Abstract
MicroRNAs (miRNAs) originate from stem-loop-containing precursors (pre-miRNAs, pri-miRNAs) and mature by means of the Drosha and Dicer endonucleases and their associated factors. The let-7 miRNAs have prominent roles in developmental differentiation and in regulating cell proliferation. In cancer, the tumor suppressor function of let-7 is abrogated by overexpression of Lin28, one of several RNA-binding proteins that regulate let-7 biogenesis by interacting with conserved motifs in let-7 precursors close to the Dicer cleavage site. Using in vitro assays, we have identified a binding site for short modified oligoribonucleotides (‘looptomirs’) overlapping that of Lin28 in pre-let-7a-2. These looptomirs selectively antagonize the docking of Lin28, but still permit processing of pre-let-7a-2 by Dicer. Looptomirs restored synthesis of mature let-7 and inhibited growth and clonogenic potential in Lin28 overexpressing hepatocarcinoma cells, thereby demonstrating a promising new means to rescue defective miRNA biogenesis in Lin28-dependent cancers.
INTRODUCTION
The biogenesis of microRNAs (miRNAs) begins with the Pol II-mediated transcription of a primary miRNA (pri-miRNA) containing a characteristic stem-loop structure (1,2). The terminal loop region (TLR) of miRNA precursors varies in length typically between 12 and 40 nts. For some precursors, this may reflect their role as docking sites for auxiliary factors, i.e. RNA-binding proteins (RBPs) that bind to this sequence and regulate biogenesis (3). Whereas short terminal loop regions can form conformationally-restricted stable structures, the longer loops may have properties more resembling single-stranded RNAs. The primary transcript is cleaved to a shorter hairpin (pre-miRNA) by the nuclear microprocessor complex and then exported to the cytoplasm where Dicer excises its TLR. The remaining duplex is incorporated into the miRISC complex where one of the strands is selected. The loaded complex targets sites in the 3′ untranslated regions (UTRs) of messenger RNAs (mRNAs), and represses gene expression (2). The regulation of miRNA biogenesis occurs at transcriptional and post-transcriptional levels. For example, several RBPs are known to bind selectively and competitively to conserved sites in miRNA precursors and to elicit a variety of regulatory effects (3,4) (see references in (5)).
Let-7 was originally identified as a miRNA regulating developmental timing in Caenorhabditis elegans and in several organisms its expression is absent during the early stages of development (6). The let-7 family is highly conserved and in humans, 10 let-7 family members are expressed from 13 loci (6). Let-7 miRNAs are important suppressors of cell growth, and their targets include K-RAS, MYC and HMGA-2. Expression of let-7′s is frequently lost in tumors and correlates with poor prognosis in patients (6,7).
Lin28 is a small RBP expressed during embryonic development (8). In humans, there are two highly similar isoforms—LIN28 (Lin28A) and LIN28B (Lin28B)—which differ mainly in the sequences of their 3′UTRs. Lin28 is prominent for its ability to reprogram fibroblasts into induced pluripotent stem cells and for its pleiotropic functions that arise through interactions with mRNAs (9,10). Lin28A and Lin28B were shown to bind and suppress synthesis of let-7 by distinct mechanisms (11–16). Furthermore, since Lin28′s mRNA is a direct target of human let-7, these components are controlled in a double-negative feedback loop (17). This RNA-RBP relationship plays a prominent role in tumorigenesis (7), including the maintenance of self-renewal and the differentiation of cancer stem cells (CSCs) (18). Both Lin28A and Lin28B are oncogenes and as such promote cellular transformation. Indeed, many tumors of different histology that overexpress Lin28 show reduced levels of let-7 (7) and redressing this balance with Lin28A and Lin28B RNAi or let-7 overexpression inhibits tumor growth. Thus, the Lin28/let-7 interaction is a potentially interesting drug target: an antagonist that would block Lin28 access to let-7 precursors, without hindering the other elements of biogenesis, is expected to de-repress let-7 synthesis and rescue its growth-inhibitory function. Lin28 binds to single or multiple sites on let-7 precursors (19–21). It inhibits Drosha processing of pri-let-7 in the nucleus (11,12), as well as processing of pre-let-7 by Dicer in the cytoplasm (13,22). It also mediates degradation of pre-let-7 initiated by terminal uridyl transferases (14,23,24). One or combinations of these mechanisms are likely to operate depending on context- and/or cell type.
The molecular features of the Lin28/let-7 interaction were clarified through combined biochemical, spectroscopic and structural efforts. Both Lin28A and Lin28B carry a cold-shock domain (CSD) and two zinc-finger motifs (ZFD) with almost identical sequence. Using nuclear magnetic resonance spectroscopy, we showed that Lin28 ZFD binds a single-stranded purine-rich NGNNG motif in pre-let-7 TLRs, at a position proximal to the Dicer cleavage site by making contacts with the H-bonding faces of the two guanines (19). Mutations in the ZFDs or the NG-dinucleotides attenuate Lin28 binding and regulation. The importance of the CSD to binding and processing of let-7 was demonstrated by a crystal structure of murine Lin28A (20), while the ZFDs reportedly contribute most of the binding affinity of the interaction (21). We recently developed a novel RNA-based enzyme-linked immunosorbent assay (ELISA) in which an array of immobilized pre-miRNAs capture RBPs from cellular lysates (5). We have used variants of this assay to demonstrate that the ZFD of human Lin28A is sufficient to recognize pre-let-7s and that the equilibrium dissociation constants (KD) of the full-length protein with the precursors range from 0.2 to 0.5 nM (5).
The principal objective of this work was to test the novel concept of re-establishing let-7a synthesis using oligonucleotides that target the TLR of pre-let-7a-2 and bypass Lin28 inhibition, but still permit processing by Dicer and Drosha enzymes. A few reports have described how antisense oligonucleotides (ASOs) bind miRNA precursors and alter miRNA biogenesis. In one example, morpholino-modified oligonucleotides targeting the stem and TLR of miR-375 were used in zebrafish to block its synthesis (25). Our group showed that the locked nucleic acid (LNA)-drug miravirsen invades miR-122 precursors in primary hepatocytes and inhibits processing (26). The Caceres group described how RBP-mediated processing of several pri-miRNAs (including pri-let-7a-1) carrying conserved TLRs was inhibited by high concentrations of loop-binding ASOs (looptomirs) in lysates (27). Here we show in a systematic study by surface plasmon resonance (SPR) of three pre-miRNAs (miR-122, miR-18a, let-7a-2) that the binding affinity of looptomirs varies greatly with target site position, length and also presumably structure of the loop. In view of the potential of let-7/Lin28 as a drug target in cancer, we further investigated looptomirs against let-7a-2. We describe the identification of a pre-let-7a-2 looptomir with exceptionally high binding affinity that antagonizes binding of Lin28A and increases let-7 synthesis and activity in Lin28-expressing cancer cells. These findings define a starting point to the development of functional looptomirs for pharmacological applications or as tools to modulate processing of specific miRNA family members.
