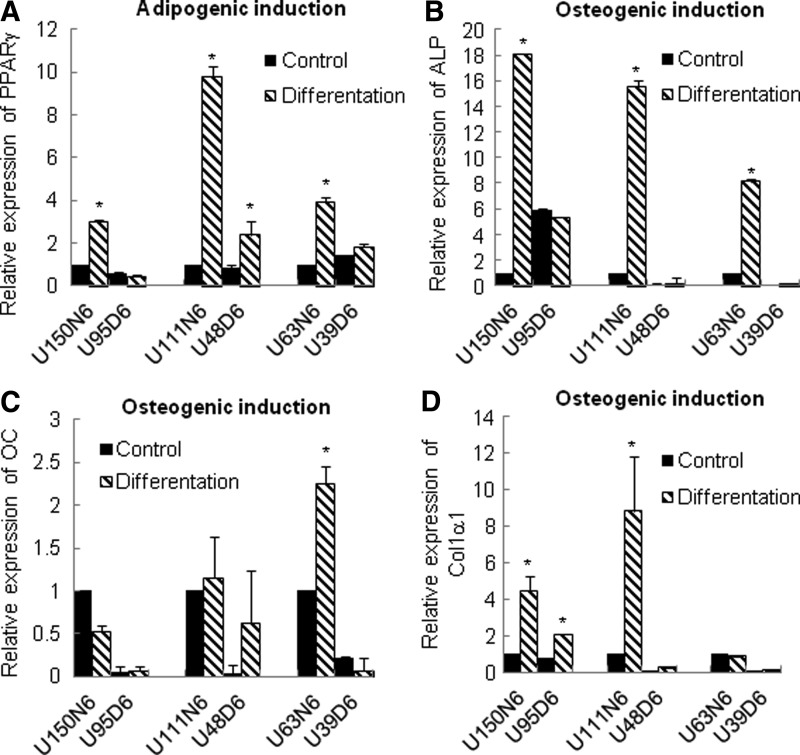
MSCs possess a multilineage differentiation potential that permits these cells to differentiate into a variety of mesodermal cell lineages such as bone, cartilage, and adipose tissue. Next, we examined the ability of GDM-UC-MSCs to undergo osteogenic and adipogenic differentiation in a differentiation-optimized medium. To evaluate the lineage-specific differentiation of these cells, marker genes PPARγ (adipogenic differentiation) and ALP, OC, and Col1α1 (osteogenic differentiation) were chosen [32,33]. As shown in Fig. 4, N-UC-MSCs from normal pregnancies showed a higher expression of all selected markers upon osteogenic and adipogenic differentiation compared with undifferentiated cells after 5 days. In contrast, induction of the differentiation markers was not observed in GDM-UC-MSCs following treatment with differentiation media. These data imply that the stem cell properties of UC-MSCs are significantly affected by GDM and that there is no direct correlation between differentiation potentials and stem cell marker expression levels in MSCs.
FIG. 4.
Stem cell differentiation potentials are largely different between normal and GDM-affected UC-MSCs. Three different cell lines of normal and GDM-affected pregnancies were cultured in a control medium or induction medium for 5 days. Upregulation of the expression of the adipogenic-specific gene PPARγ (A) and the osteogenic genes alkaline phosphatase (ALP) (B), osteocalcin (OC) (C), and collagen type 1 alpha 1 (Col1α1) (D) was evaluated by real-time RT-PCR and normalized to GAPDH. All assays were performed in triplicate; bars denote SD (*P<0.05).
