Abstract
In the current model of DNA SSBR, PARP1 is regarded as the sensor of single-strand breaks (SSBs). However, biochemical studies have implicated LIG3 as another possible SSB sensor. Using a laser micro-irradiation protocol that predominantly generates SSBs, we were able to demonstrate that PARP1 is dispensable for the accumulation of different single-strand break repair (SSBR) proteins at sites of DNA damage in live cells. Furthermore, we show in live cells for the first time that LIG3 plays a role in mediating the accumulation of the SSBR proteins XRCC1 and PNKP at sites of DNA damage. Importantly, the accumulation of LIG3 at sites of DNA damage did not require the BRCT domain-mediated interaction with XRCC1. We were able to show that the N-terminal ZnF domain of LIG3 plays a key role in the enzyme's SSB sensing function. Finally, we provide cellular evidence that LIG3 and not PARP1 acts as the sensor for DNA damage caused by the topoisomerase I inhibitor, irinotecan. Our results support the existence of a second damage-sensing mechanism in SSBR involving the detection of nicks in the genome by LIG3.
INTRODUCTION
Protecting the integrity of DNA is pivotal in maintaining cellular homeostasis. However, cellular DNA is continually damaged by intracellular and extracellular agents such as reactive oxygen species, ionizing radiation, and genotoxic chemicals. These agents cause various forms of DNA insults, and accordingly, living cells possess a large repertoire of proteins that function in the repair of DNA in damage-specific pathways (1). One of the most frequently encountered forms of DNA damage is DNA single-strand breaks (SSBs). SSBs can arise as a direct consequence of exposure to endogenous or exogenous DNA damaging agents and are also generated during the base excision repair (BER) pathway (indirect SSBs) (2). SSBs are defined as either short gaps (breaks involving loss of nucleotides) or nicks (breaks in the sugar-phosphate backbone with no missing nucleotides) that compromise the integrity of the DNA backbone. In this work, we aimed to provide cellular insights into SSB repair (SSBR) with a major emphasis on the SSB sensing step.
Based on biochemical studies, the current model for SSBR incorporates four distinct steps. The first step is SSB sensing mediated by PARP1 through its zinc finger (ZnF) domains (F1–F2 domains) (3). In response to SSB detection, Poly(ADP-ribose) polymerase 1 (PARP1) catalyzes poly(ADP-ribosyl)ation (PARylation) of itself as well as other acceptor proteins. Poly(ADP-ribose) (PAR) residues serve two main functions (i) chromatin relaxation, which permits access of SSBR proteins, and (ii) generating a PAR scaffold that can bind and retain proteins near the damage site. Usually, DNA damage is associated with ends that are incompatible with gap filling and ligation steps, and therefore the step that follows damage sensing is end processing, which is catalyzed by various enzymes, such as polynucleotide kinase/phosphatase (PNKP), that are specific to the type of damaged termini resulting from DNA insult (4). After restoration of correct DNA ends, gap filling proceeds, which is mediated by DNA polymerase β (polβ) (5). Finally, the resulting nick is sealed by DNA ligase III (LIG3) (6). An integral component in the SSBR cascade is the scaffold protein X-ray repair cross-complementing protein 1 (XRCC1), which orchestrates the steps from end processing to ligation (7).
Previous biochemical and live cell work indicated that PARP1 is the only cellular SSB sensor and that the recruitment of SSBR core proteins, particularly XRCC1, to sites of DNA damage is PARP1 dependent (8–11). Contradicting these observations, it was also shown that recruitment of SSBR core proteins, XRCC1, polβ and PNKP, to sites of DNA damage was PARP1 independent (12). Intriguingly, PARP1 knockout mouse embryonic fibroblasts (MEFs) repair SSBs and damaged bases efficiently in a manner similar to wild type (WT) MEFs (13,14). Collectively, the controversial involvement of PARP1 as a sensor in SSBR/BER suggests the possible existence of an alternative sensor. PARP1 binds damaged DNA through its ZnF domain, which shows a substrate preference for gaps over nicks (15,16). On the basis of in vitro experiments, Mackey et al. postulated that among the other SSBR proteins, LIG3 uniquely has a bona fide damage sensing module ascribed to its ZnF domain at the N-terminus, which is homologous to that of PARP1 (17). Additionally, the LIG3 ZnF, in contrast to that of PARP1, shows a substrate preference for nicks over gaps (18). The latter study demonstrated that the ZnF domain of LIG3 cooperates with a downstream DNA binding domain (DBD) within LIG3 to comprise a nick sensing module. This module, together with another nick sensing module involving the catalytic core, orchestrates a dynamic switch between the initial nick sensing and the subsequent sealing events in a ‘jack knife’ fashion. However, these two studies were performed using the LIG3β isoform and not the ubiquitously expressed LIG3α (18,19). A notable difference between the two LIG3 isoforms, apart from the differences in expression patterns, is the interaction with XRCC1. It was shown that LIG3α and not LIG3β exists in a complex with XRCC1 and this interaction is required for LIG3 stability and optimal catalytic activity (19,20). Previous work alluded to the possibility of LIG3 being involved in early damage sensing steps of SSBR. Importantly, biochemical studies indicated that LIG3α inhibits PARP1 catalytic activity upon encountering DNase I-treated DNA (21), implying that both proteins can bind independently at strand breaks. Consistent with the possibility of LIG3 being a damage sensor, it was shown that among the three different DNA ligases implicated in DNA damage response, LIG3 shows a very rapid recruitment to sites of DNA damage introduced by laser micro-irradiation (22). However an involvement of LIG3 ZnF in damage sensing was not demonstrated in this study (22).
Based on these observations, we hypothesized a role for LIG3α in sensing SSBs. Accordingly, we studied the early steps of SSBR in live cells. Here we show that PARP1 is dispensable for the recruitment and binding of SSBR proteins to sites of DNA damage. Furthermore, we identified a novel role for LIG3α as an independent sensor for DNA damage that helps in regulating the accumulation of SSBR core machinery to DNA repair sites. We also demonstrate that LIG3α can accumulate at sites of nuclear DNA damage independent of its BRCA1 C-terminus (BRCT) domain-mediated interaction with XRCC1. We elucidated the mechanism by which LIG3α (hereon referred to as LIG3) is recruited to damage sites and that the ZnF domain is required for the very rapid recruitment of LIG3 to damaged DNA, and indeed functions as a damage sensor in SSBR in live cells. Importantly, we provide cellular evidence that LIG3 is the sensor of nicks introduced by treatment of cells with the chemotherapeutic agent irinotecan (IRI).
MATERIALS AND METHODS
Cell culture and transfection
Human HeLa cells were obtained from Dr David Murray (University of Alberta) and cultured in Dulbecco's modified Eagle's medium (DMEM)-F12 media supplemented with 10% fetal calf serum (FCS). Chinese Hamster Ovary EM9 cells were kindly provided by Dr Keith Caldecott (University of Sussex) and cultured in DMEM supplemented with 10% FCS. PARP1+/+ (F20) and PARP1−/− (A1) mouse embryo fibroblasts (MEFs) were kindly provided by Dr Zhao-Qi Wang (Jena University, Germany) and cultured in DMEM low glucose media supplemented with 10% FCS. For PARP-1−/− MEFs, growth media contained neomycin at a final concentration of 600 μg/ml. For transfection, cells were plated in 35-mm glass bottom dishes (MatTek Corporation, Ashland, MA, USA) and allowed to attach over 24 h. Then, cells were transfected with DNA constructs of interest using Turbofectin 8.0 (OriGene, Rockville, MD, USA) according to the manufacturer's protocol. Cells were used for live cell imaging 24–48 h post-transfection.