MATERIALS AND METHODS
Synthesis of oligoribonucleotides
All RNAs were synthesized under standard conditions on controlled pore glass solid (CPG) support. Biotinylated pre-miR-122 and pre-miR-18a were synthesized according to (26). The synthesis of the truncated and biotinylated pre-let-7a-2 and the Cy5-labeled L29-13 was done as reported in (28). 2′-O-Me-RNA molecules were synthesized under standard conditions. All crude oligonucleotides (Supplementary Table S1) were subjected to high-performance liquid chromatography (HPLC) (Agilent 1200 Series; Agilent Technologies, Santa Clara, USA) using a C18 column (XBridge OST, particle size 2.5 μm; Waters, Milford, USA). Purified samples were analyzed on an Agilent 6130 Series Quadrupole LC/MS (Agilent Technologies, Santa Clara, USA) with electron spray ionization. Purity and yields were determined by HPLC and Nanodrop, respectively.
SPR measurements
The Biacore measurements of looptomirs and controls against the hairpin of pre-let-7a-2 and pre-miR-122 were performed using a Biacore T-100 machine. A Tris buffer was used as running buffer (20 mM trishydroxymethylaminomethane, 5 mM MgCl2·6H2O, 2 mM CaCl2·6H2O, 2 mM KCl, 140 mM NaCl and 0.05% TWEEN 20 in sterile, desalted and rigorously degassed water). For the analysis, a Series S Sensorchip SA was used. Chemically synthesized biotinylated pre-let-7a-2 (a generous gift of P. Wenter, Eurofins MWG Operon) and pre-miR-122 in Tris buffer were injected twice for 24 s over flow cell 2 at a flow rate of 5 μl/min at 25°C to yield 158 RU for pre-let-7a-2 and 993.7 RU for pre-miR-122 overall response. Flow cell 1 was left empty for referencing. The measurements were performed at flow rates of 50 μl/min at 25°C. The sensorgrams of the pre-let-7a-2 analytes were obtained by injections of 225 μl (contact time 270 s) of 100 nM solutions with five 3-fold dilutions (down to 0.412 nM) in Tris-buffer followed by a dissociation phase of 600 s. For a regeneration solution, a 2 mM HCl solution (pre-let-7a-2) or a 0.5 mM HCl solution (pre-miR-122) was injected. The analysis was performed with EVILFIT (pre-let-7a-2) (29–31) and Scrubber2.0 (pre-miR-122).
The sensorgrams of the pre-miR-122 analytes were obtained by injections of 300 μl of 342 nM solutions with 2-fold dilutions in Tris buffer followed by a dissociation phase of 330 s. For a regeneration solution, we used a 1:1:1 mixture of water, an acid regeneration solution (equal volumes of oxalic acid, H3PO4, formic acid and malonic acid, each at 0.15 M, mixed and adjusted to pH 3.0 with HCl) and an ionic regeneration solution (0.46 M KSCN, 1.83 M MgCl2, 0.92 M urea and1.83 M guanidine HCl, pH 2). The fitting of the sensorgrams was performed using Scrubber2.0 (BioLogic Software, http://www.biologic.com.au) using a 1:1 binding model accounting for mass-transport limitation.
The measurements of looptomirs and controls against the hairpin of pre-miR-18a were performed using a SPR2 machine (Sierra Sensors, GmbH, Hamburg). A Tris buffer was used as running buffer. An amine chip pre-immobilized with streptavidin was coated with chemically synthesized biotinylated pre-miR-18a to yield 10 RU overall response. The measurements were performed at a flow rate of 25 μl/min at 25°C. For a regeneration solution a 10 mM ethylenediaminetetraacetic acid (EDTA) solution was injected. The analysis was performed with Scrubber2.0.
ELISA assay
White microtiter plates (96-well plates, NUNC, Maxisorp) were coated with streptavidin (2 μg/ml in PBS) and blocked with a 1% solution of a gelatin derivative (Top-Block, Sigma) in 25 mM HEPES, 0.05% Tween pH 7 for 1 h. After washing with water (used for all subsequent washing steps), a chemically synthesized 48-nt-long truncated pre-let-7a-2 containing the loop was allowed to bind to the surfaces for 3 h at a concentration of 2.5 nM in 25 mM HEPES pH 7. After washing, looptomirs in varying concentrations were incubated in binding buffer for 1 h in the wells. Myc-tagged Lin28A from HEK 293T cells or hnRNPA1-containing Hela cell lysate (harvested in buffer containing 50 mM Tris, 0.1 mM ZnCl2, 200 mM MgCl2, 0.5 mM Triton-X-100 (Fluka), 0.5 mM TCEP [tris(2-carboxyethyl)phosphine hydrochloride, neutralized] (Pierce, Cat. No. 77720) and 1x protease inhibitor (Complete EDTA-free, Roche Applied Science, Cat. No. 11873580001)) was diluted 500-fold in binding buffer and then added to the wells. This binding buffer contained 300 mM NaCl (for Lin28) or 250 mM NaCl (for hnRNPA1), 25 mM HEPES pH 7.2, 10 μM ZnCl2, 1% Top-Block, 0.05% Tween 20, 0.5 mM TCEP. Following incubation for 30 min at 4°C, the plate was completely emptied (without washing) and exposed to 50 μl of a fixation solution (0.5% formaldehyde in 300 mM NaCl, 20 mM sodium phosphate buffer, pH 7) for 10 min. The plate-bound Lin28 or hnRNPA1 was measured by antibodies specific for the Myc-tag (sc-40, clone 9E10; Santa Cruz) or hnRNPA1 (clone 4B10; Santa Cruz) at 0.1 μg/ml in 150 mM NaCl, 25 mM HEPES pH7.2, 10 μM ZnCl2, 1% Top-Block, 0.05% Tween 20. Incubation was for 1 h at room temperature. Bound primary antibody was detected by a secondary peroxidase conjugated anti-mouse IgG antibody (# 074-1806, KPL, Gaithersburg), 1:3000 diluted, with an incubation of 45 min at room temperature. Peroxidase activity was measured in a microtiter plate reader (Mithras 940; Berthold) using a chemiluminescent substrate (BM reagent; Roche Applied Science, Cat. no. 11 582 950 001). The data were fitted to a logistic equation (Y = B0/(1+([competitor]/IC50)sl)+BG) using the Solver feature of Excel. Y is the chemiluminescence measured in the assay, B0 corresponds to the signal of the protein without inhibitor, BG is the background signal, and IC50 and sl (slope) the parameters to be determined. A similar application of this method is described in (5,19).
Liquid chromatography-mass spectrometry (LC-MS) Dicer assay
T7 in vitro-transcribed pre-let-7a-2 (final concentration 2.5 μM) was incubated with 1 Unit of recombinant Dicer (T520002; Genlantis, San Diego) and looptomir (final concentration 10 μM) in a buffered aqueous solution (30 mM Tris–HCl, 3 mM MgCl2, 60 mM NaCl, 1 mM DTT) for 2.5 h at 37°C. The reactions were stopped by addition of 0.5 μl of 0.5 M EDTA pH 8 and kept on ice prior to analysis on a C18 RP column equilibrated at 70°C with detection set to 260 nm. Elutions were performed in aqueous buffer (0.4 M HFIP, 16.2 mM TEA) with a three-step MeOH gradient: 12–16% MeOH for 1 min, 16–17% MeOH for 8 min and 17–25% MeOH for 11 min.