GDM-UC-MSCs display mitochondrial dysfunction
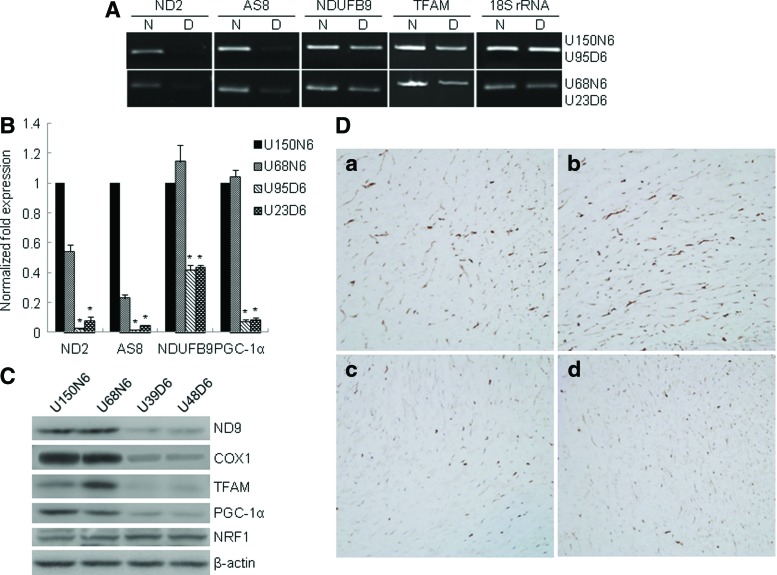
Mitochondrial dysfunction has roles in insulin resistance in the elderly [34]. An age-associated reduction in mitochondrial number and function of oxidative phosphorylation (OXPHOS) activities has been reported [35]. To further investigate the mechanism of premature aging of GDM-UC-MSCs, we evaluated the mRNA levels of mitochondria-related genes by semiquantitative PCR and qRT-PCR [21,36]. Compared with N-UC-MSCs, GDM-UC-MSCs showed significantly reduced mRNA levels of complex I subunit NADH-ubiquinone (ND2) and complex V subunit (AS8), both of which are encoded by mitochondrial DNA (Fig. 5A). Meanwhile, three nuclear-encoded proteins-TFAM, a central regulator of mitochondrial transcription and replication; PGC-1α, a regulator of mitochondria biogenesis; and NDUFB9, an OXPHOS complex I subunit-exhibited moderately decreased mRNA expression levels in GDM-UC-MSCs (Fig. 5A, B). Immunoblotting confirmed the decreased levels of two other OXPHOS subunits (ND9 and COX1) as well as TFAM and PGC-1α proteins in GDM-UC-MSCs. However, the expression of another mitochondria regulator, NRF-1, was unaltered (Fig. 5C). We further examined the immunoreactive patterns of TFAM in the umbilical cords by immunohistochemistry. The intensity and extent of TFAM immunostaining in UC-MSCs of normal term deliveries (Fig. 5D[a], [b]) were more prominent than in those from GDM patients (Fig. 5D[c], [d]). These data imply that mitochondrial functions in GDM-UC-MSCs are impaired, which could be a major cause of decreased cell proliferation and premature senescence.
FIG. 5.