Expression plasmids
pCMV6-AC-mGFP (OriGene), pCMV6-AC-mRFP (OriGene) and pCMV6-AN-mGFP (OriGene) destination vectors for mammalian expression were used to generate the fluorescent-tagged versions of both human PNKP and XRCC1. For both cDNAs forward and reverse primers were flanked with SgfI and MluI restriction sites. Primer sequences for PNKP were 5′-GAG GCG ATC GC ATG GGC GAG GTG GAG GCC-3′ (forward) and 5′-GCG ACG CGT GCC CTC GGA GAA CTG GCA GTA-3′ (reverse). Primer sequences for XRCC1 were 5′-GAG GCG ATC GCC ATG CCG GAG ATC CGC CTC-3′ (forward) and 5′-GCG ACG CGT GGC TTG CGG CAC CAC CCC ATA-3′ (reverse). Phusion high fidelity polymerase (Fermentas, Burlington, ON, USA) was used for polymerase chain reactions (PCRs) and cycling conditions were according to the manufacturer's recommendations. pEGFP-C1-ZnF and pEGFP-C1-ZnF-DBD constructs were generated from pEGFPC1 LIG3 (kind gift from Dr Heinrich Leonhardt, Ludwig-Maximilians University, Germany). pEGFP-N3-hOGG1 plasmid was a kind gift from Dr Akira Yasui (Institute of Development, Aging and Cancer, Tohoku University, Japan). ZnF and ZnF-DBD domains were cloned with forward and reverse primers flanked by SalI and BamHI restriction sites. For ZnF, the primer sequences were 5′-AGA GGC GTC GAC ATG TCT TTG GCT TTC AAG AT-3′ (forward) and 5′-ACG CGC GGA TCCCTA TCTTCT CTT TCT TCT CAG GAA-3′ (reverse). The same forward primer for cloning ZnF was used in cloning ZnF-DBD and the reverse primer sequence for ZnF-DBD was 5′-ACG CGC GGA TCC CTA CTC CTT GGT GAG CTT GGA CA-3′. pEGFPC1-ΔZnF LIG3 and pmRFPC1 LIG3 were kindly provided by Dr Heinrich Leonhardt, and pmRFP-ΔZnF LIG3 was generated by replacing EGFP with mRFP. Generated constructs were sequence-verified and tested in different cell lines for expression of appropriate sizes of fusion proteins by western blots (for PNKP, XRCC1, LIG3 and ΔZnF LIG3) (Supplementary Figure S1). pEGFPC1-hPARP1 has been previously described (23). For a schematic representation of different constructs generated and used refer to Supplementary Figure S1. Hush plasmid pGFP-V-RS (OriGene, cat. no. TG311735), supplied with the control plasmid encoding for scrambled shRNA (scr Hush), was used to transfect HeLa cells for the transient knockdown of LIG3, which was then confirmed by western blot analysis.
Site-directed mutagenesis
To generate the L360D XRCC1 mutant, substitution was carried out according to overlap extension PCR previously described in (24) using the following mutagenic primers 5′-CGGGACAGCACGCACGACATCTGTGCCTTTGC-3′(forward) and 5′-GCAAAGGCACAGATGTCGTGCGTGCTGTCCCG-3′(reverse). Subsequently, L360D was cloned into pCMV6-AN-mGFP plasmid using the restriction enzymes AscI and RsrII. To generate the ZnF mutant, ZnF R31I, we used QuikChange II site-directed mutagenesis kit (Stratagene) and followed the manufacturer's protocol using the mutagenic primers Sense 5′-GGGCACCACTTTGCCAATTATGCATACGCCCTTCACAATC-3′ and Antisense 5′-GATTGTGAAGGGCGTATGCATAATTGGCAAAGTGGTGCCC-3′. Finally, the mutants were sequence validated.
Whole cell extract preparation and western blot analysis
Whole cell extracts were prepared using RIPA buffer (25-mM Tris.HCl pH 7.6, 150-mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulphate protease inhibitor cocktail and phenylmethylsulfonyl fluoride (PMSF)). Cells were washed twice with 1X cold phosphate buffered saline (PBS). Then cold RIPA buffer was added to cells on ice for 5 min with occasional swirling. The lysate was then collected and spun at ∼14 000×g for 15 min at 4ºC to remove the cell debris. For western blots 50 μg of protein was added to each well. The antibodies used included mouse monoclonal anti-DNA ligase 3 (cat. no. 611876, BD Transduction Labs, Mississauga, ON, USA), goat polyclonal anti-actin (sc-1616, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA).
Measurement of SSBR
Cells were treated with 100 μM hydrogen peroxide in PBS for 40 min on ice, then washed twice with PBS, and then growth medium was added. Finally, cells were harvested after 15-, 60-, 120- and 240-min incubation at 37ºC. For sample preparation for the single-cell gel electrophoresis (comet) assay, we followed the kit manufacturer's (Trevigen) protocol. Briefly, cells were trypsinized and washed twice with PBS. Then the cell count was adjusted to 2 × 105 cells/ml in ice cold PBS. Twenty-five microliter cell suspension was mixed with 250 μl molten LMP (low melting point) agarose, and 75 μl of mixture was spread on each comet slide. Slides were kept in the dark for 60 min (gelling time) then immersed in ice cold alkaline lysis buffer. Samples were kept overnight in the dark at 4ºC and the next day immersed in freshly prepared cold electrophoresis buffer for 45 min in the dark, and then transferred to a horizontal electrophoresis chamber. Electrophoresis was carried out at 1 V/cm and 300 mA for 30 min. Slides were then immersed in 70% ethanol for 5 min, air dried and stained with SYBR Green (1:3000 dilution). For scoring, slides were visualized with epifluorescence using a fluorescein isothiocyanate filter with 10X objective, and the analysis was carried out using AutoComet software (TriTek, Sumerduck, VA, USA).
Immunofluorescence and microscopy
Cells were plated on 35-mm glass bottom dishes at ∼70–80% confluency. For immunofluorescence cells were fixed and permeabilized using ice cold methanol:acetone (1:1) mixture for 20 min. Then cells were rehydrated with PBS at room temperature for 15 min. For 8-oxo-dG staining, cells were treated with 2N HCl for 10 min at 37ºC, then washed with PBS (three 5-min washes) prior to incubation with primary antibody. Subsequently cells were blocked with 5% bovine serum albumin for 1 h and incubated with primary antibodies for 1 h at room temperature or overnight at 4ºC. After incubation, cells were washed once with 0.1% Triton-X-100 in PBS, then twice in PBS. Finally, cells were incubated with appropriate secondary antibodies for 30–45 min at room temperature, and then washed as described above. In the final wash, 4′,6-diamidino-2-phenylindole (DAPI) was added for nuclear DNA staining and left for 15 min then washed with PBS. Primary antibodies used included mouse monoclonal anti-8-oxo-dG (cat. no. 4354-MC-050, Trevigen, Gaithersburg, MD, USA) (1:250 dilution), mouse monoclonal anti-γH2AX (cat. no. 05–636, Millipore, Billerica, MA, USA) (1:5000 dilution), mouse monoclonal anti-53BP1 (Millipore) (1:400 dilution), mouse monoclonal anti-XRCC1 (ab1838–250, Abcam, Cambridge, MA, USA) (1:500 dilution), rabbit polyclonal anti-DNA Ligase 3 (GTX103197, GeneTex, Irvine, CA, USA) (1:250 dilution) and mouse monoclonal anti-poly(ADP-ribose) (ab14459, Abcam) (1:5000 dilution). Secondary antibodies used were Alexa Fluor 555 goat anti-rabbit (A-21430, Invitrogen, Burlington, ON, USA) (1:200 dilution) and Alexa Fluor 488 goat anti-mouse (A-11017, Invitrogen) (1:200) conjugated antibodies. For image acquisition, fixed and stained cells were placed on the stage of a Zeiss confocal LSM 710 microscope. Images were acquired using either 40X or 63X objectives as 12 bit grayscale images, and then exported as Tiff 16 bit grayscale images for processing using ImageJ software. For immunofluorescence experiments, three independent experiments were carried out. In each experiment 15–20 cell images were acquired and analyzed.
Laser micro-irradiation (two-photon laser micro-irradiation and 405-nm diode)
For two-photon laser micro-irradiation, cells were grown on 35-mm glass bottom dishes. Before laser micro-irradiation, cells were incubated with Hoechst 33258 (Sigma, cat. no. 94403) to a final concentration of 1 μg/ml for 20 min and then fed with fresh growth medium for 10 min. Where indicated, cells were incubated with either 1- or 2-μM AG14361 (IC50 = 29 nM, Selleckchem, cat. no. S2178), or with 2.5- and 10-μM PJ-34 (IC50 = 20 nM, Enzo Life Sciences, cat. no. ALX-270–289), for 1–2 h prior to micro-irradiation. Subsequently, cells were placed on a 37ºC-heated stage of a Zeiss LSM510 NLO laser-scanning confocal microscope. Micro-irradiation was carried out using a near-infrared titanium sapphire laser. To introduce damage within nuclei of individual cells, a 1.2-μm-wide region was pre-defined and subsequently micro-irradiated with 10 iterations of a 750-nm (50 mW) laser line at 10% power using a Plan-Neofluar 40X/1.3 NA oil immersion objective. For immunofluorescence of endogenous proteins and protein modifications, cells were fixed right after laser micro-irradiation and counterstained with antibodies of interest. For time lapse experiments of mRFP-tagged proteins, the fluorescent signal was recorded using excitation with a 543-nm He–Ne laser and a 559–634-nm band-pass emission filter. Similarly, for mGFP- and EGFP-tagged proteins, the signal was recorded after excitation with a 488-nm argon laser and a 515–540-nm band-pass emission filter. Cells with low to medium expression levels of fluorescent proteins were selected and accumulation of fluorescently tagged protein in micro-irradiated areas was quantified and compared to that in unirradiated regions. After background subtraction as previously described (25), the intensity was normalized so that the total cell intensity remained constant throughout the experiment. This process compensates for photobleaching during acquisition (25). Images were then realigned using ImageJ software and fluorescence signals of the exported Tiff images were subsequently quantified using Metamorph software 6.0 (Molecular Devices). Plotted results of recruitment kinetics represent averages of three independent experiments. For each experiment 10–12 cells were analyzed (total 30–36 cells).