Cell cultures and transfections
HEK 293-T cells and Huh-7 cells (ATCC) were cultured as monolayers in DMEM GlutaMAXTM-I (31966-021, Gibco®; Life Technologies, Carlsbad) supplemented with 10% of FBS (fetal bovine serum). HepG2 cells were maintained as a monolayer in DMEM/F-12 GlutaMAXTM (10565-018, Gibco®; Life Technologies) supplemented with 10% of FBS. Transfections were performed according to the manufacturer's protocol with Oligofectamine 2000 (12252-011, Invitrogen, Life Technologies, Carlsbad) for looptomirs and siRNAs and JetPEI (101-10; Polyplus transfections, Illkirch) was used for plasmid DNA.
Luciferase assays
Cells were seeded in opaque white 96-well-plates (136101; Nunc, Roskilde) in 80 μl medium/10′000 cells per well, and transfected according to the experimental setup with the reagents described above. Reporter plasmids were transfected at 20 ng. The assay was performed with the Dual-Glo® Luciferase Assay System (E2980; Promega, Fitchburg). The cell culture medium was removed and 30 μl of a solution composed of luciferase buffer with dissolved luciferase substrate and water in a ratio of 1:1 was added to each well. After 10 min, the luminescence was measured on a Mithras LB 940 plate reader (Berthold Technologies, Bad Wildbad). Fifteen microliter of a 1% solution of Stop & Glo® substrate in Stop & Glo® buffer was added to each well and luminescence was again measured after 10 min. Each time luminescence was measured for 1 s per well. For each well Renilla luminescence counts were normalized to firefly luminescence counts, and then average values were computed and normalized to the mock sample.
psiCHECK-2 reporter constructs
The target sequences were amplified from synthetic DNA using oligonucleotides (Supplementary Table S1) from Microsynth (Balgach) and cloned into the NotI and XhoI restriction sites of psiCHECK-2 plasmid (C8021; Promega, Fitchburg). The primers used for cloning as well as the inserted sequences in the psiCHECK2 vector are reported in the supplementary material. The ligated fragment was transformed into Subcloning Efficiency™ DH5α™ Competent E. coli (18265-017; Life Technologies, Carlsbad), plated on LB agar with 100 μg/ml Amp and incubated for at least 16 h at 37°C. The single colonies were picked and resuspended in water. Clones were screened by a Colony polymerase chain reaction (PCR) for the desired plasmid. Positive clones were used to make glycerol stocks and DNA minipreps were sequenced by GATC (Cologne). All the PCR purifications were performed following the protocol of MinElute® PCR Purification Kit (250) (28006; Qiagen, Venlo). Minipreps were performed following the protocol of QIAprep® Spin Miniprep Kit (250) (27106; Qiagen, Venlo). Digestions were performed following the guideline of the NEB website (http://www.neb.com) using NotI and XhoI according to the supplied protocol (NEB, Ipswich). The ligations were performed with ratios of 1:3, 1:4 (vector to insert) using the dephosphorylated psiCHECK-2 vector with T4 DNA Ligase (M1804; Promega, Fitchburg).
Quantitative reverse transcriptase-polymerase chain reaction
Total RNA was extracted using the RNeasy kit (74104, Qiagen, Venlo). TaqMan® quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) was performed using standard reagents from Life Technologies (TaqMan® MicroRNA Assays: hsa-let-7a: 000377, hsa-let-7c 000379, hsa-let-7g: 002282, RNU44: 001094). The RT was performed using the TaqMan® primers from MicroRNA Assays and the TaqMan® MicroRNA Reverse Transcription Kit (4366596, Life technologies, Carlsbad) with 25 ng total RNA. The PCR was performed in a LightCycler 480 instrument (Roche, Penzberg) with GoTaq® Probe qPCR Master mix (A6102, Promega, Fitchburg) according to the manufacturer's protocol. Each reaction was carried out in four technical replicates. Ct values were calculated for each and averaged.
Western blot
Cells were lysed in lysis buffer (10 mM Hepes, 400 mM NaCl, 3 mM MgCl2, 0.5%Triton X-100, 1 mM DTT and 10% glycerol) for protein detection. Protein concentrations were determined using a BCA assay (Thermo Fisher Scientific 23225); 15 μg of protein was mixed with equal quantities of sodium dodecyl sulphate (SDS) loading buffer (100 mM Tris–HCl, 4% SDS, 20% glycerol, 0.2% bromophenol blue). Samples were heated at 95°C for 5 min, separated on SDS gels and transferred to polyvinylidene difluoride membranes. Non-specific membrane binding was blocked for 40 min at room temperature with 5% milk in phosphate-buffered saline containing 0.05% Tween 20. Membranes were incubated overnight at 4°C with appropriate primary antibodies against Lin28B (#A4196; Cell Signaling Technology, Danvers), HMGI-C (#sc-30223; Santa Cruz Biotechnology, Dallas) or β-actin (#A5316; Sigma Life Science, St. Louis). After washing, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature in blocking buffer and washed again. Signals generated by the chemiluminescent substrate (ECL(+); GE Healthcare, Little Chalfont) were captured by a cooled CCD camera (Bio-Rad, Hercules). Protein bands were quantified by densitometry using the analysis software Fiji. Samples were normalized against β-actin protein and then on the corresponding protein level after treatment with the negative control sequence Lcon.
Sulforhodamine B cell proliferation assay
Cells (Huh7 at 5000 cells/well; HepG2 at 6000 cells/well) were seeded in transparent 96-well-plates (92096, TPP, Trasadingen) in 80 μl medium per well for 4 h before transfection as described above. Cell proliferation was assessed every 12 h from the first day until the third day after transfection. Medium of cells was removed prior to fixing in 10% ice cooled trichloroacetic acid (TCA). Plates were washed with tap water and air dried at room temperature. Sulforhodamine B (SRB) (0.057%) (S1402-1G; Sigma Life Science, St. Louis) was used for staining prior to washing the plates three times in 1% acetic acid. SRB was dissolved in 10 mM trisbase solution (pH 10.5). After SRB solubilization, plate OD at 530 nm was measured on Mithras LB 940 plate reader.
Colony formation assay
For clonogenicity analysis, cells (Huh7: 100 cells/well; HepG2: 1000 cells/well) were seeded in transparent 24-well-plates (92024, TPP, Trasadingen) in 500 μl medium per well 6 h prior to transfection as described above. After 10 days of cell growth, cells were fixed with methanol and stained with crystal violet 1%. Colonies were assessed by manual counting and using the analysis software Fiji.