UC-MSCs from GDM patients reveal decreased mitochondria-related gene expression. Genes involved in mitochondrial oxidative phosphorylation function, ND2, AS8, COX1, and NDUFB9, and mitochondria biogenesis, PGC-1α and TFAM, were analyzed. (A) Semiquantitative RT-PCR was performed using the primers detailed in Table 1 and analyzed by 1.2% agarose gel electrophoresis. The RT-PCR products for 18S rRNA were used as a control. (B, C) For quantitative analysis, the levels of ND2, AS8, NDUFB9, and PGC-1α transcripts were evaluated by qRT-PCR and western blotting. The mean value obtained from three independent experiments is shown in each column; bar denotes SD (*P<0.05 determined by t test). (D) Comparison of immunohistochemical staining of TFAM protein in Wharton's jelly tissues. The intensity and extent of TFAM immunostaining in the umbilical cords from normal term deliveries (a, b) are more prominent than those from GDM patients (c, d). The photo was taken at 200×magnification. Color images available online at www.liebertpub.com/scd
Mitochondrial activity profile of GDM-UC-MSCs reveals defects in OXPHOS activity
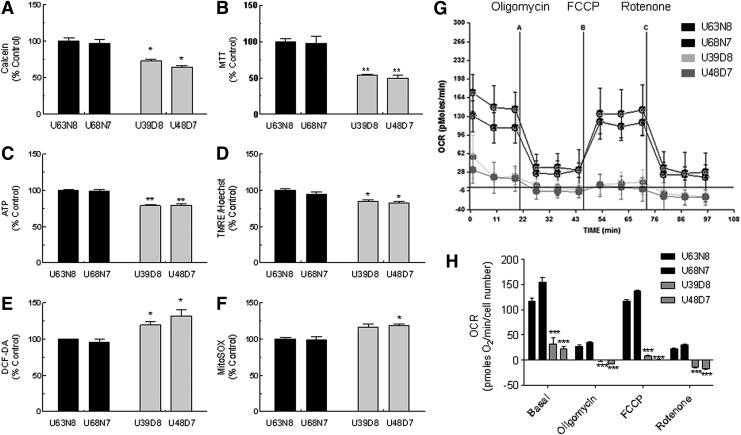
To analyze mitochondrial functions of GDM-UC-MSCs, we employed a 96-well cell-based mitochondrial functional assay system [21]. The parameters analyzed were cell membrane integrity using a calcein assay, complex I activity of mitochondrial respiratory chain system (NADH dehydrogenase activity) using tetrazolium precipitation of MTT, measurement of intracellular ATP contents, and measurement of mitochondrial membrane potential using a TMRE assay, which are positive mitochondrial functional markers. In contrast, the measurement of total ROS generation using DCF-DA and mitochondrial superoxide generation using MitoSOX is a negative mitochondrial functional marker. As shown in Fig. 6, GDM-UC-MSCs exhibited ∼30% lower levels of the positive functional markers, calcein, MTT, ATP, and TMRE (Fig. 6A–D) and ∼20%–30% higher levels of the negative functional markers DCF-DA and MitoSOX (Fig. 6E, F) compared with N-UC-MSCs.
FIG. 6.
GDM-UC-MSCs display mitochondrial dysfunction. Mitochondrial function was analyzed using a cell-based mitochondrial activity profiling system. (A) Calcein for cell viability, (B) MTT for NADH dehydrogenase (complex I activity), (C) intracellular ATP contents using the luciferase–luciferin reaction, (D) TMRE assay for mitochondrial membrane potential, (E) DCF-DA fluorescence assay for ROS generation, and (F) MitoSOX fluorescent assay for mitochondria-derived superoxide generation. Data obtained from three independent experiments are shown in each column; bar denotes SD (*P<0.05, **P<0.01 determined by t test). (G) Endogenous cellular oxygen consumption rate (OCR) curves for four UC-MSC lines. Curves were generated using a Seahorse XF-24 Analyzer. The ATP synthase inhibitor oligomycin, the mitochondrial uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and the respiratory complex I inhibitor rotenone were added at the indicated time points (A, B, and C, respectively). Representative traces are shown. Every point represents an average of six different wells. (H) The OCR data were normalized to the protein concentration after each experiment. The mean of six different wells is shown in each column, and bars correspond with the SD (***P<0.001). Basal, endogenous rate; oligomycin, ATP synthase-inhibited OCR; FCCP, maximal uncoupled rate; rotenone, nonmitochondrial rate; DCF-DA, 2′,7′-dichlorofluorescin diacetate; ROS, reactive oxygene species.