For the 405-nm laser micro-irradiation, we applied the same settings as described by Dinant et al. (26). Briefly, cells transiently expressing a fluorescent protein-tagged protein of interest were pre-sensitized with Hoechst dye for 20 min, at a final concentration of 0.5 μg/ml, and then the media was replaced prior to micro-irradiation. Cells were placed on a 37ºC-heated stage of a Zeiss LSM710 NLO laser-scanning confocal microscope. To introduce damage within nuclei of individual cells, a 1.2-μm-wide region was pre-defined and subsequently micro-irradiated with 30 iterations of a 405-nm (30 mW) laser line at 60% power using a Plan-Neofluar 63X/1.3 NA oil immersion objective. For time lapse experiments of EGFP/mGFP-tagged proteins, the signal was recorded after excitation with a 488-nm argon laser and a 515–540-nm band-pass filter. Cells with low to medium expression levels of fluorescent proteins were selected and analyzed. For quantification, analyses were performed as previously described (25).
Fluorescence recovery after photo-bleaching
Cells were placed on a 37ºC-heated stage of a Zeiss LSM 510 confocal microscope. Fluorescence recovery after photo-bleaching (FRAP) was carried out on 1.5-μm strips across the width of the nuclei of cells under investigation using a 488-nm Argon laser line set at 100% intensity for 30 iterations using a 40X 1.3 N.A. objective. Laser power used for scanning during post-bleach time lapses was 3% to minimize photobleaching during the acquisition of the time lapse data. For data quantification, fluorescence intensities were measured in the bleached region, the entire nucleus and extracellular background using LSM image browser software. Each image was normalized for total fluorescence intensity relative to the first image collected after photobleaching to correct for any photobleaching that occurred during the collection of the post-bleach time series (25). Data on FRAP curves were plotted based on readings of 20–25 cells that were scanned over two independent experiments for each curve. For drug treatments, AG14361 was added at the indicated concentrations 90–120 min before FRAP experiments, and 10 mM H2O2 was added immediately prior to data acquisition. For irinotecan (IRI) treatment, cells were treated with IRI, diluted to a final concentration of 5 mM, for 30 min. Then FRAP analysis was performed as described above.
Statistical analysis
A two-tailed Student's t-test was used to calculate statistical significance. Calculations were performed using Microsoft Excel 2010.
RESULTS
Establishment of a micro-irradiation system that specifically activates SSBR not BER
An inherent problem with studying SSBR proteins (PARP, XRCC1, PNKP and LIG3) in real time is their participation in the BER pathway; therefore, the establishment of a technique that clearly discriminates between both pathways would be pivotal to our work. It is known that different laser micro-irradiation systems enable the analysis of different DNA repair pathways in real time (26,27). However, a drawback can be the creation of multiple types of DNA damage, including SSBs and damaged bases. Accordingly, to study the behavior of SSBR factors we established conditions that primarily activate the SSBR machinery rather than the BER machinery. We compared two different laser micro-irradiation systems, namely the 405-nm laser diode and the two-photon 750-nm Ti-Sapphire laser, and studied the nature of DNA damage introduced by both of them. Cells expressing XRCC1-mRFP were micro-irradiated by both systems, and 3–5 min after micro-irradiation cells were fixed and stained for 8-oxo-dG, which is one of the predominant base lesions that serve as substrates for the BER machinery. Whereas XRCC1 showed robust recruitment to sites of DNA damage following irradiation under both conditions, we found that 8-oxo-dG was produced by the 405-nm laser diode system but not the two-photon 750-nm laser (Figure 1). To further confirm our observation, we studied the impact of both micro-irradiation systems on the recruitment of the BER protein OGG1 and the SSBR/BER scaffold protein, XRCC1, in real time. Consistent with previous work, OGG1 showed robust recruitment to sites of DNA damage introduced by the 405-nm micro-irradiation, but only minimal accumulation at damage sites generated by the two photon micro-irradiation using 750-nm light (Figure 2A). Importantly, XRCC1 recruited to sites of DNA damage generated by both systems, consistent with its roles in both BER and SSBR (Figure 2B). Finally, it was previously shown that the L360D mutant of XRCC1 recruits specifically to sites of BER and not SSBs (11). Accordingly, we examined the recruitment of the mGFP-XRCC1 L360D mutant, in cells co-expressing WT XRCC1-mRFP, to sites of DNA damage introduced by both systems. Consistent with our observations, L360D mutant showed very limited accumulation at sites of DNA damage generated by the two-photon 750-nm laser, however it showed marked accumulation at sites generated by the 405-nm laser system (Figure 2C). Collectively, our observations indicate that the two-photon 750-nm laser system generates SSBs with minimal activation of the BER machinery. Consequently, and distinct from other live cell studies of SSBR that were performed using the 405-nm laser micro-irradiation, we relied on the two-photon laser micro-irradiation system for studying SSBR in live cells.
Figure 1.
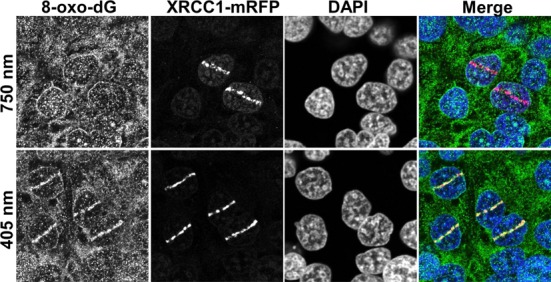
Comparative induction of base damage and strand breaks by different laser micro-IR conditions. Laser micro-irradiation was performed on HeLa cells using either 750-nm multi-photon excitation or a 405-nm laser diode. The production of base damage was gauged on the basis of production of 8-oxo-dG, while XRCC1 recruitment was used as a marker of strand break induction.
Figure 2.
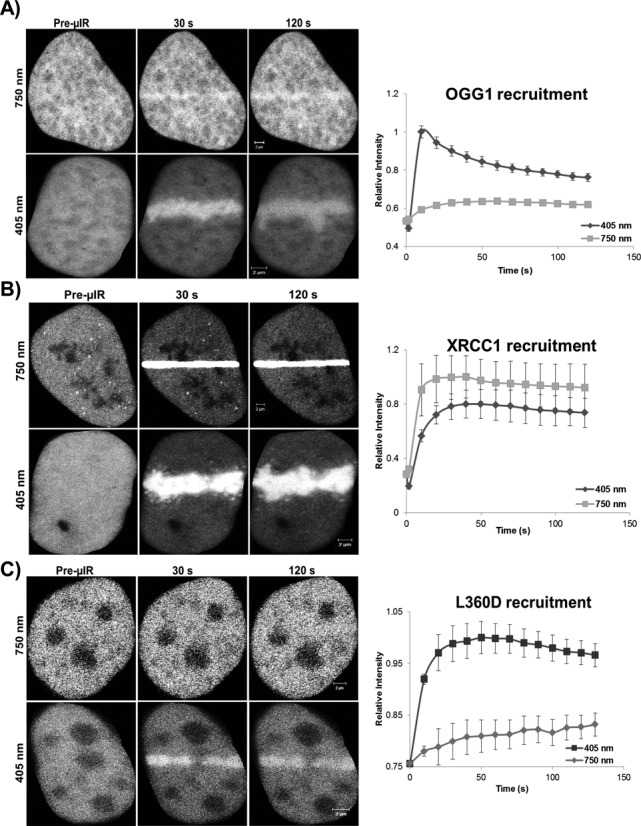
Recruitment of OGG1 and XRCC1 under different laser conditions. Recruitment kinetics of (A) the BER protein OGG1, (B) BER/SSBR scaffold protein XRCC1 and (C) XRCC1 mutant L360D were compared following irradiation of HeLa cells expressing pEGFP-OGG1 or wild type or mutant XRCC1-mGFP with either 750-nm multi-photon excitation or 405-nm laser excitation. The recruitment of the XRCC1 mutant, L360D, was tested in cells co-expressing WT XRCC1-mRFP (shown in Supplementary Figure S2). Error bars represent SEM from three independent experiments each analyzing 12 cells (i.e. n = 36).