Sphere cultures
HepG2 cells (40′000 cells/well) were seeded in six-well-plates (92006, TPP, Trasadingen) in 2 ml medium per well 6 h prior to transfection as described above. Twelve hours after transfection, HepG2 cells from monolayer culture were collected and washed to remove serum and then suspended in serum free DMEM/F-12 GlutaMAXTM (10565-018, Gibco®; Life Technologies) supplemented with 1x B-27® Supplement (Gibco; Life technologies, Carlsbad), 100 μg/ml penicillin-streptomycin (Gibco; Life technologies, Carlsbad), 20 ng/ml recombinant human EGF (AF-100-15; PeproTech, Rocky Hill) and 10 ng/ml recombinant human FGF-basic 154 amino acids (100-18B, PeproTech, Rocky Hill) prior to seeding 1000 cells/ml on ultra-low attachment culture dishes (Corning, Tewksbury). Sphere formation was assessed 6 days after treatment before spheres reached a diameter of 100 μm.
Imaging methods
For live imaging of L29-13 transfection efficacy Huh7 cells (4′000 cells/well) were seeded in eight well chambered # 1.0 Borosilicate Coverglass System dishes (Thermo Fisher Scientific, Nunc, Rochester) in 250 μl medium per well 6 h prior to transfection with L29-13 bearing a Cy5-label. Twelve hours later stacks of images were taken by a Zeiss Spinning Disk Fluorescence and Brightfield Confocal Microscope. One middle slice was chosen and post-processed in Fiji using ‘unsharpen mask’ command. Fluorescence image stacks of immunostained HepG2 and Huh7 cells were taken by a Leica Confocal SP2 Microscope and images were merged using Z-projection. Pictures were post-processed in Fiji using ‘unsharpen mask’ command.
Statistics
Each experiment was repeated at least three times. All statistical analyses were performed by ANOVA using Dunnett's post-test, comparing against the lowest dose in each group or the negative control treatment. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. All statistics were run with GraphPad Prism 6.
RESULTS
A few studies have described the binding properties of short ASOs (including looptomirs) complementary to sites on RNA hairpins derived from mRNAs (32) and pre-miRNAs (33). In one early study of a loop-forming region of Ha-ras mRNA (32), most of the ASOs tested were shown to bind with much lower affinity to the hairpin than to length-matched single-stranded complements, reflecting a strong negative influence of the RNA structure on hybridization. Interestingly however, one of the sequences bound to the 5′ side of the loop with higher affinity than to its single-strand complement, suggesting that its binding site was favored for duplex formation. Our objective was to identify such a privileged binding site for a looptomir on the let-7a-2 TLR, which would prevent Lin28 from docking, while leaving the hairpin structure intact so that other miRNA-processing factors, e.g. Dicer, or Drosha would not be impeded.
In view of the lack of guidelines in the literature on how to predict optimal binding sites, we resorted to empirical testing, similar to the approach of Mandir (33) but using oligonucleotides in solution. Before addressing looptomirs against a let-7 member, we selected two pre-miRNAs—miR-122 and miR-18a—to investigate how the affinity of a series of fixed-length looptomirs was affected by binding site location. For pre-miR-122, the most stable Mfold-predicted structure suggests that it has a stable stem-loop junction with a loop of 12 nt (Figure 1a). On the other hand, for pre-miR-18a, mismatches and bulges close to the sites of Dicer cleavage suggest that its loop may ‘breathe’ (Figure 1b). We synthesized pre-miR-122 and pre-miR-18a with terminal biotin groups and anchored them to streptavidin-coated chips for SPR spectroscopy, using a protocol we described previously (26,34). We then measured systematically the binding affinities of 2′-O-methyl (Me) looptomirs (length: 10-nt) by ‘walking’ around the TLRs. Looptomir-binding affinities were all in the nanomolar range (Figure 1a and b). For both cases however, at positions close to the 3′-side of the loop, shifts in the looptomir sequence by a single nucleotide resulted in large changes of affinity. Thus, for looptomirs against pre-miR-122, the affinity dropped by factors of more than 3-fold around the tightest binding sequence SP25. The low affinity of SP27 to 29 can be explained by the energy cost of opening the predicted stem. The 5′ terminus of looptomir SP25 bound to the last unpaired base before the stem-loop junction, possibly stacking co-axially on the stem. In the case of pre-miR-18a, the step from SP26 to SP27 was accompanied by an 18-fold increased affinity, hardly explained by the energy gain of an additional GC pair. Looptomirs starting near the 5′ terminal of the loop were clearly disfavored. Taken together, the data indicated that high affinity binding sites for looptomirs with 5′ ends close to the 3′ side of the loops may be a general feature of (pre-miRNA) stem-loops. The results of this study encouraged us to scan for optimal looptomirs on the pre-let-7a-2 loop with the additional requirement of preservation of the Dicer cleavage site. Pre-let-7a-2 has an unusually long TLR in which the binding site for Lin28 is located on the 3′ side of the loop (Figure 1c).
Figure 1.
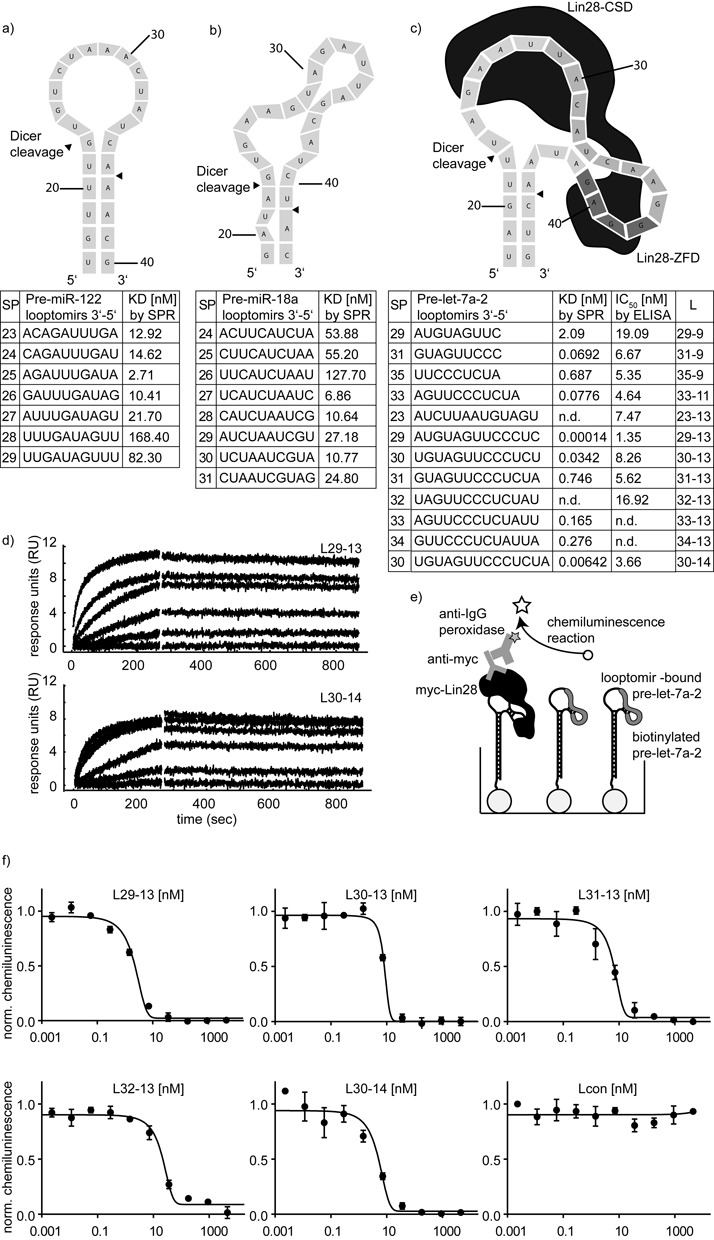
