Abstract
Clustered regularly interspaced short palindromic repeat (CRISPR) loci and CRISPR-associated (Cas) proteins form an adaptive immune system that protects prokaryotes against plasmids and viruses. The Cmr complex, a type III-B effector complex, uses the CRISPR RNA (crRNA) as a guide to target RNA. Here, we show that the Cmr complex of Pyrococcus furiosus cleaves RNA at multiple sites that are 6 nt apart and are positioned relative to the 5′-end of the crRNA. We identified Cmr4 as the slicer and determined its crystal structure at 2.8 Å resolution. In the crystal, Cmr4 forms a helical filament that most likely reflects its structural organization in the Cmr complex. The putative active site is located at the inner surface of the filament where the guide and substrate RNA are thought to bind. The filament structure of Cmr4 accounts for multiple periodic cleavage sites on the substrate. Our study provides new insights into the structure and mechanism of the RNA-targeting Cmr complex.
INTRODUCTION
Bacteria and archaea are constantly challenged by viruses and plasmids. One important defense system that protects prokaryotes from these mobile genetic elements is composed of the clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (Cas) genes (1–4). CRISPR loci serve as memory of previous infections and consist of identical direct repeats (30–50 nt) separated by variable spacer sequences of similar size derived from invader DNA (5–7). The Cas genes are located in proximity of CRISPR loci (8,9) and their products function in various stages of CRISPR-mediated defense.
The spacer sequence is acquired from invader DNA and inserted at the leading end of the CRISPR array. The mechanism of the so-called “adaptation” process remains poorly understood. The CRISPR array is transcribed and processed into short CRISPR RNAs (crRNAs) that contain a spacer sequence flanked with partial repeat sequences at one or both ends. At the interference stage, crRNAs associate with Cas proteins into effector complexes that target DNA or RNA for degradation.
CRISPR–Cas systems are categorized into three major types and 10 subtypes (10). Type I, II and III systems are characterized by the signature proteins Cas3, Cas9 and Cas10, respectively, and substantially differ at the interference stage. The CRISPR associated complex for antiviral defense (Cascade) in Escherichia coli, which is a type I-E effector complex, consists of five proteins Cse1, Cse2, Cas7, Cas5e and Cas6e with stoichiometry of 1:2:6:1:1 and a 61 nt crRNA (11,12). The crRNA in Cascade pairs with one strand of target DNA, leading to the formation of an R-loop structure and degradation of the unpaired DNA strand by the Cas3 nuclease (13–18). The electron microscopy (EM) and recent crystallographic studies of Cascade reveal an extended structure with Cas7 forming a spiral backbone (19–22). In type II systems, the crRNA associates with a single Cas9 protein and a trans-activating crRNA (tracrRNA) to form the effector complex that cleaves both strands of target DNA with the dual nuclease domains of Cas9 (23–28). Type III-A systems have been proposed to target DNA; however, these biochemical activities have not been demonstrated in vitro (29,30).
The type III-B effector complex is unique in that it cleaves RNA, rather than DNA. The first type III-B effector complex, also known as the Cmr complex, was purified from Pyrococcus furiosus (Pf) and was found to contain six proteins (Cmr1–6) and a crRNA of 39 or 45 nt in length (31). The two types of crRNA contain a common 8 nt 5′-tag and a spacer of various length and are generated through Cas6-mediated endonucleolytic cleavage at the repeat region of pre-crRNA, followed by exonucleolytic processing of the 3′-end (31–35). The two crRNAs guide the cleavage of target RNA at different sites, leading to the proposal that the cleavage site is determined at 14 nt upstream of the 3′-end of the crRNA via a 3′ ruler mechanism (31). Crystal structures have been determined for Cmr5, Cmr2, Cmr3 and Cmr1 (36–42). The recently determined EM structure of the entire Pf Cmr complex shows three Cmr4–Cmr5 pairs form a helical backbone that is capped by Cmr2 and Cmr3 at the 5′-end of crRNA and by Cmr6 and Cmr1 at the 3′-end of crRNA (43). The Cmr complex from Thermus thermophilus (Tt) is highly similar to the Pf Cmr complex in terms of protein composition and structural organization (44). However, the Tt Cmr complex cleaves RNA at multiple sites with 6 nt intervals via a 5′ ruler mechanism. The Cmr complex from Sulfolobus solfataricus contains seven Cmr subunits (Cmr1-7) and cleaves the UA dinucleotide in a sequence-dependent manner (45). Although Cmr complexes from different species all target RNA, their mechanisms of action appear to be remarkably different. Additionally, the nuclease responsible for target cleavage remains elusive.