Rapid recruitment of SSBR machinery to DNA damage sites
With the aim of studying early events of the SSBR cascade, we first examined the accumulation of SSBR core machinery proteins at sites of laser-induced nuclear DNA damage. We employed laser micro-irradiation of HeLa cells that transiently expressed fluorescently tagged versions of PARP1, LIG3, XRCC1 and PNKP (a schematic representation of the fluorescent-tagged proteins used in this work is shown in Supplementary Figure S1). We observed rapid accumulation (t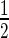 ≤ 5.3 s) of the proteins at damage sites (Figure 3). It is also clear that the retention of the proteins is longer than expected for conventional SSBR, which typically is regarded to have a t
≤ 5.3 s) of the proteins at damage sites (Figure 3). It is also clear that the retention of the proteins is longer than expected for conventional SSBR, which typically is regarded to have a t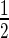 ∼2 min (28). Others have similarly observed a long retention time of XRCC1 at laser micro-irradiation tracks (29). This may be due to the generation of complex damage in the laser track, and indeed we observed the formation of DSB (using γH2AX as a marker) under our irradiation conditions (Supplementary Figure S4A and B), although others found that XRCC1 is rapidly released from DNA damage induced by high LET radiation (30).
∼2 min (28). Others have similarly observed a long retention time of XRCC1 at laser micro-irradiation tracks (29). This may be due to the generation of complex damage in the laser track, and indeed we observed the formation of DSB (using γH2AX as a marker) under our irradiation conditions (Supplementary Figure S4A and B), although others found that XRCC1 is rapidly released from DNA damage induced by high LET radiation (30).
Figure 3.
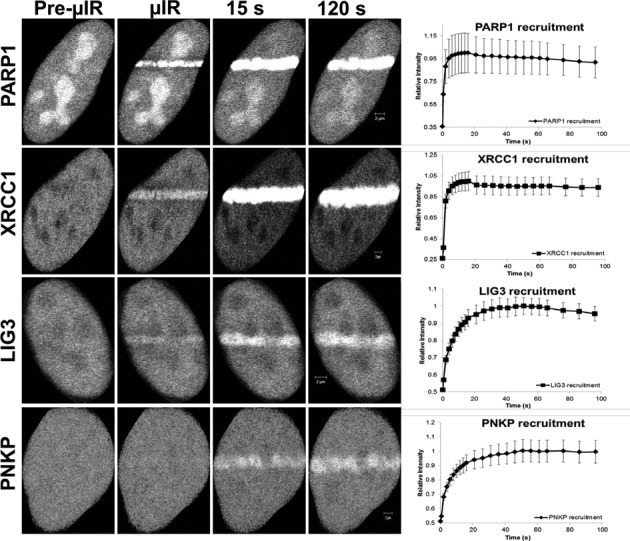
Recruitment and retention of SSBR proteins following multi-photon 750-nm laser micro-irradiation. EGFP-PARP1, XRCC1-mGFP and EGFP-LIG3 show near instantaneous recruitment to sites of DNA damage, and PNKP-mGFP is also rapidly recruited. Laser micro-irradiation using multi-photon 750 nm was carried out as outlined in the Materials and Methods section using HeLa cells expressing fluorescently tagged versions of indicated proteins. Recruitment curves show quantification of signals over the observed time scale starting at the time when the damage is introduced by the laser (t = 0). Error bars represent SEM from three independent experiments for a total of 36 individual cells.
PARP1-mediated poly(ADP-ribosyl)ation accelerates the initial recruitment of SSBR core proteins (XRCC1, LIG3 and PNKP) to sites of DNA damage but is not required for retention
It is known that once PARP1 binds to damaged DNA it rapidly undergoes a conformational change that stimulates its catalytic activity (31), leading to the formation of PAR polymers that mediate the recruitment of downstream repair factors. To examine the effect of PARP1 on the accumulation of SSBR proteins, we studied the recruitment kinetics of XRCC1 and PNKP to sites of DNA damage in PARP1−/− and PARP1+/+ MEFs (Figure 4). We observed that the extent of accumulation of both XRCC1 and PNKP in a PARP1−/− background was comparable to that in PARP1+/+ MEFs (Figure 4B and C). We confirmed the results observed in the PARP1−/− and PARP1+/+ MEFs by specific inhibition of PARP1 in HeLa cells. We made use of two chemically unrelated small molecule inhibitors of PARP, AG14361 (32) and PJ-34 (33), and tested their effects on the recruitment kinetics of SSBR core machinery. We observed that at concentrations of 1 and 2 μM, AG14361 markedly inhibited the DNA damage-triggered poly(ADP-ribosyl)ation (Supplementary Figure S4A). To delineate the site of damage, cells were also stained with anti-γH2AX. We then tested the effect of the inhibitor, AG14361, on the recruitment profiles of XRCC1, PNKP and LIG3 in real time (Figure 5A–C). All three proteins showed only a briefly delayed recruitment to sites of DNA damage in response to PARP1 inhibition, but displayed a similar accumulation to untreated cells at later time points. We confirmed the results observed with AG14361 using PJ-34 (Supplementary Figure S4B). Our results in PARP1−/− MEFs and in cells treated with PARP1 inhibitors raise the possibility of the presence of an additional sensor or sensors of DNA SSBs apart from PARP1. Therefore we tested the possibility of LIG3 in fulfilling such a role.
Figure 4.
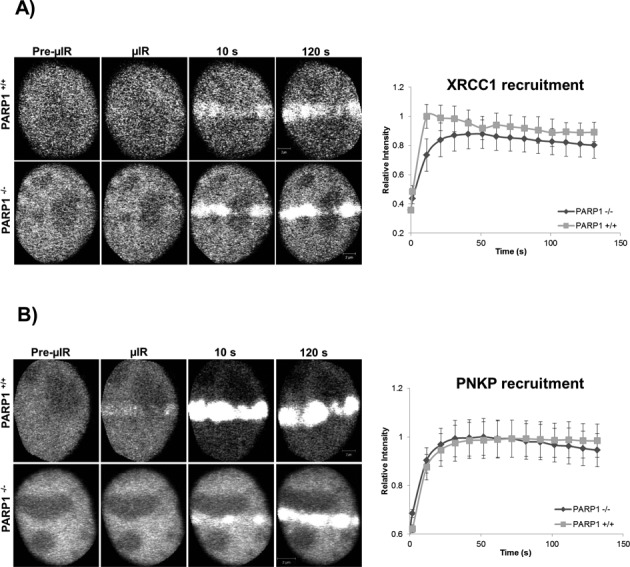
Recruitment of XRCC1 and PNKP in PARP1 WT and KO cells. The recruitment of SSBR proteins was monitored in PARP+/+ and PARP−/− MEFS expressing (A) XRCC1-mGFP and (B) PNKP-mGFP subjected to 750-nm multiphoton micro-irradiation. Error bars represent SEM from three independent experiments for a total of 36 individual cells.
Figure 5.
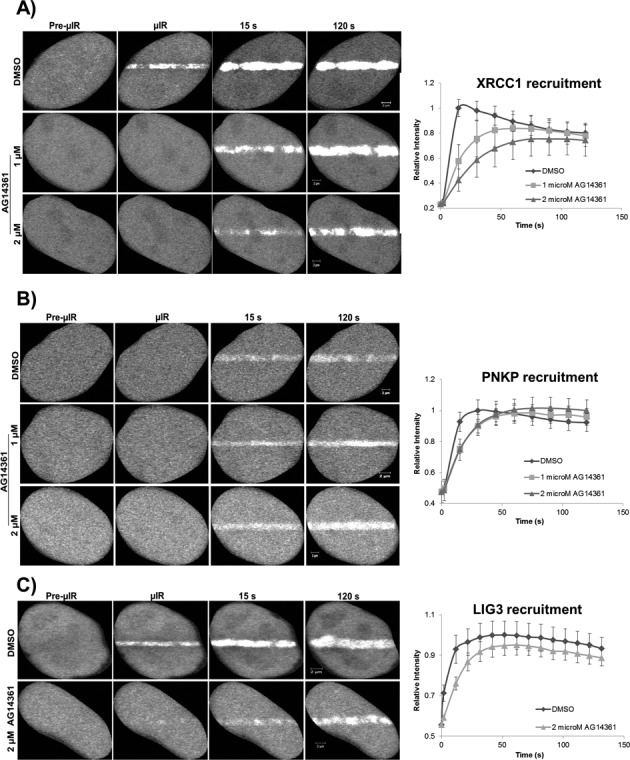
PARP1 inhibition and the recruitment of SSBR proteins to sites of DNA damage. HeLa cells expressing fluorescently tagged SSBR proteins were treated with the PARP inhibitor, AG14361, and then subjected to laser micro-irradiation. PARP inhibition only affected early recruitment events of (A) XRCC1, (B) PNKP and (C) LIG3 with almost no effect on the late events of accumulation of all the proteins at sites of DNA damage. For recruitment curves, error bars represent SEM from three independent experiments for a total of 36 individual cells.