Looptomirs targeting the TLR of pre-let-7a-2 inhibit the binding of Lin28A. (a) Predicted structure (Supplementary Figure S9) of pre-miR-122 TLR and binding affinity of 7 10-nt 2′-O-Me-looptomirs measured for binding to biotinylated pre-miR-122 bound to the surface of a streptavidin-coated biosensor by SPR spectroscopy. (SP: position of the looptomir 3′ position). (b) Predicted structure (Supplementary Figure S9) of pre-miR-18a TLR and binding affinity of 8 10-nt 2′-O-Me-looptomirs measured by SPR spectroscopy (SP: position of the looptomir 3′ position). (c) Upper: Predicted structure (Supplementary Figure S9) of the TLR of pre-let-7a-2 showing the sites of interaction of Lin28 zinc finger (Lin28-ZFD)- and cold shock-domains (Lin28-CSD) and Dicer cleavage sites (arrowheads); Lower: looptomirs were assayed for binding to a truncated biotinylated pre-let-7a-2 by SPR spectroscopy (Lx-y refers to a 2′-O-Me looptomir of length y nt, binding with its 3′ terminal nt at position x); binding data of selected looptomirs to pre-let-7a-2 from RNA-based ELISA. (d) SPR sensorgrams from the two strongest binders L29-13 (upper panel) and L30-14 (lower panel) are shown (sensorgrams were analyzed with EVILFIT, as described previously (26)). (e) Principle of the RNA-based ELISA, which measures the IC50 of looptomir oligonucleotides for inhibition of Lin28 binding to immobilized pre-let-7a-2. Full-length Myc-tagged Lin28A from HEK 293 T cell lysate was added to wells and the fraction of bound Lin28A was determined by immunostaining of its Myc-tag. (f) RNA ELISA curves for six selected looptomirs complementary to the TLR depicted in (c).
We synthesized a variety of 2′-O-Me looptomirs including a series of 13-nt sequences (Figure 1c and supplementary information). Some of these partially covered the Lin28-CSD and the Lin28-ZFD binding sites on the 5′ and 3′ sides of the TLR, respectively (Figure 1c), but all were sufficiently short so as not to invade and avoid perturbing the stem region close to the sites of Dicer cleavage. We focused on the 3′ side of the loop because of the observations made with pre-miR-122 and pre-miR-18a, but also because it hosted the binding site for Lin28. Most of the sequences bound with high picomolar to low nanomolar affinities (Figure 1c, Supplementary Figure S1). Notably, in some pairwise comparisons, increasing the length of the looptomir did not always increase its binding affinity (L31-9 versus 31-13; L33-11 versus L33-13), suggesting that conformational restrictions in the loop structure prevented some looptomirs from binding throughout their whole length. Consistent with the observations made with pre-miR-122 and pre-miR-18a, one looptomir, L29-13, showed a particularly high affinity and shifting its sequence by a single nucleotide (to L30-13) drastically reduced its binding strength. We selected the two most potent sequences for follow-up studies: L29-13 and the 14 nt L30-14. Their affinities were in the sub-nanomolar range and showed very slow rates of dissociation (Figure 1d), though L29-13 was clearly the stronger binder of the two.
Looptomir sequences were then tested for their ability to inhibit a Lin28A-Myc-tagged protein from HEK 293T cell lysate from binding to pre-let-7a-2 in the RNA-based ELISA (Figure 1e, 1f, Supplementary Figure S2). An unrelated negative control sequence Lcon was inactive. Rankings of the inhibitory potency of the Kd values in the RNA-based ELISA and in the SPR-assay, respectively correlated rather well (Figure 1c). The two lead sequences (L29-13: IC50 = 1.35 nM; L30-14: IC50 = 3.66 nM) were particularly effective, with the shorter sequence being approximately 3-fold more active (Figure 1c). Other RBPs are known to interact with let-7 precursors. HnRNPA1 was shown to bind to nuclear pri-let-7a-1 and inhibit its processing by displacing the positive regulator KSRP (35). We showed previously that hnRNPA1 bound most pre-let-7 family members with nanomolar affinity (5). We therefore examined whether L29-13 and L30-14 would also inhibit hnRNPA1 from binding to pre-let-7a-2: incubation of the looptomirs with immobilized pre-let-7a-2, did not prevent hnRNPA1 from binding (Supplementary Figure S3), possibly because the binding sites of the looptomir and hnRNPA1 are not coincident.
Having identified two looptomirs which antagonize sequence-selectively the Lin28A/pre-let-7a-2 interaction, we then examined their potential effects on the processing of pre-let-7a-2 by recombinant Dicer in vitro using a reverse-phase high-performance liquid chromatography (RP-HPLC)-based assay (Figure 2a). Dicer cleavage of pre-miRNAs in cells and in vitro can be influenced by various attributes of the hairpin structure leading to cleavage sites spanning several nucleotides (36,37). Hence, incubation of recombinant Dicer with pre-let-7a-2 produced 5′-phosphorylated RNAs identified by their masses, corresponding to let-7a-5p and let-7a-3p sequences reported in miRBase (www.mirbase.org) (Figure 2b–e). At least two sequences differing by one nt for let-7a 5p- and 3p- miRNAs were identified, accompanied by a large number of minor fragments and unprocessed starting material. Next, pre-let-7a-2 was incubated separately with equal concentrations of looptomirs L29-13 and Lcon. The assays produced highly similar cleavage patterns with no discernible differences in residual intact precursor, strongly suggesting that L29-13 does not disturb Dicer processing of pre-let-7a-2 (Figure 2d). As L29-13 binds strongly to pre-let-7a-2, at least in vitro (Figure 1c), we cannot be certain whether Dicer, which also binds pre-miRNAs with nanomolar affinity (38), processes the pre-miRNA in its L29-13-bound or -unbound state (Figure 2d).
Figure 2.
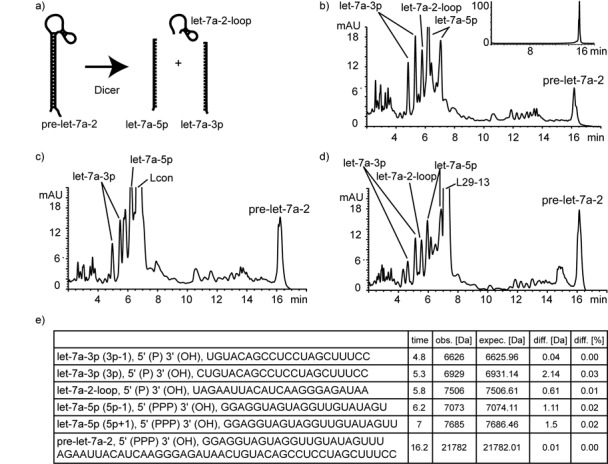
Pre-let-7a-2 is cleaved by Dicer into mature miRNAs in the presence of let-7 looptomirs. (a) Schematic cleavage of pre-miRNAs by Dicer into mature miRNA 5p and 3p strands. (b) In vitro-transcribed pre-let-7a-2 incubated with recombinant Dicer for 150 min: reaction mixtures were analyzed by RP-HPLC, and peaks were identified by MS (inset: HPLC-chromatogram of pre-let-7a-2 used in the study). Pre-let-7a-2 incubated under identical conditions with recombinant Dicer and Lcon (c) and L29-13 (d). Products were analyzed by RP-HPLC. (e) Masses observed and calculated for expected miRNA products from (b).
