Abstract
Distinct structural regions have been found in several globular proteins composed of single polypeptide chains. The existence of such regions and the continuity of peptide chain within them, coupled with kinetic arguments, suggests that the early stages of three-dimensional structure formation (nucleation) occur independently in separate parts of these molecules. A nucleus can grow rapidly by adding peptide chain segments that are close to the nucleus in aminoacid sequence. Such a process would generate three-dimensional (native) protein structures that contain separate regions of continuous peptide chain. Possible means of testing this hypothesis are discussed.
Keywords: protein structure, chain continuity, independent regions, self-assembly
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
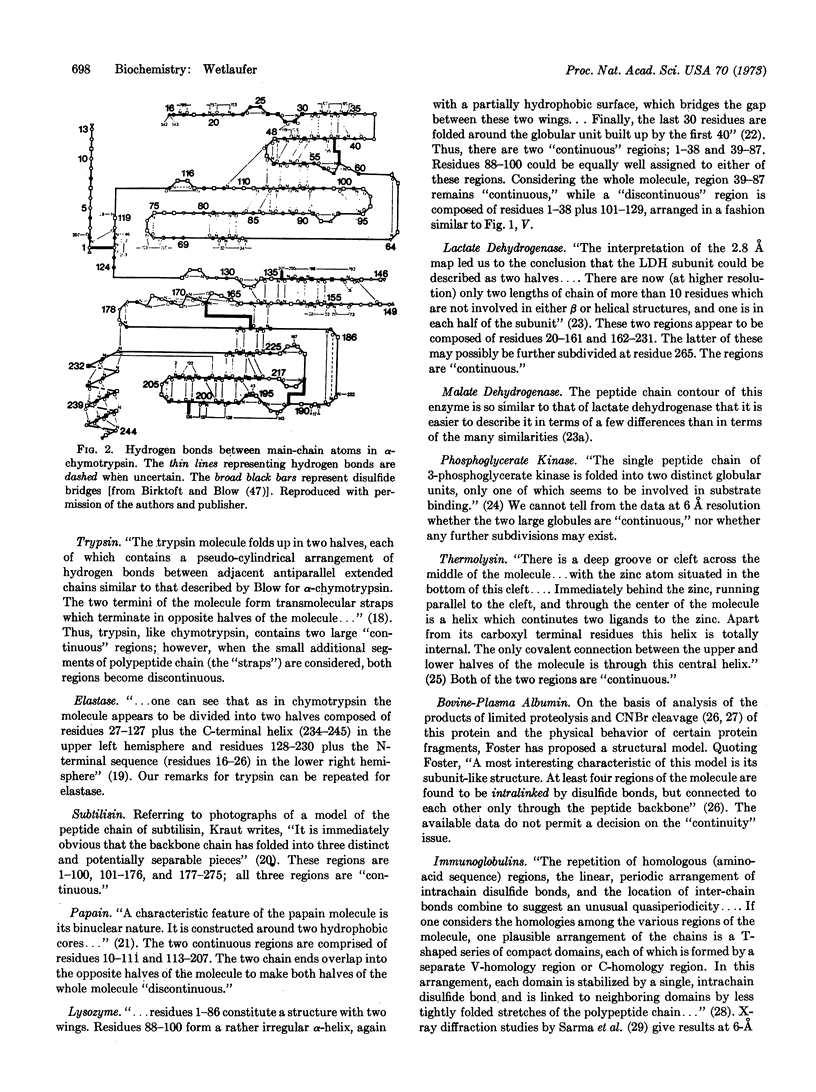
- Birktoft J. J., Blow D. M. Structure of crystalline -chymotrypsin. V. The atomic structure of tosyl- -chymotrypsin at 2 A resolution. J Mol Biol. 1972 Jul 21;68(2):187–240. doi: 10.1016/0022-2836(72)90210-0. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Evans P. R., Scopes R. K. Structure of horse-muscle phosphoglycerate kinase at 6 angstrom resolution. Nat New Biol. 1972 Feb 16;235(59):195–198. doi: 10.1038/newbio235195a0. [DOI] [PubMed] [Google Scholar]
- Bodanszky A., Ondetti M. A., Mutt V., Bodanszky M. Synthesis of secretin. IV. Secondary structure in a miniature protein. J Am Chem Soc. 1969 Feb 12;91(4):944–949. doi: 10.1021/ja01032a026. [DOI] [PubMed] [Google Scholar]
- Brown J. E., Klee W. A. Helix-coil transition of the isolated amino terminus of ribonuclease. Biochemistry. 1971 Feb 2;10(3):470–476. doi: 10.1021/bi00779a019. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Bier C. J., Day V. W., Hazen E. E., Jr, Larsen S. Some aspects of the structure of staphylococcal nuclease. I. Crystallographic studies. Cold Spring Harb Symp Quant Biol. 1972;36:243–249. doi: 10.1101/sqb.1972.036.01.032. [DOI] [PubMed] [Google Scholar]
- Drenth J., Hol W. G., Jansonius J. N., Koekoek R. A comparison of the three-dimensional structures of subtilisin BPN' and subtilisin novo. Cold Spring Harb Symp Quant Biol. 1972;36:107–116. doi: 10.1101/sqb.1972.036.01.016. [DOI] [PubMed] [Google Scholar]
- Epand R. M. Conformational properties of cyanogen bromide-cleaved glucagon. J Biol Chem. 1972 Apr 10;247(7):2132–2138. [PubMed] [Google Scholar]
- Epstein H. F., Schechter A. N., Chen R. F., Anfinsen C. B. Folding of staphylococcal nuclease: kinetic studies of two processes in acid renaturation. J Mol Biol. 1971 Sep 28;60(3):499–508. doi: 10.1016/0022-2836(71)90184-7. [DOI] [PubMed] [Google Scholar]
- Goodman M., Verdini A. S., Toniolo C., Phillips W. D., Bovey F. A. Sensitive criteria for the critical size for helix formation in oligopeptides. Proc Natl Acad Sci U S A. 1969 Oct;64(2):444–450. doi: 10.1073/pnas.64.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRISON S. C., BLOUT E. R. REVERSIBLE CONFORMATIONAL CHANGES OF MYOGLOBIN AND APOMYOGLOBIN. J Biol Chem. 1965 Jan;240:299–303. [PubMed] [Google Scholar]
- Herriott J. R., Sieker L. C., Jensen L. H., Lovenberg W. Structure of rubredoxin: an x-ray study to 2.5 A resolution. J Mol Biol. 1970 Jun 14;50(2):391–406. doi: 10.1016/0022-2836(70)90200-7. [DOI] [PubMed] [Google Scholar]
- Huber R., Kukla D., Rühlmann A., Steigemann W. Pancreatic trypsin inhibitor (Kunitz). I. Structure and function. Cold Spring Harb Symp Quant Biol. 1972;36:141–148. doi: 10.1101/sqb.1972.036.01.019. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kartha G., Bello J., Harker D. Tertiary structure of ribonuclease. Nature. 1967 Mar 4;213(5079):862–865. doi: 10.1038/213862a0. [DOI] [PubMed] [Google Scholar]