The mitochondrial physiological activity was estimated by measuring the OCR and extracellular acidification using an XF-24 bioenergetic assay. Oligomycin blocks the Fo proton channel of ATP synthase and prevents state 3 (phosphorylation) respiration. In our normal N-UC-MSCs, oligomycin treatment revealed the amount of oxygen consumption needed for ATP synthesis (oligomycin-mediated ATP turnover rate) [37]. The uncoupler FCCP disrupts ATP synthesis by diverting protons from an ATP synthase proton channel and shuttles them across the mitochondrial membrane. Thus, FCCP treatment induced membrane potential dissipation and rapid oxygen consumption to maintain the cellular energy balance. We defined this OCR as the FCCP-induced respiratory capacity. Finally, rotenone, a mitochondrial respiratory chain complex I inhibitor, blocked mitochondrial respiration completely, demonstrating nonmitochondrial respiration. As shown in Fig. 6G and H, the basal OCR of GDM-UC-MSCs was only ∼30% that of N-UC-MSCs. Moreover, the FCCP-induced respiratory capacity and oligomycin-mediated ATP turnover rate of GDM-UC-MSCs were almost negligible. We also noticed that N-UC-MSCs exhibited maximum OCR in their basal state, and even 2–3-fold increased FCCP-inhibited OCR compared with GDM-UC-MSCs. This result suggests that there may be no spare respiratory capacity in GDM-UC-MSCs. Taken together, these data support the theory that GDM-UC-MSCs are programmed to mimic the metabolic and ROS signaling properties of GDM patients.
Discussion
In this study, we report for the first time that UC-MSCs derived from GDM patients have premature senescence phenotypes, decreased differentiation potential, and mitochondrial dysfunction. We found a significant difference in the proliferation of UC-MSCs derived from GDM patients relative to those derived from healthy pregnant women. GDM-UC-MSCs did not commonly grow above passage 6 or 7, whereas one of the N-UC-MSC lines from normal term pregnancies grew until passage 37 in our culture conditions (unpublished data). These results are consistent with the report of Liu et al., which stated that MSCs isolated from the sternal bone marrow of a DM patient group exhibit lower growth than those isolated from a control group [38]. Gene expression profiling of DM MSCs revealed increased expression of genes related to tumor necrosis factor-related apoptosis pathways and decreased expression of antiapoptotic genes such as Bcl-2. In our results, cell cycle regulators p53 and p16Ink4a were markedly increased in GDM-UC-MSCs compared with N-UC-MSCs. Of note, p53 proteins were only detected in GDM-UC-MSCs at passage 7–10. p16Ink4a inhibits cyclin-dependent kinase, leading to cell cycle arrest, and plays a major role in regulating the decline in proliferation and regenerative capacity in certain organs when they age [39–41]. These results raise a question as to whether decreased growth of GDM-UC-MSCs is due to increased apoptotic cell death or a premature aging process. We believe that the data showing p53 and p16 accumulation and earlier senescence account for the decreased growth of GDM-UC-MSCs, rather than cytostatic growth arrest or cell death. In addition, it is important to emphasize that although premature senescence in GDM-UC-MSCs is similar to that in cells with damaged DNA, as with prolonged p53 expression, the mechanism behind this could differ markedly. We observed minimal p53 (Ser15) phosphorylation, reflecting the DNA damage response [31], in the presence of distinctly elevated p53 protein levels in GDM-UC-MSCs. Therefore, accumulating DNA damage might not be a senescence-causing factor in GDM-UC-MSCs.
Many studies have shown a relationship between mitochondrial DNA defects and human stem cell aging [42]. Mitochondrial DNA mutations and respiratory chain deficiency accumulate in aging human mitotic tissues, such as the colon and small intestine, leading to a decrease in crypt cell number and cell proliferation [43,44]. In fact, mice with a homozygous D257A mutation in the proofreading domain of mitochondrial DNA polymerase γ display severe respiratory chain deficiency and a premature aging phenotype [45,46]. We showed that two major regulators of mitochondrial biogenesis, TFAM and PGC-1α, are downregulated in GDM-UC-MSCs. Although there is a common consensus that an age-associated decline in mitochondrial function triggers insulin resistance in the elderly, the detailed mechanism of mitochondrial dysfunction in GDM-UC-MSCs needs to be further elucidated.