Having identified two let-7 looptomirs that satisfied our preliminary requirements as selective antagonists of Lin28A/pre-let-7a-2 in vitro, we examined their activities in several cellular systems. We chose HEK 293T and hepatocellular carcinoma (HCC)-derived Huh7 and HepG2 cells, all of which reportedly (14,16,39) express Lin28A and B. Treatment of Huh7 cells with a Cy-5-labeled L29-13 analog complexed with Oligofectamine indicated that the single-stranded short oligonucleotide readily accumulated in the cytoplasm (Supplementary Figure S4). Furthermore, in HepG2 and Huh7 cells immunofluorescence showed clearly an accumulation of endogenous Lin28B in the cell cytoplasm and cell nucleus, as reported previously (16,39) (Supplementary Figure S5).
Transfection of both L29-13 and L30-14 into Huh7, HEK 293T (Figure 3a) and HepG2 (Supplementary Figure S6) cells elevated endogenous let-7a levels up to 4-fold as measured by real-time RT-PCR (RT-qPCR). L29-13 was more effective than L30-14 in HEK 293T, consistent with its higher potency in the in vitro assays. L29-13 was also potent several days post-transfection in a cancer sphere-forming assay, which is characteristic of CSCs and relates to stemness of hepatoma cells in vitro (40,41). Under sphere-forming conditions in HepG2 cells, L29-13 increased let-7a levels 8-fold after 6 days. For comparison, a pre-characterized Lin28B siRNA induced let-7 levels more than 20-fold, presumably by allowing processing of all expressed let-7 family members in parallel (Figure 3b).
Figure 3.
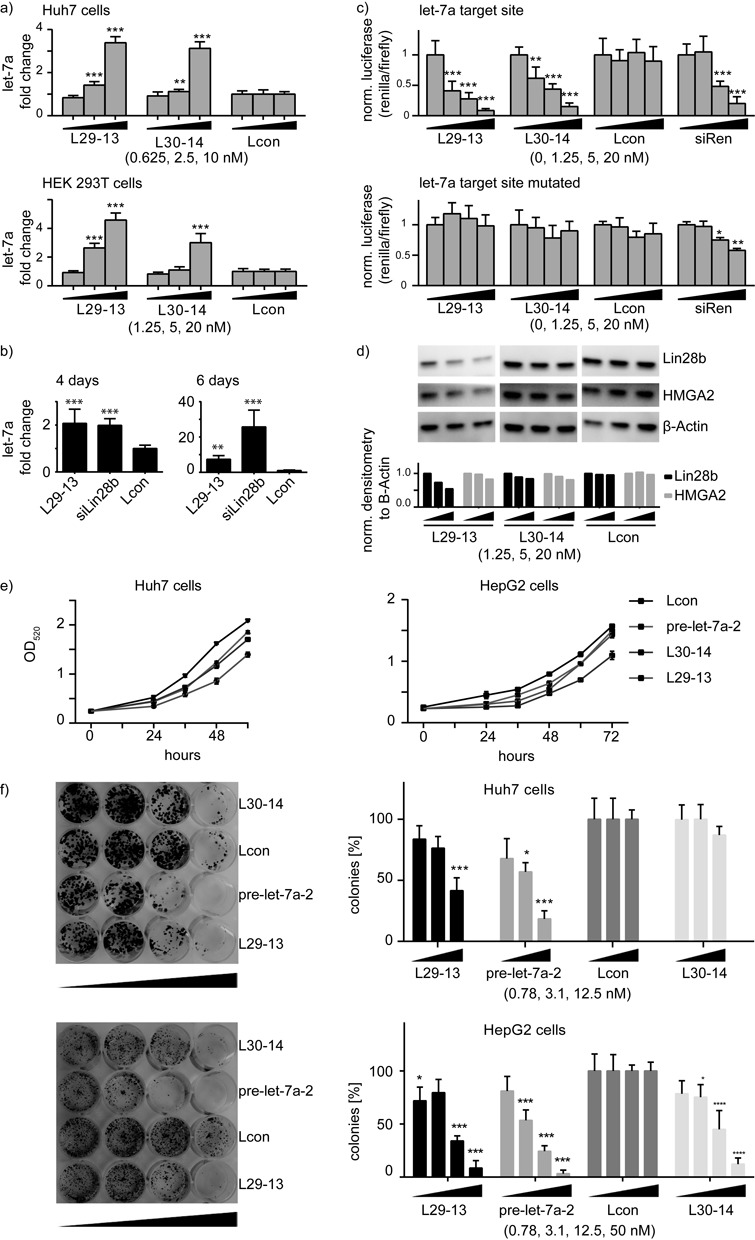
Looptomirs L29-13 and L30-14 induce the expression of functional let-7a. (a) Endogenous levels of mature let-7a 48 h after transfections of L29-13, L30-14 and Lcon in Huh7 and HEK 293T cells, measured by TaqMan RT-qPCR (normalized to RNU44 and Lcon); similar results were found in HepG2 cells (Supplementary Figure S6). (b) Activity of L29-13 and siLin28b in sphere-forming HepG2 cells: TaqMan RT-qPCR of cancer stem-like cells measuring endogenous let-7a levels 4 (left panel) and 6 days (right panel) after transfection of L29-13 at 50 nM concentration. (c) Looptomirs and controls were co-transfected with dual-luciferase vectors into Huh7 cells to measure activity of endogenous let-7. Upper panel: a ‘sensor’ vector containing one let-7a fully complementary target site; lower panel: the same construct mutated at 3 positions in the seed sequence. (d) Western blots showing levels of let-7 target proteins Lin28B and HMGA2 after treatment of Huh7 cells with let-7 looptomirs. (e) Huh7 (left panel) and HepG2 (right panel) cells were transfected with 50 nM of L30-14 and L29-13, as well as pre-let-7a-2 and Lcon controls. OD of stained cells was measured over a 4-day period: OD was normalized to the Lcon treatments. Error bars are SD; ***: P < 0.001. (f) Huh7 (upper panels) and HepG2 cells (lower panels) were transfected with 0.8, 3.1, 12.5 and 50 nM of looptomirs and controls. Colonies were stained with crystal violet and counted after 10 days; cell numbers were normalized to Lcon treatments (toxicity was observed in the 50 nM treatment of Huh-7 cells). Error bars are SD; ***: P < 0.001.