- King T. P., Spencer M. Structural studies and organic ligand-binding properties of bovine plasma albumin. J Biol Chem. 1970 Nov 25;245(22):6134–6148. [PubMed] [Google Scholar]
- Kretsinger R. H., Nockolds C. E., Coffee C. J., Bradshaw R. A. The structure of a calcium-binding protein from carp muscle. Cold Spring Harb Symp Quant Biol. 1972;36:217–220. doi: 10.1101/sqb.1972.036.01.029. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D. Tertiary structure in carboxypeptidase. J Am Chem Soc. 1972 Nov 29;94(24):8568–8572. doi: 10.1021/ja00779a046. [DOI] [PubMed] [Google Scholar]
- LINDERSTRØM-LANG K. Structure and enzymatic break-down of proteins. Cold Spring Harb Symp Quant Biol. 1950;14:117–126. doi: 10.1101/sqb.1950.014.01.016. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lewis P. N., Momany F. A., Scheraga H. A. Folding of polypeptide chains in proteins: a proposed mechanism for folding. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2293–2297. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky A. E., Pauling L. On the Structure of Native, Denatured, and Coagulated Proteins. Proc Natl Acad Sci U S A. 1936 Jul;22(7):439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. M., Foster J. F. Subtilisin cleavage of bovine plasma albumin. Reversible association of the two primary fragments and their relation to the structure of the parent protein. Biochemistry. 1969 Jun;8(6):2357–2365. doi: 10.1021/bi00834a016. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Adams M. J., Buehner M., Ford G. C., Hackert M. L., Lentz P. J., Jr, McPherson A., Jr, Schevitz R. W., Smiley I. E. Structural constraints of possible mechanisms of lactate dehydrogenase as shown by high resolution studies of the apoenzyme and a variety of enzyme complexes. Cold Spring Harb Symp Quant Biol. 1972;36:179–191. doi: 10.1101/sqb.1972.036.01.025. [DOI] [PubMed] [Google Scholar]
- SCHELLMAN J. A. The stability of hydrogen-bonded peptide structures in aqueous solution. C R Trav Lab Carlsberg Chim. 1955;29(14-15):230–259. [PubMed] [Google Scholar]
- Sarma V. R., Davies D. R., Labaw L. W., Silverton E. W., Terry W. D. Crystal structure of an immunoglobulin molecule by x-ray diffraction and electron microscopy. Cold Spring Harb Symp Quant Biol. 1972;36:413–419. doi: 10.1101/sqb.1972.036.01.053. [DOI] [PubMed] [Google Scholar]
- Saxena V. P., Wetlaufer D. B. Formation of three-dimensional structure in proteins. I. Rapid nonenzymic reactivation of reduced lysozyme. Biochemistry. 1970 Dec 8;9(25):5015–5023. doi: 10.1021/bi00827a028. [DOI] [PubMed] [Google Scholar]
- Scheraga H. A. Theoretical and experimental studies of conformations of polypeptides. Chem Rev. 1971 Apr;71(2):195–217. doi: 10.1021/cr60270a003. [DOI] [PubMed] [Google Scholar]
- Shearer W. T., Brown R. K., Bryce G. F., Gurd F. R. Reversible disruption by cupric ions of a helical conformation of a polypeptide derived from ribonuclease. J Biol Chem. 1966 Jun 10;241(11):2665–2671. [PubMed] [Google Scholar]
- Stroud R. M., Kay L. M., Dickerson R. E. The crystal and molecular structure of DIP-inhibited bovine trypsin at2.7Angstrom resolution. Cold Spring Harb Symp Quant Biol. 1972;36:125–140. doi: 10.1101/sqb.1972.036.01.018. [DOI] [PubMed] [Google Scholar]
- Sund H., Weber K. The quaternary structure of proteins. Angew Chem Int Ed Engl. 1966 Feb;5(2):231–245. doi: 10.1002/anie.196602311. [DOI] [PubMed] [Google Scholar]
- Tsong T. Y., Baldwin R. L. A sequential model of nucleation-dependent protein folding: kinetic studies of ribonuclease A. J Mol Biol. 1972 Feb 14;63(3):453–469. doi: 10.1016/0022-2836(72)90440-8. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Tsernoglou D., Hanson A. W., Knox J. R., Lee B., Richards F. M. The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 A. J Biol Chem. 1970 Jan 25;245(2):305–328. [PubMed] [Google Scholar]