In this study, we revisited the RNA-guided nuclease activity of the Pf Cmr complex and demonstrate that it cleaves the target RNA at multiple periodic sites via a 5′, rather than a 3′, ruler mechanism. This finding reconciles the apparent difference between the Pf and Tt Cmr complexes. We further identified Cmr4 as the slicer and determined its crystal structure. Cmr4 forms a helical filament in the crystal that most likely reflects its physiological organization in the Cmr complex.
MATERIALS AND METHODS
Cloning, expression and purification of Cmr proteins
The Cmr1–6 genes were amplified from P. furiosus genomic DNA and cloned into modified pET28 (Novagen) vectors. Cmr1, Cmr2, Cmr4 and Cmr5 were fused to a C-terminal His6-tag. Cmr3 and Cmr6 were fused to an N-terminal His6-SMT3 tag. Mutations were introduced with the QuikChange method and were confirmed by sequencing.
All Cmr proteins were individually expressed in the BL21-Gold(DE3) strain. Cell cultures were grown to an OD600 of 0.7 and protein expression was induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside overnight at 18°C. The cells were pelleted, resuspended in 20 mM HEPES–Na, pH 7.0 and 500 mM NaCl, and broken with a JN-3000 homogenizer (JNBIO), and the cell lysate was clarified by centrifugation. The supernatant was heated at 75°C for 15 min and clarified again. The supernatant was applied to a HisTrap column, followed by washing with 20 mM HEPES–Na, pH 7.0, 500 mM NaCl and 20 mM imidazole. The target protein was eluted with 20 mM HEPES-Na, pH 7.0, 500 mM NaCl, and 500 mM imidazole. Following HisTrap affinity chromatography, the His6-SMT3 tags of Cmr3 and Cmr6 were removed by ULP1. All Cmr proteins were buffer exchanged into 20 mM HEPES-Na, pH 7.0, 500 mM NaCl and 5 mM MgCl2 using Superdex 200 gel filtration columns (GE Healthcare).
RNA preparation
The crRNA and substrate RNA were flanked by a 5′ hammerhead ribozyme and a 3′ hepatitis delta virus ribozyme (46). The coding sequences were assembled with oligonucleotides, cloned into a pUC18 vector and confirmed by sequencing. RNAs were prepared by in vitro transcription using standard conditions. Both ribozymes were automatically self-cleaved, resulting in a 5′ hydroxyl in the target RNA. The target RNA was purified using denaturing polyacrylamide gel electrophoresis and electroelution.
The substrate RNA was 5′-labeled with [γ-32P]ATP using T4 polynucleotide kinase (NEB). For 3′-labeling, the substrate was incubated with T4 polynucleotide kinase to remove 3′-phosphoryl groups, followed by phenol extraction and ethanol precipitation. The substrate was then labeled with [5’-32P]pCp using T4 RNA ligase. Labeled RNAs were purified with illustra MicroSpin G-25 columns (GE Healthcare).
Activity assay
The Cmr–crRNA complex was assembled at a 1:1:1:4:4:1:0.1 molar ratio of Cmr1:Cmr2:Cmr3:Cmr4:Cmr5:Cmr6:crRNA at 70°C for 30 min. The labeled substrate (∼0.2 nM) was incubated with 50 nM Cmr–crRNA complex in reaction buffer containing 20 mM HEPES–Na, pH 7.0, 500 mM NaCl and 5 mM MgCl2 at 70°C for 30 min. The RNA was purified by phenol extraction and ethanol precipitation and resolved using 10% denaturing polyacrylamide gel electrophoresis followed by phosphorimaging.