The incidence of metabolic diseases has been increasing for decades, and metabolic syndrome affects the physiology of several organs. The findings of recent studies strongly suggest that metabolic disorders in adults and children originate in the fetal period and are connected with maternal nutrition during pregnancy and the subsequent fetal programming [47]. GDM increases the risk of chronic diseases in offspring by fetal metabolic programming. Ruchat et al. [48] have shown differences in placental and cord blood methylomes between newborns exposed to GDM and newborns delivered by healthy mothers. They found that GDM has epigenetic effects in metabolic disease pathways. Nomura et al. [18] have also found changes in global methylation in the placenta in pregnancies with GDM, preeclampsia, and obesity. Hence, fetal programming seems to have multiple facets. We demonstrated mitochondrial dysfunction of GDM-UC-MSCs and propose that the changes in the biological properties of GDM-UC-MSCs in this study are an additional feature of fetal metabolic programming.
Strategies to develop cellular therapeutics using human MSCs have been based on the application of either autologous or allogeneic MSCs. More than 200 clinical trials involving human MSCs are globally underway, with promising results [49]. However, safety issues and concerns about long-term consequences of therapies using MSCs persist [50]. For example, transplanted human MSCs into NOD/SCID mice with CCL4-induced liver injury showed profibrogenic effects and hepatocytic differentiation was rare [51]. Pathological conditions of hosts also affect the biological properties of MSCs. Klinkhammer et al. [52] found premature senescence and decreased proliferation in bone marrow-derived MSCs obtained from rats with chronic kidney disease. These cells lost the beneficial effects on anti-Thy1.1 nephritis seen in MSCs obtained from healthy subjects. Limited regenerative potential of MSCs obtained from older donors was also associated with both morphological and functional changes in mitochondria [53,54]. hTERT-immortalized bone marrow-derived MSCs from patients with Parkinson's disease showed decreased mitochondrial complex I, II, and IV activities [55]. All these observations by different investigators are in line with our finding in GDM-UC-MSCs that the host condition is one of the major determinants of the biological properties and potential of MSCs.
In summary, the findings of the present study strongly suggest that studies of UC-MSCs may have profound implications for understanding the biology of the human fetus and also that crucial biological properties of human UC-MSCs differ according to obstetrical conditions. The findings also underscore the importance of maternal–fetal conditions in biological studies of hUC-MSCs and the development of future therapeutic strategies using hUC-MSCs.
Acknowledgments
This work was supported by grants NRF-2013R1A1A2062598 and SRC 2008-0061888 from the National Research Foundation of Korea and by a grant from the Asan Institute for Life Sciences 2013-560.
Author Disclosure Statement
All the authors agree that there are no conflicts of interest.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. and Jones JM. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147 [DOI] [PubMed] [Google Scholar]
- 2.de Lazaro I, Yilmazer A. and Kostarelos K. (2014). Induced pluripotent stem (iPS) cells: a new source for cell-based therapeutics? J Control Release 185c:37–44 [DOI] [PubMed] [Google Scholar]
- 3.Weiss ML, Anderson C, Medicetty S, Seshareddy KB, Weiss RJ, VanderWerff I, Troyer D. and McIntosh KR. (2008). Immune properties of human umbilical cord Wharton's jelly-derived cells. Stem Cells 26:2865–2874 [DOI] [PubMed] [Google Scholar]