Next, we investigated whether the levels of let-7a elevated by looptomirs in HCC cells were functional. A sensitive means to measure the activity of miRNAs are Renilla reporter gene assays (42). We prepared a vector containing a single complementary target site for let-7a, as well as a control vector in which the site was mutated at three nucleotides. Co-transfection into Huh7 cells of the reporter plasmid together with a previously characterized positive-control siRNA (siRen) (42) led to a strong reduction in Renilla luminescence (Figure 3c). Looptomirs L29-13 and L30-14 repressed Renilla efficiently, with the former being approximately 2-fold more active. No effects were observed on the mutated reporter or from treatment with Lcon (Figure 3c). Taken together with the in vitro experiments (Figures 1 and 2), the results confirmed that let-7 looptomirs re-established the expression and function of let-7, at least partly through selective antagonism of Lin28, likely in the cell's cytoplasm. We then examined whether let-7a-2 looptomirs would silence endogenous let-7 targets in cells. For well-validated targets of let-7, which are highly expressed in HCC cells, we selected HMGA2 (43) and Lin28B (17), both of which are confirmed oncogenes. Lin28B is highly expressed in HCC patient samples, and is associated with coordinate repression of all let-7 family members (7). Huh7 cells were transfected with looptomirs and protein was isolated for western blotting using antibodies for HMGA2 and Lin28B. Whereas L30-14 showed little or no activity (Figure 3d and Supplementary Figure S7), the more potent L29-13 reduced levels of Lin28B protein significantly, with a minor effect on HMGA2. Similar results were obtained with protein samples of L29-13-transfected HepG2 cells (Supplementary Figures S7 and S8). Although the inhibition of the two targets is relatively weak under these conditions compared to the direct effects from a Lin28B siRNA, these can be considered as strong effects from a 13 nt 2′-O-Me oligoribonucleotide (ORN) acting indirectly by a distinct mechanism. Reports describing similar approaches using short 2′-O-Me ORNs are rare (see reference (44) for an example). Although the 2′-O-Me modification of ORNs exhibits generally increased thermodynamic binding affinity for RNA, as well as increased nuclease resistance, several higher affinity-binding looptomirs would be expected to demonstrate more potent effects.
To determine whether L29-13 was able to inhibit cellular proliferation through increased let-7, we monitored growth of treated Huh7 and HepG2 cells over time. We employed exogenously-delivered pre-let-7a-2 as a positive control for this assay as we have shown previously that pre-miRNAs are effective miRNA mimics (42). Although it is probably also a substrate for endogenous Lin28, there is sufficient hairpin to produce mature let-7a (19). In Huh7 cells, delivery of L29-13, pre-let-7a-2 and L30-14 inhibited significantly cell proliferation, with L29-13 showing a comparable effect to that of pre-let-7a-2 (Figure 3e). Similar results were obtained in HepG2 cells, although L30-14 showed a surprisingly strong effect after 60 h, which may be mechanism-independent (Figure 3e). Finally, a colony formation assay in Huh7 and HepG2 cells provided an additional clear-cut demonstration of the effects of the pre-let-7a and L29-13 treatments on clonogenic potential. Both reagents reduced colony formation at the concentration of 12.5 nM, whereas Lcon and L30-14 were not, or less effective (Figure 3f).
DISCUSSION
In summary, although chemically-modified ORNs are highly useful ligands to inhibit the activity of messenger and non-coding RNAs, their use to modulate well-characterized RNA-RBP interactions is rare. In this context, the Lin28/pre-let-7 interaction is an exciting drug target for the subset of cancers in which Lin28 and let-7 share a mechanistically founded inverse relationship of expression and function. At least four RBPs are known to bind to let-7 TLR during regulation of its biogenesis: Dicer, hnRNPA1 (35), KSRP (45) and Lin28. In this study we used our recently developed in vitro binding assay to scan the pre-let-7a-2 TLR for a high-affinity looptomir. Previous studies have suggested that high affinity sites for complementary ASOs in highly structured RNAs (e.g. hairpins) cannot be rationalized on the basis of simple base-pairing thermodynamics, i.e. looptomir length and sequence (32). In addition, they showed that large swings in binding affinity for ASOs occur on shifting by one or a few nucleotides, caused possibly by pre-organization of a binding site, or co-axial stacking onto the closing base-pair of a stem region (46). We made similar observations on scanning the TLRs of pre-miR-122, pre-miR-18a and pre-let-7a-2 with a series of looptomirs. We identified L29-13, as the most potent 13-nt sequence, which hybridizes on the 3′ side of the pre-let-7a-2 TLR, covering two key guanines important for interaction with the Lin28-ZBD. L29-13 was also the most potent inhibitor of the interaction between Lin28A and pre-let-7a-2; however, it did not prevent hnRNPA1 from interacting with pre-let-7a-2. Consistent with these observations, L29-13 selectively de-repressed Lin28 suppression of let-7 synthesis. In Lin28B-expressing HCC cells, the increased levels of let-7 suppress endogenous targets that drive proliferation and clonogenic potential.
Modified oligonucleotides represent an exciting new class of drugs, but their development is complicated by their large size. It is striking that potent inhibitory activity is obtained from a short 13-nt looptomir antagonist of the Lin28/pre-let-7a-2 complex. The effects of L29-13 may, to some degree, be amplified in this case by the activation of the Lin28/let-7 double-negative feedback loop (with prolonged inhibition of Lin28 function). Bypassing Lin28 activity with the help of a substrate specific oligonucleotide represents a new approach to addressing Lin28-dependent cancers in addition to double-stranded siRNAs or mimics. Thanks to the double-negative feedback loop, let-7 looptomirs, let-7 mimics and Lin28 siRNAs may show overlapping biological effects, despite their targeting of the loop at different points. There will likely be advantages and disadvantages to all three approaches; however, short high-affinity looptomirs may hold important advantages over their larger double-stranded counterparts. For example, the size and chemical structure of double-stranded siRNAs and miRNA mimics cannot be greatly modified without abrogating their ability to activate RISC (47). In contrast, single-stranded oligonucleotide drugs that function by a target-occupancy mechanism can be shortened and highly modified to optimize both their pharmacodynamics and pharmacokinetics. Indeed, recent work has demonstrated the superiority of shorter oligonucleotides in terms of unassisted cellular uptake and in vivo efficacy (48). Furthermore, current data from a related study indicate that off-target effects inherent to siRNAs may be less of a concern when using fully-modified single-stranded oligonucleotides (49). Based on the data in the manuscript, it is not possible to predict which of the three aforementioned reagent classes would be pharmacologically superior. This will be determined only by empirically testing head-to-head large numbers of modified siRNAs, mimics and looptomirs.
As our knowledge concerning the regulation of miRNA biogenesis increases, we anticipate that our approach to developing and using looptomirs may be of value in three areas: uncovering new functional roles of RBPs, as unique tools to modulate the processing of specific miRNA family members and as pharmacological agents in disease-related mechanisms, for example Lin28/let-7 in cancer, the processing of miR-1 in myotonic dystrophy (50) or the targeting of miR-122 biogenesis for hepatitis C infections (26).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Andreas Brunschweiger for synthesis of biotinylated pre-let-7a-2, Luca Gebert for preliminary Dicer assays and F. Loughlin for Myc-Lin28 expressing plasmid.