Purification of Cmr4 for crystallization
Cmr4-expressing E. coli cells were lysed using a JN-3000 homogenizer (JNBIO) in buffer A (20 mM HEPES–Na pH 7.6, 500 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol and 5% v/v glycerol). After centrifugation, the supernatant was loaded onto a HisTrap column (GE Healthcare), followed by washing with buffer A. Cmr4 was eluted with 300 mM imidazole in buffer A. The protein that eluted at higher concentrations of imidazole primarily formed aggregates. The eluted sample was exchanged to buffer Q (20 mM Tris–HCl, pH 8.0 and 100 mM NaCl) using a HiTrap desalting column and was loaded onto a HiTrap Q column (GE Healthcare). The target protein was eluted with a 0.1–0.5 M linear gradient of NaCl, concentrated and further purified with a Superdex 200 column in buffer S (20 mM HEPES–Na, pH 7.6 and 200 mM NaCl). Purified Cmr4 protein was concentrated to ∼15 mg/ml using ultrafiltration and stored at −80°C. The selenomethionine (SeMet)-labeled protein was prepared in M9 medium by blocking methionine biosynthesis (47) and purified in the same way as the native protein, except that the protein sample was supplemented with 5 mM dithiothreitol after the HisTrap step.
Crystallization, data collection and structure determination
Initial crystallization screens for Cmr4 were carried out using the sitting-drop vapor-diffusion method. The native crystal was obtained in 100 mM sodium citrate, 7% (w/v) PEG 3350 at 20°C and the SeMet-substituted Cmr4 crystal was grown under similar conditions. The native and SeMet-substituted Cmr4 crystals were cryoprotected with the reservoir solution containing 25% (v/v) glycerol and flash frozen in liquid nitrogen. All data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U and were processed with HKL2000 (48). The structure was determined by single-wavelength anomalous diffraction (SAD) using a SeMet derivative dataset collected at the peak wavelength of Se to 3.2 Å resolution. PHENIX was used for structural determination, automatic model building and refinement (49). Manual model adjustment was conducted in Coot (50). The final structure was refined against a 2.8 Å native dataset and contained two copies of Cmr4. Residues 17–27, 70–79, 123–126 and 205–227 were not modeled due to missing electron density. Ramachandran plot analysis showed that 97.3% of the residues are in most favorable regions and 2.7% in allowed regions. Structural figures were produced with PyMOL and Chimera (51,52).
RESULTS
The Pf Cmr complex cleaves the substrate at multiple periodic sites
The Pf Cmr complex that is assembled with 39 or 45 nt crRNA has been reported to cleave substrates at a single site located 14 nt upstream of the 3′-end of the crRNA (31). To verify this observation, we measured the nuclease activity for the Cmr complex that was reconstituted from six recombinant Cmr proteins and a 45 nt crRNA encoded by CRISPR 7.01. The substrate was a 5′-32P-labeled 37 nt RNA complementary to the spacer region (Figure 1A). Surprisingly, we detected three, rather than one, distinct 5′-products in the denaturing gels. These products were 14, 20 and 26 nt in length compared with RNA ladders and products of other crRNAs (Supplementary Figure S1). The corresponding cleavage sites are referred to as sites 31, 25 and 19, respectively, according to their distances to the 5′-end of the crRNA. This designation follows a 5′ ruler mechanism as demonstrated below. Notably, these cleavage sites are separated with a period of 6 nt.
Figure 1.