- 4.Troyer DL. and Weiss ML. (2008). Wharton's jelly-derived cells are a primitive stromal cell population. Stem Cells 26:591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DW, Staples M, Shinozuka K, Pantcheva P, Kang SD. and Borlongan CV. (2013). Wharton's jelly-derived mesenchymal stem cells: phenotypic characterization and optimizing their therapeutic potential for clinical applications. Int J Mol Sci 14:11692–11712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian Q, Lye E, Suan Yeo K, Khia Way Tan E, Salto-Tellez M, Liu TM, Palanisamy N, El Oakley RM, Lee EH, Lim B. and Lim SK. (2007). Derivation of clinically compliant MSCs from CD105+, CD24- differentiated human ESCs. Stem Cells 25:425–436 [DOI] [PubMed] [Google Scholar]
- 7.Anzalone R, Lo Iacono M, Loria T, Di Stefano A, Giannuzzi P, Farina F. and La Rocca G. (2011). Wharton's jelly mesenchymal stem cells as candidates for beta cells regeneration: extending the differentiative and immunomodulatory benefits of adult mesenchymal stem cells for the treatment of type 1 diabetes. Stem Cell Rev 7:342–363 [DOI] [PubMed] [Google Scholar]
- 8.Weiss ML. and Troyer DL. (2006). Stem cells in the umbilical cord. Stem Cell Rev 2:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss ML, Medicetty S, Bledsoe AR, Rachakatla RS, Choi M, Merchav S, Luo Y, Rao MS, Velagaleti G. and Troyer D. (2006). Human umbilical cord matrix stem cells: preliminary characterization and effect of transplantation in a rodent model of Parkinson's disease. Stem Cells 24:781–792 [DOI] [PubMed] [Google Scholar]
- 10.Ma S, Liang S, Jiao H, Chi L, Shi X, Tian Y, Yang B. and Guan F. (2014). Human umbilical cord mesenchymal stem cells inhibit C6 glioma growth via secretion of dickkopf-1 (DKK1). Mol Cell Biochem 385:277–286 [DOI] [PubMed] [Google Scholar]
- 11.Gauthaman K, Yee FC, Cheyyatraivendran S, Biswas A, Choolani M. and Bongso A. (2012). Human umbilical cord Wharton's jelly stem cell (hWJSC) extracts inhibit cancer cell growth in vitro. J Cell Biochem 113:2027–2039 [DOI] [PubMed] [Google Scholar]
- 12.Harris DT. (2013). Umbilical cord tissue mesenchymal stem cells: characterization and clinical applications. Curr Stem Cell Res Ther 8:394–399 [DOI] [PubMed] [Google Scholar]
- 13.Lakshmy R. (2013). Metabolic syndrome: role of maternal undernutrition and fetal programming. Rev Endocr Metab Disord 14:229–240 [DOI] [PubMed] [Google Scholar]
- 14.Thornburg KL, O'Tierney PF. and Louey S. (2010). Review: the placenta is a programming agent for cardiovascular disease. Placenta 31Suppl:S54–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrara A. (2007). Increasing prevalence of gestational diabetes mellitus: a public health perspective. Diabetes Care 30Suppl 2:S141–S146 [DOI] [PubMed] [Google Scholar]
- 16.Schwartz R, Gruppuso PA, Petzold K, Brambilla D, Hiilesmaa V. and Teramo KA. (1994). Hyperinsulinemia and macrosomia in the fetus of the diabetic mother. Diabetes Care 17:640–648 [DOI] [PubMed] [Google Scholar]
- 17.Pedersen J. (1952). Diabetes and pregnancy; blood sugar of newborn infants during fasting and glucose administration. Nord Med 47:1049. [PubMed] [Google Scholar]
- 18.Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, Manaster I, Huynh N, Finik J, et al. (2014). Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci 21:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzger BE, Gabbe SG, Persson B, Buchanan TA, Catalano PA, Damm P, Dyer AR, Leiva A, Hod M, et al. (2010). International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 33:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redline RW. (2008). Placental pathology: a systematic approach with clinical correlations. Placenta 29Suppl A:S86–S91 [DOI] [PubMed] [Google Scholar]
- 21.Piao Y, Kim HG, Oh MS. and Pak YK. (2012). Overexpression of TFAM, NRF-1 and myr-AKT protects the MPP(+)-induced mitochondrial dysfunctions in neuronal cells. Biochim Biophys Acta 1820:577–585 [DOI] [PubMed] [Google Scholar]