The influence of LIG3 on the recruitment of SSBR core machinery
To test for the possible involvement of LIG3 in mediating the accumulation of other SSBR proteins to DNA repair sites, we monitored the recruitment kinetics of PNKP and XRCC1 in response to DNA damage under conditions of reduced LIG3 expression (Supplementary Figure S5). To knock down LIG3, we made use of the shRNA plasmids that co-express a GFP reporter, facilitating the identification of knockdown cells within the population. In contrast to the effect of PARP1 inhibition, the transient knockdown of LIG3 decreased the level of PNKP and XRCC1 recruited to sites of DNA damage over the time frame examined (Figure 6A and B). We then determined if LIG3 and PARP1 are redundant SSB sensors by examining the effect of simultaneous inhibition of PARP1 and depletion of LIG3 on the accumulation of PNKP (Figure 6C). As noted above, PARP1 inhibition alone caused an initial deceleration in the recruitment of PNKP at sites of DNA damage while reduced LIG3 expression alone resulted in a sustained decrease of the level of PNKP that accumulated at sites of DNA damage. The simultaneous lack of PARP1 activity and lowered LIG3 expression had an additive effect on the observed decreased accumulation of PNKP. This indicates that PARP1 and LIG3 function in a non-redundant manner, possibly because they recognize different subsets of SSBs. To further confirm that there is no redundancy between PARP1 and LIG3, we monitored the accumulation of endogenous LIG3 at sites of DNA damage in response to PARP1 inhibition using two different DNA damage treatments, laser micro-irradiation and H2O2. Following laser micro-irradiation, endogenous LIG3 showed robust accumulation at tracks of induced DNA damage despite the substantial reduction in PAR production as a result of AG14361 treatment. Similarly, endogenous XRCC1 accumulated at tracks of DNA damage in AG14361-treated cells (Supplementary Figure S6A). We were also able to demonstrate that in response to H2O2-induced DNA damage, LIG3 exhibited a unique pattern of distribution that was not affected by the efficient inhibition of PARP1 catalytic activity (Supplementary Figure S6B). To rule out the possibility that this pattern of LIG3 might be attributed to its recently discovered role in DSBR (34), we examined whether LIG3 would colocalize or not with the different DSB markers γH2AX and 53BP1 under conditions of PARP1 inhibition. As shown in (Supplementary Figure S6C and D), LIG3 did not colocalize with either γH2AX or 53BP1 indicating that LIG3 distribution in response to H2O2 is not associated with DSBR.
Figure 6.
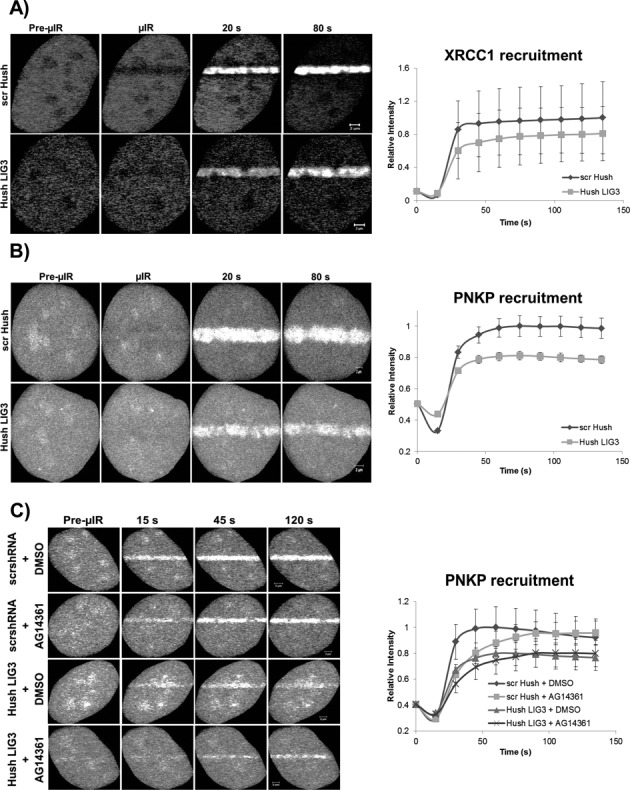
LIG3 knockdown and the recruitment of XRCC1 and PNKP to sites of DNA damage. Laser micro-irradiation was performed on HeLa cells expressing reduced levels of LIG3 (see Supplementary Figure S5) and (A) XRCC1-mRFP or (B) PNKP-mRFP. Reduced background levels of LIG3 lead to decreased overall recruitment of XRCC1 and PNKP to sites of DNA damage. (C) Simultaneous inhibition of PARP1 (using AG14361) and knockdown of LIG3 showed an additive effect on the reduction of the amount of PNKP recruited to sites of DNA damage. For recruitment curves, error bars represent SEM from three independent experiments for a total of 36 individual cells. Note that the mRFP photobleaches during laser micro-irradiation resulting in an initial loss of fluorescence at the damage sites.
ZnF domain, a nick sensor in LIG3
We extended our investigation of LIG3 to identify which domain(s) is required for its SSB sensor function. Previous biochemical studies have suggested that the N-terminal ZnF domain of LIG3 might be a candidate for this role (17,18). To test the importance of this domain in the recruitment of LIG3 in live cells, we directly compared the recruitment kinetics of full-length LIG3 and LIG3 lacking the ZnF domain (ΔZnF-LIG3) to sites of DNA damage induced by laser micro-irradiation. In contrast to previous work that showed no difference in the recruitment to sites of DNA damage between both forms of LIG3 (22), both fluorescent-tagged versions (EGFP and mRFP) of ΔZnF-LIG3 exhibited a significantly decreased level (P < 0.05) of recruitment compared to full-length LIG3 (Figure 7A) under our experimental conditions. The residual recruitment of ΔZnF-LIG3 to sites of DNA damage might reflect the BRCT domain-mediated interaction of LIG3 with XRCC1, which was observed to be required for the final nick sealing event in SSBR (35). To test this, we expressed both full-length LIG3 and ΔZnF-LIG3 in EM9 cells (a CHO cell line devoid of XRCC1) and compared the recruitment profiles of both proteins in the absence of XRCC1. Consistent with our hypothesis, the ΔZnF-LIG3 showed severely reduced recruitment at laser-damaged DNA tracks (Figure 7B) while full-length LIG3 accumulated at laser-damaged tracks in the absence of XRCC1. The latter observation was surprising because it implies that recruitment of LIG3 to strand breaks can occur independent of XRCC1.
Figure 7.
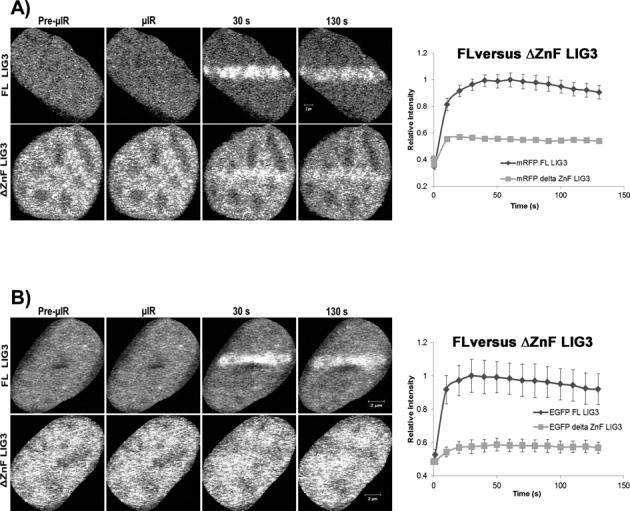
Comparison between the recruitment of full-length LIG3 and LIG3 lacking the zinc finger to sites of DNA damage. (A) HeLa cells or (B) EM9 cells expressing full-length (FL) LIG3 or mutant ΔZnF LIG3 were subjected to laser micro-irradiation. In HeLa cells, FL LIG3 was robustly recruited to sites of DNA damage while ΔZnF LIG3 was recruited less efficiently. Furthermore, FL-LIG3 was recruited to sites of DNA damage even in the absence of XRCC1 (EM9 cells) while ΔZnF-LIG3 could not. For recruitment curves, error bars represent SEM; n = 36. Both cell lines were tested with mRFP- and EGFP-tagged proteins and the tags were shown not to influence the result.