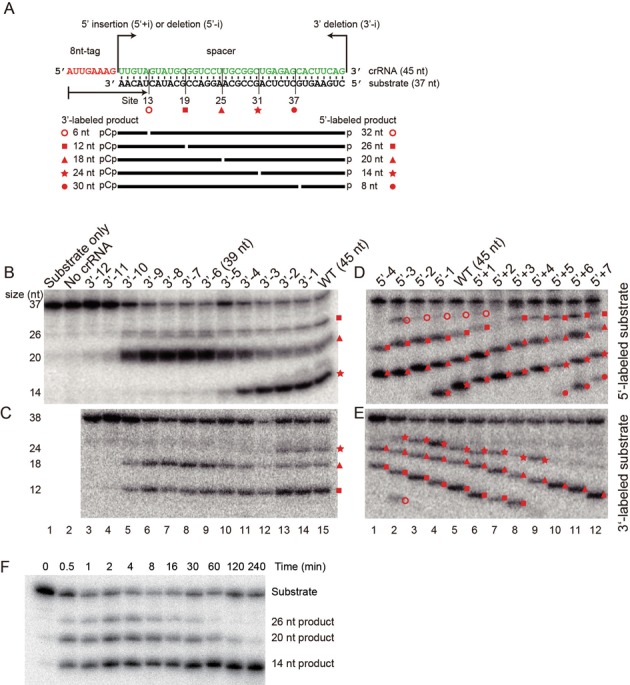
RNA cleavage activity of the Pf Cmr complex. (A) The sequence of the crRNA and substrate used in the activity assay. The 5′ tag and spacer sequences of the 45 nt crRNA are colored red and green, respectively. Five detected cleavage sites are marked with their distances to the 5′-end of the crRNA and by specific symbols. Substrates were 5′-labeled with 32P to detect 5′-products or 3′-labeled with 32pCp, which would increase the RNA length by 1 nt, to detect 3′-products. Products for the 45 nt crRNA are marked by their lengths. (B–E) Nuclease activity assay. The Cmr complex was loaded with a crRNA whose spacer was modified at the 3′- (B, C) 5′-end (D, E) and incubated with 5′- (B, D) or 3′-labeled (C, E) substrates for 30 min at 70°C. RNAs were separated with denaturing gels and visualized by phosphoimaging. Negative controls contain only substrate or crRNA-free Cmr complexes. Products are labeled with symbols indicative of the cleavage sites. (F) Time course of cleavage on the 5′-labeled substrate by the Cmr complex bound with the 45 nt crRNA.
If the site of cleavage is measured from the 3′ end of the crRNA, truncation at the 3′ end of the crRNA would lead to a shift in the cleavage site and longer 5′-products. To test this model, we shortened the crRNA sequentially from its 3′ end by 1 to 12 nt (named as 3′-1 to 3′-12). Surprisingly, the size of all products remained unchanged (Figure 1B). Identical results were observed when the 3′ pCp-labeled substrates were used to detect 3′-products (Figure 1C). Thus, the cleavage site is not measured from the 3′-end of the crRNA.
Although the position of the cleavage site is not related to the 3′ end of the crRNA, whether cleavage occurs at a site did depend on the crRNA length. Cleavage at site 31 was significantly reduced when the crRNA was truncated by five or more nucleotides, and cleavage at all sites was abolished for the 3′-11 (34 nt) and shorter crRNAs.
We wondered whether the cleavage site is determined from the 5′-end of the crRNA, namely, using a 5′ ruler mechanism. To test this idea, we removed 1 to 4 nt from (named as 5′-1 to 5′-4) or inserted 1–7 nt into (named as 5′+1 to 5′+7) the 5′-end of the spacer (Figure 1D–E). Upon successive extension of the spacer, all 5′-products increased in length in 1-nt increments and all 3′-products accordingly decreased in length. Truncation of the 5′ end of the spacer led to opposite changes in product sizes. Importantly, the distances between cleavage sites and the 5′ end of the crRNA remained constant for these crRNA variants. These results clearly indicate that the 5′-end of the crRNA serves as a reference point for determination of the cleavage site.
We clearly observed new cleavage events at site 37 when the spacer was extended by five or more nucleotides at the 5′-end (Figure 1D–E, lanes 10–12) and at site 13 for the 5′-3 crRNA (Figure 1D–E, lane 2). Low levels of 5′-products that resulted from cleavage at site 13 were also discernible for other crRNAs (Figure 1D–E, lanes 3–6). In total, we found five cleavage sites at positions 13, 19, 25, 31 and 37, all following a 6 nt period.
It is notable that all possible cleavage products for a specific crRNA are not produced at identical levels. Short cleavage products are generally more abundant than long cleavage products, although extremely short RNA products would be difficult to detect in the gel due to diffusion. This phenomenon suggests additional processing of intermediate products. Indeed, the time course of target RNA cleavage showed that long products were converted into short products over time, although the kinetic of secondary cleavage appears to be rather slow (Figure 1F).