- 22.Lee JS, Lee MO, Moon BH, Shim SH, Fornace AJ., Jr. and Cha HJ. (2009). Senescent growth arrest in mesenchymal stem cells is bypassed by Wip1-mediated downregulation of intrinsic stress signaling pathways. Stem Cells 27:1963–1975 [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Sun Z, Han Q, Liao L, Wang J, Bian C, Li J, Yan X, Liu Y, Shao C. and Zhao C. (2009). Human mesenchymal stem cells inhibit cancer cell proliferation by secreting DKK-1. Leukemia 23:925–933 [DOI] [PubMed] [Google Scholar]
- 24.Koo HJ, Piao Y. and Pak YK. (2012). Endoplasmic reticulum stress impairs insulin signaling through mitochondrial damage in SH-SY5Y cells. Neurosignals 20:265–280 [DOI] [PubMed] [Google Scholar]
- 25.Birket MJ, Orr AL, Gerencser AA, Madden DT, Vitelli C, Swistowski A, Brand MD. and Zeng X. (2011). A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J Cell Sci 124:348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh M. and Piekorz RP. (2013). Senescence-associated lysosomal alpha-L-fucosidase (SA-alpha-Fuc): a sensitive and more robust biomarker for cellular senescence beyond SA-beta-Gal. Cell Cycle 12:1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim WY. and Sharpless NE. (2006). The regulation of INK4/ARF in cancer and aging. Cell 127:265–275 [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Lin AW, McCurrach ME, Beach D. and Lowe SW. (1997). Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602 [DOI] [PubMed] [Google Scholar]
- 29.Jones CJ, Kipling D, Morris M, Hepburn P, Skinner J, Bounacer A, Wyllie FS, Ivan M, Bartek J, Wynford-Thomas D. and JA Bond. (2000). Evidence for a telomere-independent “clock”. limiting RAS oncogene-driven proliferation of human thyroid epithelial cells. Mol Cell Biol 20:5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbig U, Jobling WA, Chen BP, Chen DJ. and Sedivy JM. (2004). Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 14:501–513 [DOI] [PubMed] [Google Scholar]
- 31.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR. and Campisi J. (2009). Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol 11:973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng L, Jia Z, Yin X, Zhang X, Liu Y, Chen P, Ma K. and Zhou C. (2008). Comparative analysis of mesenchymal stem cells from bone marrow, cartilage, and adipose tissue. Stem Cells Dev 17:761–773 [DOI] [PubMed] [Google Scholar]
- 33.Ode A, Schoon J, Kurtz A, Gaetjen M, Ode JE, Geissler S. and Duda GN. (2013). CD73/5′-ecto-nucleotidase acts as a regulatory factor in osteo-/chondrogenic differentiation of mechanically stimulated mesenchymal stromal cells. Eur Cell Mater 25:37–47 [DOI] [PubMed] [Google Scholar]
- 34.Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW. and Shulman GI. (2003). Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300:1140–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HY, Choi CS, Birkenfeld AL, Alves TC, Jornayvaz FR, Jurczak MJ, Zhang D, Woo DK, Shadel GS, et al. (2010). Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab 12:668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dimauro S. and Davidzon G. (2005). Mitochondrial DNA and disease. Ann Med 37:222–232 [DOI] [PubMed] [Google Scholar]
- 37.Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW. and Ferrick DA. (2010). Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp (46):pii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Li Z, Liu T, Xue X, Jiang H, Huang J. and Wang H. (2013). Impaired cardioprotective function of transplantation of mesenchymal stem cells from patients with diabetes mellitus to rats with experimentally induced myocardial infarction. Cardiovasc Diabetol 12:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S. and Sharpless NE. (2006). p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443:453–457 [DOI] [PubMed] [Google Scholar]