ZnF domain is sufficient for the initial rapid recruitment of LIG3 to sites of DNA damage
We next asked whether the ZnF domain is sufficient for recruitment to sites of DNA damage. Biochemical evidence indicated that LIG3 possesses two distinct SSB-sensing modules, the first of which mediates sensing distortions in the DNA backbone (early sensing function) and is comprised of the ZnF and a DBD. In vitro, these two domains within this module have been shown to function cooperatively to promote efficient SSB sensing/DNA binding and then the second SSB-sensing module (the catalytic core) mediates subsequent repair of SSBs (18). Therefore, we examined the behavior of the domains of the early SSB-sensing function in live cells. Accordingly, we designed EGFP constructs encoding the ZnF and the ZnF-DBD (Figure 8A) and tested the recruitment of these domains to sites of DNA damage introduced by laser micro-irradiation. We observed that both ZnF alone and the tandem module ZnF-DBD are rapidly recruited to sites of DNA damage in live cells similar to that shown by full-length LIG3, providing evidence that the in vitro SSB-sensing functions can also operate in vivo. However, the steady accumulation of the ZnF and ZnF-DBD domains after the initial response is considerably lower than full-length LIG3. This is consistent with protein–protein interactions occurring outside the ZnF and DBD domains being responsible for generating most of the binding sites responsible for the retention of LIG3. Nonetheless, the results clearly indicate that the ZnF domain is capable of recognizing and binding the damage site independent of other domains within LIG3. To examine whether ZnF recruitment to damaged DNA is mediated by direct DNA binding or not, we examined the recruitment of a ZnF mutant that was reported to lose its DNA binding without impacting the conformation of the ZnF, ZnF R31I (36). Compared to WT ZnF, the ZnF domain R31I mutant showed a significantly reduced recruitment (P < 0.05) to sites of DNA damage introduced by laser micro-irradiation (Figure 8B).
Figure 8.
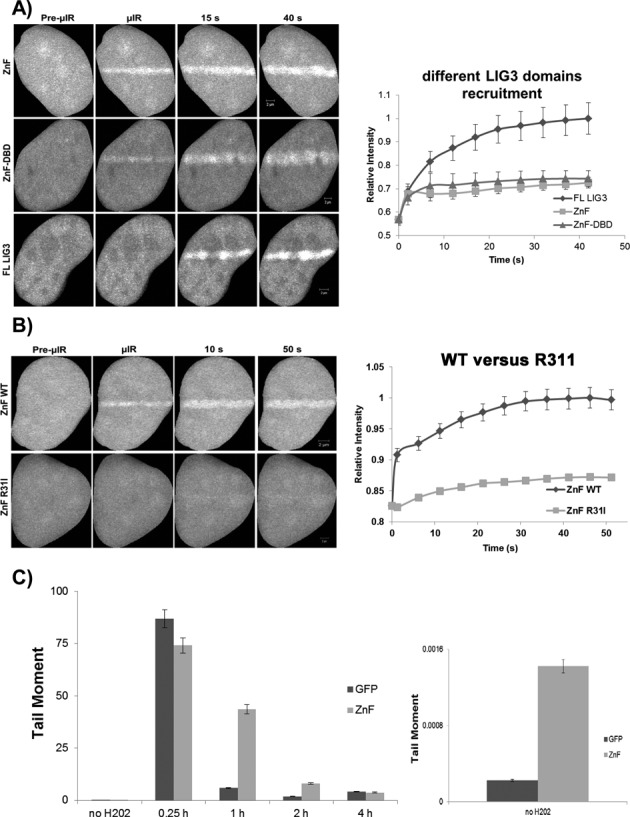
The ZnF domain is required for the damage sensing function of LIG3. (A) Comparison of the recruitment of fluorescently tagged full-length (FL) LIG3 and the ZnF and ZnF-DBD domains of LIG3 to micro-irradiated DNA in HeLa cells. (B) Comparison of recruitment of wild type (WT) and the DNA binding mutant of ZnF-R31I to micro-irradiated DNA in HeLa cells. For recruitment curves shown in (A) and (B), bars represent SEM; n = 36. (C) Expression of the ZnF domain of LIG3 retards single-strand break repair. HeLa cells expressing either the GFP-ZnF or GFP alone (control) were treated with 100 μM hydrogen peroxide for 40 min on ice and then strand break repair was monitored by the alkaline comet assay and quantification of tail moments at the indicated time points as described in the Materials and Methods section. Expanding the ordinate (plot on the right-hand side) showed that even in the absence of the hydrogen peroxide the ZnF expressing cells exhibit a slightly higher background level of damage.
If the ZnF functions in damage sensing, then overexpression of this domain alone may slow the kinetics of SSBR by competing with the endogenous machinery without being able to support the protein–protein interactions necessary to repair the break. To test this, we carried out an alkaline comet assay on cells overexpressing GFP (control vector) or GFP-ZnF (Figure 8C). Control cells showed rapid repair after 1-h recovery after damage. Clearly, repair was impaired in cells overexpressing ZnF, as judged by the tail moments at 1- and 2-h recovery time points, indicating that this domain can function in a dominant negative manner to impede SSBR.
Binding kinetics of SSBR proteins to damaged DNA is PARP1 independent
Having demonstrated that PARP1 catalytic activity is dispensable for the recruitment of SSBR core machinery proteins to sites of DNA damage and that LIG3 appears to be an alternative SSB sensor, we asked if PARP1-mediated poly(ADP-ribosyl)ation (PARylation) might affect the binding kinetics of LIG3, PNKP and XRCC1 to damaged DNA in live cells. We therefore carried out FRAP experiments to study the mobility of fluorescently tagged versions of these proteins in cells treated with H2O2 in the absence and presence of 2 μM AG14361. Consistent with their roles in the SSBR pathway, all three proteins showed reduced mobility in response to H2O2 treatment. In these experiments, a significant reduction (P < 0.05) in mobility arises when the fluorescent molecules bind to substrates that are essentially immobile on the time scale of minutes. Consequently, these experiments detect binding to damaged DNA. Surprisingly, inhibition of PARP1 catalytic activity using 2 μM AG14361 did not significantly impact (P-values >0.05) the mobility of the SSBR factors during the ongoing repair process (Figure 9A–C). The latter observation implies that the core SSBR proteins under study bind directly to the damaged DNA independent of the formation of PAR polymer. To further validate our hypothesis and previous results regarding the role of the ZnF domain of LIG3, we compared the mobility of full-length LIG3 and ΔZnF-LIG3. Both proteins showed reduced mobility in the presence of DNA damage, although ΔZnF-LIG3 recovered more rapidly than that of full-length LIG3 (Supplementary Figure S7). This result indicates that both the ZnF and domains outside of the ZnF contribute to the retention of LIG3 at SSBs.
Figure 9.
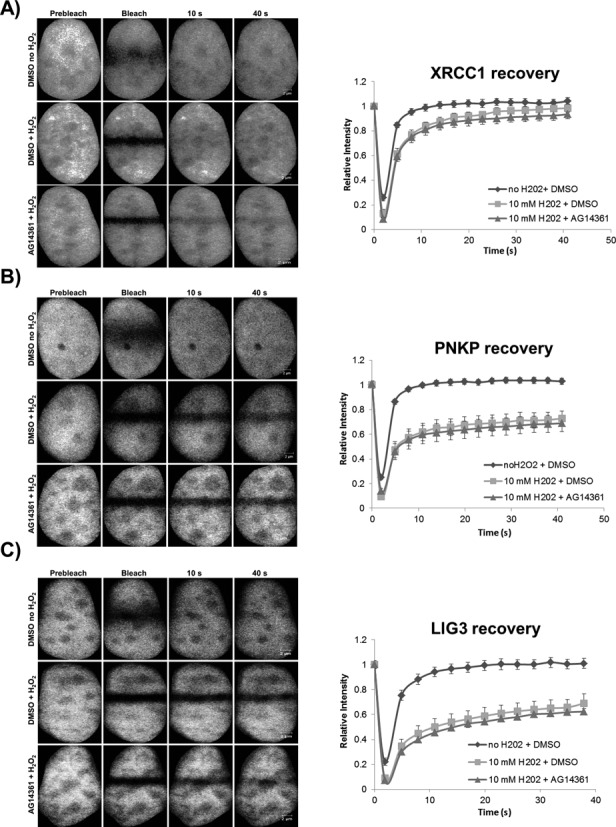
PARP1 inhibition and the retention of SSBR proteins at sites of DNA damage. FRAP analysis on HeLa cells expressing GFP-tagged (A) XRCC1, (B) PNKP and (C) LIG3, respectively, before and after DNA damage with 10 mM hydrogen peroxide in the absence and presence of 2 μM AG14361 as described in the Materials and Methods section. ‘Prebleach’ indicates no photobleaching and ‘Bleach’ is the 0-s time point. For recovery curves, error bars represent SEM; n = 24.