The 5′ tag is essential for crRNA function
The Pf crRNA contains 8 invariant nucleotides (AUUGAAAG) at the 5′-end that are derived from the repeat sequence. We examined the role of the 5′-tag by mutagenesis using a 39 nt crRNA (Figure 2A). Removal of the entire 5′-tag abolished the cleavage activity, indicating that the tag is essential for crRNA function, as shown previously (35). Substitution of each of the 5′-tag nucleotides with its complementary sequence identified U2 as a key nucleotide. Additional mutagenesis showed that position 2 can accommodate a pyrimidine U or C, but not a purine A or G. These results show that the sequence of the 5′-tag is essential for crRNA function and is likely recognized by Cmr proteins.
Figure 2.

Requirements for cleavage activity of the Cmr complex. (A) Key nucleotides in the 5′-tag of crRNA. The Cmr complex was loaded with a 39 nt crRNA with the indicated mutation and assayed for cleavage activity using a 5′-32P labeled substrate. (B) Activity of the Cmr complex with individual subunit omitted. The 45 nt crRNA and 5′-labeled substrate were used. (C) Key residues of Cmr4. The Cmr complex loaded with a 39 nt crRNA and containing no, wild-type (WT) or the indicated mutant Cmr4 was assayed for cleavage activity using a 5′-32P labeled substrate.
Cmr4 is the slicer in the Cmr complex
Among the six Cmr proteins, all except Cmr5 have been shown to be required for the cleavage activity (31). However, we found that omission of each of the Cmr2, Cmr3, Cmr4 and Cmr5 subunit completely abolished the target RNA cleavage (Figure 2B). Removal of Cmr1 or Cmr6 reduced, but did not abolished, the cleavage activity, indicating that they are needed for the optimal activity of the Cmr complex (Figure 2B).
It remains unknown which Cmr protein functions as the slicer that cleaves the substrate. Our observation of multiple cleavage sites suggests that the Cmr complex contains multiple copies of the slicer. The recently determined EM structures of the Cmr complex reveal that multiple Cmr4 subunits align along the path of the crRNA and suggest that Cmr4 may be the slicer (43,44). Because the nuclease activity of Cmr complexes is dependent on divalent ions such as Mg2+ (31), the active site of the slicer probably harbors acidic residues that are essential for metal ion coordination and catalysis. We identified several conserved acidic residues based on multiple sequence alignment of Cmr4 (Figure 3) and tested their importance for activity with alanine mutation (Figure 2C). The D26A mutation completely abolished cleavage of the substrate RNA, and the D86A mutation substantially reduced the activity. Mutations of other residues (H15, E32, E151, E152, E227 and E228) caused no apparent effect. These results strongly suggest that Cmr4 is the slicer in the Cmr complex and that D26 may act as a catalytic residue.
Figure 3.
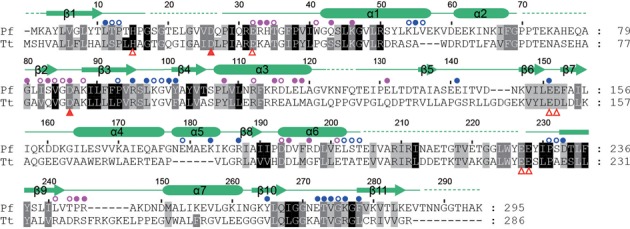
Sequence alignment of Cmr4 homologs. A total of 303 Cmr4 sequences were aligned and only the Pyrococcus furiosus (Pf) and Thermus thermophilus (Tt) Cmr4 are shown. The residues that are conserved in 95, 80 and 60% of all aligned sequences are shaded in black, gray and light gray, respectively. Amino acid similarity groups are defined as follows: S and T; D and E; K and R; L, I, V, M, F, Y and W. The secondary structures observed in the crystal structure are displayed on the top. Dashed lines denote disordered regions. Residues that are buried by 10–30 Å2 or >30 Å2 surface area as a result of oligomerization are labeled with open and closed circles, respectively (blue for subunit A, magenta for subunit B). The analyzed mutations that affect or do not affect the activity are labeled with solid or open triangles below the alignment.