- 40.Yu KR. and Kang KS. (2013). Aging-related genes in mesenchymal stem cells: a mini-review. Gerontology 59:557–563 [DOI] [PubMed] [Google Scholar]
- 41.Sousa-Victor P, Gutarra S, Garcia-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, Jardi M, Ballestar E, Gonzalez S, et al. (2014). Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506:316–321 [DOI] [PubMed] [Google Scholar]
- 42.Baines HL, Turnbull DM. and Greaves LC. (2014). Human stem cell aging: do mitochondrial DNA mutations have a causal role?. Aging Cell 13:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nooteboom M, Johnson R, Taylor RW, Wright NA, Lightowlers RN, Kirkwood TB, Mathers JC, Turnbull DM. and Greaves LC. (2010). Age-associated mitochondrial DNA mutations lead to small but significant changes in cell proliferation and apoptosis in human colonic crypts. Aging Cell 9:96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein C, Facista A, Nguyen H, Zaitlin B, Hassounah N, Loustaunau C, Payne CM, Banerjee B, Goldschmid S, et al. (2010). Cancer and age related colonic crypt deficiencies in cytochrome c oxidase I. World J Gastrointest Oncol 2:429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417–423 [DOI] [PubMed] [Google Scholar]
- 46.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, et al. (2005). Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309:481–484 [DOI] [PubMed] [Google Scholar]
- 47.Wu G, Imhoff-Kunsch B. and Girard AW. (2012). Biological mechanisms for nutritional regulation of maternal health and fetal development. Paediatr Perinat Epidemiol 26Suppl 1:4–26 [DOI] [PubMed] [Google Scholar]
- 48.Ruchat SM, Houde AA, Voisin G, St.-Pierre J, Perron P, Baillargeon JP, Gaudet D, Hivert MF, Brisson D. and Bouchard L. (2013). Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 8:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier RP, Muller YD, Morel P, Gonelle-Gispert C. and Buhler LH. (2013). Transplantation of mesenchymal stem cells for the treatment of liver diseases, is there enough evidence?. Stem Cell Res 11:1348–1364 [DOI] [PubMed] [Google Scholar]
- 50.Breitbach M, Bostani T, Roell W, Xia Y, Dewald O, Nygren JM, Fries JW, Tiemann K, Bohlen H, et al. (2007). Potential risks of bone marrow cell transplantation into infarcted hearts. Blood 110:1362–1369 [DOI] [PubMed] [Google Scholar]
- 51.di Bonzo LV, Ferrero I, Cravanzola C, Mareschi K, Rustichell D, Novo E, Sanavio F, Cannito S, Zamara E, et al. (2008). Human mesenchymal stem cells as a two-edged sword in hepatic regenerative medicine: engraftment and hepatocyte differentiation versus profibrogenic potential. Gut 57:223–231 [DOI] [PubMed] [Google Scholar]
- 52.Klinkhammer BM, Kramann R, Mallau M, Makowska A, van Roeyen CR, Rong S, Buecher EB, Boor P, Kovacova K, et al. (2014). Mesenchymal stem cells from rats with chronic kidney disease exhibit premature senescence and loss of regenerative potential. PLoS One 9:e92115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mantovani C, Raimondo S, Haneef MS, Geuna S, Terenghi G, Shawcross SG. and Wiberg M. (2012). Morphological, molecular and functional differences of adult bone marrow- and adipose-derived stem cells isolated from rats of different ages. Exp Cell Res 318:2034–2048 [DOI] [PubMed] [Google Scholar]
- 54.Geissler S, Textor M, Kuhnisch J, Konnig D, Klein O, Ode A, Pfitzner T, Adjaye J, Kasper G. and Duda GN. (2012). Functional comparison of chronological and in vitro aging: differential role of the cytoskeleton and mitochondria in mesenchymal stromal cells. PLoS One 7:e52700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moon HE, Yoon SH, Hur YS, Park HW, Ha JY, Kim KH, Shim JH, Yoo SH, Son JH, et al. (2013). Mitochondrial dysfunction of immortalized human adipose tissue-derived mesenchymal stromal cells from patients with Parkinson's disease. Exp Neurobiol 22:283–300 [DOI] [PMC free article] [PubMed] [Google Scholar]