LIG3 and not PARP1 functions as a nick sensor in live cells
To generate SSB comprising only nicks, as opposed to gaps, cells were treated with irinotecan (IRI), a topoisomerase 1 poison, which generates abortive topoisomerase-1 cleavage complexes in the DNA. The resolution of such abortive complexes requires the action of TDP1, which removes the covalently bound topoisomerase from the DNA leaving nicks with 3′-phosphate and 5′-OH termini (37). Following exposure of the cells to IRI, we compared the mobility in FRAP experiments of both PARP1 and LIG3. Surprisingly, PARP1 mobility was not significantly reduced (P > 0.05) in response to IRI treatment, but, LIG3 mobility was substantially retarded (P < 0.05) (Figure 10). This result indicates that PARP1 may not recognize nicks but LIG3 clearly does, which is consistent with our prediction that LIG3 functions as a nick sensor in cells.
Figure 10.
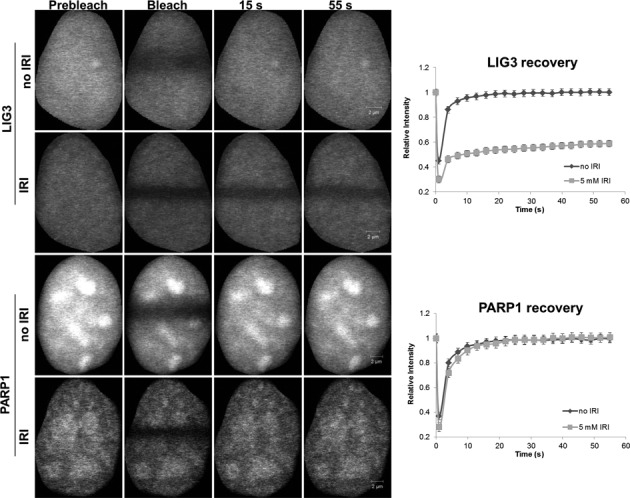
LIG3 is an in vivo nick sensor for irinotecan-induced DNA damage. FRAP analysis showing differences in binding kinetics after 5 mM irinotecan (IRI) treatment in HeLa cells expressing PARP1 and LIG3. ‘Prebleach’ indicates no photobleaching and ‘Bleach’ is the 0-s time point. Error bars represent SEM; n = 24.
DISCUSSION
Indications of an alternative SSB sensor to PARP1
The purpose of this study was to elucidate the mechanisms of recruitment and retention of the SSBR proteins, PARP1, XRCC1, PNKP and LIG3 to sites of SSBs in live cells using the combination of laser micro-irradiation and FRAP experiments. We initially established that the multi-photon excitation conditions we employed appeared to strongly activate SSBR but only weakly activate BER, as judged by the lack of production of one of the most abundant base lesions, 8-oxoguanine, and the minimal recruitment of OGG1, which is a DNA glycosylase that removes 8-oxoguanine, and the L360D mutant of XRCC1, which has a marked preference for BER over SSBR (11). In agreement with Campalans et al. (11), we observed that 405-nm micro-irradiation in the presence of dye (in their case Ro-19–8022 and in our experiments Hoechst 33258) activated the BER pathway.
A large body of data has led to the current model of the SSBR pathway, in which PARP1 plays a leading role in sensing strand breaks and signaling their presence to enhance recruitment of the other SSBR proteins (8–11,38). Surprisingly, we found that loss or inhibition of PARP1 led to only slightly delayed, rather than completely inhibited, recruitment of the other SSBR proteins.
Previous in vitro biochemical studies have demonstrated that LIG3 binds with high affinity to model DNA substrates containing SSBs (17,18). Consequently, we examined the potential of LIG3 to contribute to the recruitment of the core SSBR machinery. We found that partial knockdown of LIG3, while not eliminating XRCC1 or PNKP recruitment, did significantly reduce (P < 0.05) the accumulation of XRCC1 and PNKP at DNA damage sites. When an inhibitor of PARP1 was combined with shRNA directed against LIG3, we found that the recruitment was both delayed, which can be seen with PARP1 inhibition alone, and reduced in the total amount accumulated, which can be seen with LIG3 shRNA alone, suggesting that the two proteins act independently and additively as SSB sensors.
To further define the potential of LIG3 to act as a sensor for SSBs in vivo, we examined the ability of the LIG3 ZnF, which is homologous to ZnF2 of PARP1, and the LIG3 ZnF-DBD alone to recruit to sites of DNA damage. These have previously been shown to bind nicked DNA in vitro (18). We found that, despite being incapable of forming a complex with XRCC1, both the LIG3 ZnF and the LIG3 ZnF-DBD constructs rapidly localized to sites of DNA damage. Although both fragments of LIG3 were capable of being recruited with the same kinetics as the full-length protein, these domains did not accumulate to the same extent as the WT protein. This is consistent with two mechanisms involved in the recruitment of LIG3 to SSBs. The first mechanism, which is mediated through the ZnF or ZnF-DBD domains, is direct binding to damaged DNA (17,18). The second mechanism is likely through the established association with XRCC1. The latter mechanism seems not to play a major role in the initial recruitment of LIG3 at sites of DNA damage, as our results demonstrated that LIG3 is efficiently recruited to sites of DNA damage in EM9 cells, which lack the expression of XRCC1, but does play an important role in retention. Importantly, this result provides an explanation for how LIG3 can participate in the repair of mitochondrial DNA, which does not require XRCC1 (39). To further validate the importance of LIG3 in SSBR, we reasoned that the LIG3 ZnF should behave in a dominant negative fashion. When we overexpressed the LIG3 ZnF and then examined the rate of SSBR using the comet assay, we found that the rate of SSBR is significantly reduced. This is consistent with the ZnF binding to SSBs but not properly initiating the assembly of the SSBR machinery.
In vitro, the ZnF domain of LIG3 shows a preference for nicks over gaps (18). We therefore tested the potential of LIG3 to directly sense nicks introduced by cellular treatment with irinotecan. Using FRAP, we found that LIG3 had dramatically reduced mobility following irinotecan treatment. Surprisingly, PARP1 showed no significant change in its mobility under the same conditions. This is in contrast to treatment with hydrogen peroxide, where both PARP1 and LIG3 showed reduced mobility (Supplementary Figures S8 and S9C). The observation that PARP1 is activated in response to topoisomerase 1 poisoning by camptothecin and its analogs has led to the expectation that this is mainly due to the binding of PARP1 to nicks. This was further supported by the finding that PARP1 inhibition sensitizes cells to camptothecin treatment (40). Our findings suggest that PARP1 is not the nick sensor for breaks introduced by topoisomerase I inhibition. However, a plausible explanation for the increased sensitivity to topoisomerase I poisons upon PARP1 inhibition might lie in the finding that PARP1 null cells have lowered tyrosyl DNA-phosphodiesterase I (TDP1) activity compared to the WT cells (41) and that PARP1 activity stabilizes TDP1 protein and enhances its accumulation at sites of DNA damage (39).
Collectively, our results reveal a direct role for LIG3 in SSB sensing and recruitment of the SSBR machinery and a surprisingly more limited role for PARP1 in these same processes. When PARP1 is inhibited, the SSBR core machinery shows delayed recruitment but there is no observed reduction in recruitment once the break is detected. Because PARP1 stimulates the rate but not the abundance of SSBR proteins recruited to sites of DNA damage, the primary role of PARP1-mediated poly(ADP-ribosyl)ation may be to decondense the chromatin rather than act as a scaffold for the assembly of SSBR proteins. It remains possible that binding to PAR is responsible for generating a large number of binding sites rapidly after DNA damage but that PAR binding plays a relatively minor role in the recruitment and retention of these proteins once the SSB response has been initiated. Instead our results indicate that binding sites established downstream of the recognition of breaks by LIG3 are a major mechanism responsible for the retention of SSB proteins. Partial knockdown of LIG3 resulted in a comparable reduction in recruitment of XRCC1 and PNKP. This residual recruitment was not sensitive to PARP1/2 inhibition. Thus, whether the remaining LIG3 was sufficient for the observed recruitment or whether another protein that has not yet been identified as a SSB sensor was responsible for this recruitment remains to be determined. In either case, our results reveal that the canonical SSB sensing pathway centered around PARP1 does not explain the recruitment of SSBR proteins that we observe in living cells. Rather, we find that LIG3 can function in place of PARP1 as a sensor for SSBs, especially nicks, that is capable of initiating signaling and assembly of the SSBR (42) machinery independent of PARP1 activity (Figure 11).
Figure 11.