FUNDING
Krebsliga Schweiz [KFS-2648-08-2010]; the Swiss National Science Foundation [CRSII3_127454]; Eidgenössische Technische Hochschule Grant [ETH-01 11-2 to J.H.]. Funding for open access charge: ETH Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Castilla-Llorente V., Nicastro G., Ramos A. Terminal loop-mediated regulation of miRNA biogenesis: selectivity and mechanisms. Biochem. Soc. Trans. 2013;41:861–865. doi: 10.1042/BST20130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siomi H., Siomi M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Towbin H., Wenter P., Guennewig B., Imig J., Zagalak J.A., Gerber A.P., Hall J. Systematic screens of proteins binding to synthetic microRNA precursors. Nucleic Acids Res. 2013;41:e47. doi: 10.1093/nar/gks1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Büssing I., Slack F.J., Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Viswanathan S.R., Powers J.T., Einhorn W., Hoshida Y., Ng T.L., Toffanin S., O'Sullivan M., Lu J., Phillips L.A., Lockhart V.L., et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viswanathan S.R., Daley G.Q. Lin28: a microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 10.Jin J., Jing W., Lei X.X., Feng C., Peng S., Boris-Lawrie K., Huang Y. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman M.A., Thomson J.M., Hammond S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 14.Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Piskounova E., Viswanathan S.R., Janas M., LaPierre R.J., Daley G.Q., Sliz P., Gregory R.I. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 16.Piskounova E., Polytarchou C., Thornton J.E., LaPierre R.J., Pothoulakis C., Hagan J.P., Iliopoulos D., Gregory R.I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Lin X., Zhong X., Kaur S., Li N., Liang S., Lassus H., Wang L., Katsaros D., Montone K., et al. Double-negative feedback loop between reprogramming factor LIN28 and microRNA let-7 regulates aldehyde dehydrogenase 1-positive cancer stem cells. Cancer Res. 2010;70:9463–9472. doi: 10.1158/0008-5472.CAN-10-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shyh-Chang N., Daley G.Q. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loughlin F.E., Gebert L.F.R., Towbin H., Brunschweiger A., Hall J., Allain F.H.T. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 2012;19:84–89. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 20.Nam Y., Chen C., Gregory R.I., Chou J.J., Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desjardins A., Yang A., Bouvette J., Omichinski J.G., Legault P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2012;40:1767–1777. doi: 10.1093/nar/gkr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lightfoot H.L., Bugaut A., Armisen J., Lehrbach N.J., Miska E.A., Balasubramanian S. A LIN28-dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–7521. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagan J.P., Piskounova E., Gregory R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thornton J.E., Chang H.M., Piskounova E., Gregory R.I. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H.A. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebert L.F.R., Rebhan M.A.E., Crivelli S.E.M., Denzler R., Stoffel M., Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michlewski G., Guil S., Semple C.A., Cáceres J.F. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pradère U., Brunschweiger A., Gebert L.F.R., Lucic M., Roos M., Hall J. Chemical synthesis of mono- and bis-labeled pre-microRNAs. Angew. Chem. Int. Ed. Engl. 2013;52:12028–12032. doi: 10.1002/anie.201304986. [DOI] [PubMed] [Google Scholar]
- 29.Schuck P., Zhao H. The role of mass transport limitation and surface heterogeneity in the biophysical characterization of macromolecular binding processes by SPR biosensing. Methods Mol. Biol. 2010;627:15–54. doi: 10.1007/978-1-60761-670-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svitel J., Boukari H., Ryk D.V., Willson R.C., Schuck P. Probing the functional heterogeneity of surface binding sites by analysis of experimental binding traces and the effect of mass transport limitation. Biophys. J. 2007;92:1742–1758. doi: 10.1529/biophysj.106.094615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorshkova I.I., Svitel J., Razjouyan F., Schuck P. Bayesian analysis of heterogeneity in the distribution of binding properties of immobilized surface sites. Langmuir. 2008;24:11577–11586. doi: 10.1021/la801186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lima W.F., Monia B.P., Ecker D.J., Freier S.M. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- 33.Mandir J.B., Lockett M.R., Phillips M.F., Allawi H.T., Lyamichev V.I., Smith L.M. Rapid determination of RNA accessible sites by surface plasmon resonance detection of hybridization to DNA arrays. Anal. Chem. 2009;81:8949–8956. doi: 10.1021/ac9015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebhan M.A.E., Brunschweiger A., Hall J. Measurement by SPR of very low dissociation rates: oxidation-mediated loss of biotin-streptavidin affinity. Chembiochem. 2013;14:2091–2094. doi: 10.1002/cbic.201300468. [DOI] [PubMed] [Google Scholar]
- 35.Michlewski G., Cáceres J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J.E., Heo I., Tian Y., Simanshu D.K., Chang H., Jee D., Patel D.J., Kim V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermeulen A., Behlen L., Reynolds A., Wolfson A., Marshall W.S., Karpilow J., Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma E., MacRae I.J., Kirsch J.F., Doudna J.A. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y., Chen Y., Ito H., Watanabe A., Ge X., Kodama T., Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Visvader J.E., Lindeman G.J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 41.Uchida Y., Tanaka S., Aihara A., Adikrisna R., Yoshitake K., Matsumura S., Mitsunori Y., Murakata A., Noguchi N., Irie T., et al. Analogy between sphere forming ability and stemness of human hepatoma cells. Oncol. Rep. 2010;24:1147–1151. doi: 10.3892/or_00000966. [DOI] [PubMed] [Google Scholar]
- 42.Guennewig B., Roos M., Dogar A.M., Gebert L.F.R., Zagalak J.A., Vongrad V., Metzner K.J., Hall J. Synthetic pre-microRNAs reveal dual-strand activity of miR-34a on TNF-α. RNA. 2014;20:61–75. doi: 10.1261/rna.038968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayr C., Hemann M.T., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gude L., Berkovitch S.S., Santos W.L., Kutchukian P.S., Pawloski A.R., Kuimelis R., McGall G., Verdine G.L. Mapping targetable sites on human telomerase RNA pseudoknot/template domain using 2′-OMe RNA-interacting polynucleotide (RIPtide) microarrays. J. Biol. Chem. 2012;287:18843–18853. doi: 10.1074/jbc.M111.316596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mir K.U., Southern E.M. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- 47.Shukla S., Sumaria C.S., Pradeepkumar P.I. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. ChemMedChem. 2010;5:328–349. doi: 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- 48.Straarup E.M., Fisker N., Hedtjarn M., Lindholm M.W., Rosenbohm C., Aarup V., Hansen H.F., Orum H., Hansen J.B., Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obad S., dos Santos C.O., Petri A., Heidenblad M., Broom O., Ruse C., Fu C., Lindow M., Stenvang J., Straarup E.M., et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rau F., Freyermuth F., Fugier C., Villemin J.P., Fischer M.C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D., et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