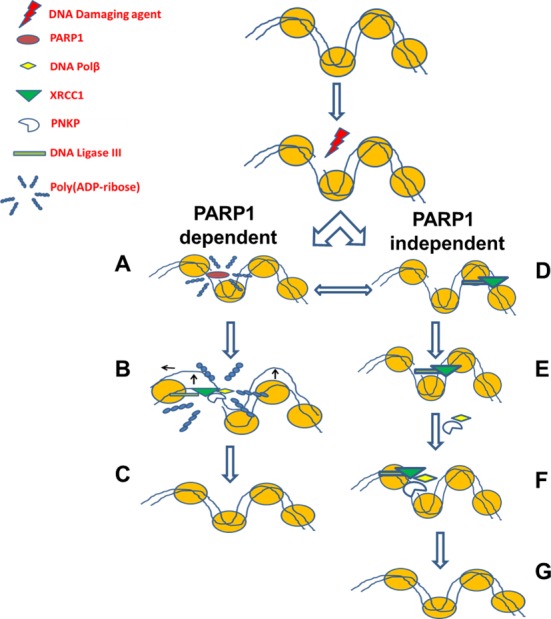
Two pathways exist for the short patch repair of SSBs. In the canonical pathway (PARP1 dependent) (A) PARP1 senses DNA damage and rapidly catalyzes the formation of PAR residues that allow for (B) chromatin expansion which in turn facilitates (C) the recruitment of downstream repair proteins. In the second pathway (PARP1 independent) (D) XRCC1–LIG3 complex continuously scans the DNA, upon sensing an interruption (via LIG3), (E) the complex is capable of causing a localized nucleosomal disruption (dependent on LIG3) (42), and the scaffold XRCC1 is capable of (F) loading downstream repair factors, PNKP and Polβ, and then repair continues as previously described (G).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
AG14361 was graciously provided by Zdenek Hostomsky of Pfizer Global. PARP-1−/− mouse embryo fibroblasts were kindly provided by Dr Zhao-Qi Wang of Jena University, Germany and EM9 CHO cells were kindly provided by Dr Keith Caldecott of the University of Sussex, UK. We thank Dr Heinrich Leonhardt of Ludwig-Maximilians University, Germany, for generously providing LIG3 constructs and Dr Akira Yasui of the Institute of Development, Aging and Cancer, Tohoku University, Japan for providing the OGG1 construct. We would like to thank Mesfin Fanta and Darin McDonald for technical assistance. We also thank Dr Xuejun Sun and Geraldine Barron of the Cell Imaging Facility at the Cross Cancer Institute (Edmonton) for providing space and support in live cell imaging experiments.
FUNDING
Canadian Institutes of Health Research [MOP 15385 to M.W., M.H.; MOP 200709 to G.G.P.]; Alberta Innovates-Health Solutions Senior Scholarship [to M.H.]; Tier 1 Canada Research Chair in Targeted Proteomics [to G.G.P.]; Alberta Cancer Foundation Graduate Studentship [ACF GSA 26283 to I.A.]. Funding for open access charge: Canadian Institutes of Health Research [MOP 15385].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Caldecott K. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 3.Rouleau M., Patel A., Hendzel M.J., Kaufmann S.H., Poirier G.G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinfeld M., Mani R.S., Abdou I., Aceytuno R.D., Glover J.N.M. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson S.H., Beard W.A, Shock D.D., Batra V.K., Cavanaugh N.A, Prasad R., Hou E.W., Liu Y., Asagoshi K., Horton J.K., et al. Base excision repair and design of small molecule inhibitors of human DNA polymerase β. Cell. Mol. Life Sci. 2010;67:3633–3647. doi: 10.1007/s00018-010-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappelli E., Taylor R., Cevasco M., Abbondandolo A., Caldecott K., Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 7.Horton J., Watson M., Stefanick D. XRCC1 and DNA polymerase β in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Khamisy S.F., Masutani, M., Suzuki, H. and Caldecott, K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortusewicz O., Amé J.-C., Schreiber V., Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godon C., Cordelières F.P., Biard D., Giocanti N., Mégnin-Chanet F., Hall J., Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campalans A., Kortulewski T., Amouroux R., Menoni H., Vermeulen W., Radicella J.P. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013;41:3115–3129. doi: 10.1093/nar/gkt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanssen-Bauer A., Solvang-Garten K., Sundheim O., Peña-Diaz J., Andersen S., Slupphaug G., Krokan H.E., Wilson D.M., Akbari M., Otterlei M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ. Mol. Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vodenicharov M.D., Sallmann F.R., Satoh M.S., Poirier G.G. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ström C.E., Johansson F., Uhlén M., Szigyarto C.A.-K., Erixon K., Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrucco S. Sensing DNA damage by PARP-like fingers. Nucleic Acids Res. 2003;31:6689–6699. doi: 10.1093/nar/gkg890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eustermann S., Videler H., Yang J.-C., Cole P.T., Gruszka D., Veprintsev D., Neuhaus D. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 2011;407:149–170. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackey Z.B., Niedergang C., Murcia J.M., Leppard J., Au K., Chen J., de Murcia G., Tomkinson A.E. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 18.Cotner-Gohara E., Kim I.-K., Tomkinson A.E., Ellenberger T. Two DNA-binding and nick recognition modules in human DNA ligase III. J. Biol. Chem. 2008;283:10764–10772. doi: 10.1074/jbc.M708175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldecott K.W., McKeown C.K., Tucker J.D., Ljungquist S., Thompson L.H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldecott K.W., Tucker J.D., Stanker L.H., Thompson L.H. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldecott K.W., Aoufouchi S., Johnson P., Shall S. XRCC1 polypeptide interacts with DNA polymerase and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortusewicz O., Rothbauer U., Cardoso M.C., Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haince J.-F., McDonald D., Rodrigue A., Déry U., Masson J.-Y., Hendzel M.J., Poirier G.G. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 24.Vallejo A.N., Pogulis R.J., Pease L.R. PCR mutagenesis by overlap extension and gene SOE. CSH Protoc. 2008;3:1–7. doi: 10.1101/pdb.prot4861. [DOI] [PubMed] [Google Scholar]
- 25.Carrero G., McDonald D., Crawford E., de Vries G., Hendzel M.J. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29:14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- 26.Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 27.Kong X., Mohanty S.K., Stephens J., Heale J.T., Gomez-Godinez V., Shi L.Z., Kim J.-S., Yokomori K., Berns M.W. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Schans G.P., Paterson M.C., Cross W.G. DNA strand break and rejoining in cultured human fibroblasts exposed to fast neutrons or gamma rays. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1983;44:75–85. doi: 10.1080/09553008314550881. [DOI] [PubMed] [Google Scholar]
- 29.Della-Maria J., Hegde M.L., McNeill D.R., Matsumoto Y., Tsai M.-S., Ellenberger T., Wilson D.M., Mitra S., Tomkinson A.E. The interaction between polynucleotide kinase phosphatase and the DNA repair protein XRCC1 is critical for repair of DNA alkylation damage and stable association at DNA damage sites. J. Biol. Chem. 2012;287:39233–39244. doi: 10.1074/jbc.M112.369975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakob B., Splinter J., Conrad S., Voss K.-O., Zink D., Durante M., Löbrich M., Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malanga M., Althaus F.R. The role of poly (ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 32.Veuger S.J., Curtin N.J., Smith G.C.M., Durkacz B.W. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene. 2004;23:7322–7329. doi: 10.1038/sj.onc.1207984. [DOI] [PubMed] [Google Scholar]
- 33.Mabley J.G., Jagtap P., Perretti M., Getting S.J., Salzman A.L., Virág L., Szabó E., Soriano F.G., Liaudet L., Abdelkarim G.E., et al. Anti-inflammatory effects of a novel, potent inhibitor of poly (ADP-ribose) polymerase. Inflamm. Res. 2001;50:561–569. doi: 10.1007/PL00000234. [DOI] [PubMed] [Google Scholar]
- 34.Simsek D., Brunet E., Wong S.Y.-W., Katyal S., Gao Y., McKinnon P.J., Lou J., Zhang L., Li J., Rebar E.J., et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor R.M., Wickstead B., Cronin S., Caldecott K.W. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 36.Taylor R.M., Whitehouse C.J., Caldecott K.W. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 2000;28:3558–3563. doi: 10.1093/nar/28.18.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caldecott K.W. Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair. 2014;19:108–113. doi: 10.1016/j.dnarep.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 39.Katyal S., McKinnon P.J. Disconnecting XRCC1 and DNA ligase III. Cell Cycle. 2011;10:2269–2275. doi: 10.4161/cc.10.14.16495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y.-W., Regairaz M., Seiler J.A., Agama K.K., Doroshow J.H., Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–3620. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pommier Y., Barcelo J., Rao V.A., Sordet O., Jobson A.G., Miao Z., Seiler J., Zhang H., Marchand C., Redon C. Repair of topoisomerase I-mediaed DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odell I.D., Barbour J.-E., Murphy D.L., Della-Maria J.A., Sweasy J.B., Tomkinson A.E., Wallace S.S., Pederson D.S. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Mol. Cell. Biol. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