Crystal structure of Cmr4
To better understand the function of Cmr4, we crystallized the full-length Cmr4 protein and solved its structure using Se-phasing. The structure was refined at 2.8 Å resolution to an Rwork of 19.6% and an Rfree of 22.3% (Table 1). Cmr4 crystallized in space group P41 with two molecules in the asymmetry unit (ASU). The structures of two protomers are nearly identical and can be superimposed with a root mean square deviation (RMSD) of 0.176 Å.
Table 1. Data collection and refinement statistics.
| Crystal form | Se-labeled | Native |
|---|---|---|
| Data collection | ||
| Space group | P41 | P41 |
| Cell dimensions | ||
| a, b, c (Å) | 63.8, 63.8, 192.7 | 64.0, 64.0, 195.3 |
| α, β, γ (º) | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.9791 | 0.9791 |
| Resolution range (Å) | 30–3.2 (3.26–3.2) | 20–2.8 (2.85–2.8) |
| Unique reflections | 12584 | 19448 |
| Redundancy | 15.0 (12.4) | 3.6 (3.1) |
| I/σ | 43.3 (6.8) | 13.7 (2.4) |
| Completeness (%) | 99.9 (99.8) | 98.8 (93.1) |
| Rmerge | 0.116 (0.455) | 0.084 (0.476) |
| Structure refinement | ||
| Resolution range (Å) | 20–2.8 (2.89–2.8) | |
| No. of reflections | 19149 | |
| No. atoms | 3668 | |
| Mean B factor (Å2) | 78.5 | |
| Rwork | 0.196 (0.263) | |
| Rfree | 0.223 (0.304) | |
| RMSD bond length (Å) | 0.003 | |
| RMSD bond angles (º) | 0.668 |
Values for the data in the highest resolution bin are shown in parentheses.
Cmr4 folds into an ellipsoid-shaped globular structure that is composed of seven α-helices and 11 β-strands (Figure 4A-B). Strands β10, β11, β1, β9, β2, β3, β4, β8 and β5 constitute the central mixed β-sheet, which is packed by α-helices α1, α2, α7 and α6 and strands β6 and β7 on one side and by α-helices α3, α4 and α5 on the other side.
Figure 4.
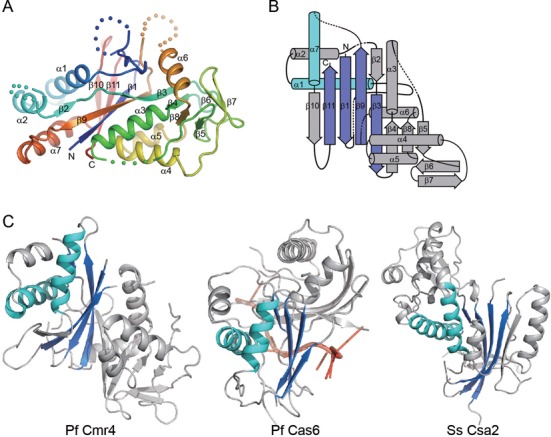
Structure of Cmr4. (A) Ribbon representation of the Cmr4 structure. The polypeptide chain is colored blue to red from the N- to C-terminus. The secondary structure elements and the N- and C-termini are labeled. Dots denote disordered loops. (B) Topology diagram of Cmr4. The α-helices and β-strands are shown as cylinders and arrows. The RAMP structural motif is highlighted in blue and cyan. (C) Structural comparison of Pf Cmr4, Pf Cas6 (3PKM) and Ss Csa2 (3PS0). These structures are aligned by their shared RAMP domains (colored blue and cyan). The Pf Cas6 structure is shown with a bound RNA molecule (red).
Cmr4 is predicted to contain a repeat associated mysterious protein (RAMP) domain (10), which is a derivative of the RNA recognition motif (RRM) or the ferredoxin fold. The structure of Cmr4 reveals a hidden RAMP domain composed of secondary structure elements β1-α1-β3-β9-α7-β11. We searched for structural homologs of Cmr4 using the DALI server and found RAMP-type Cas proteins that display high Z-scores, including P. horikoshii Cas6 (Z-score = 7.8, PDB code 3QJL), P. furiosus Cas6 (Z-score = 5.9, 3PKM), Streptococcus pyogenes Cas5d (Z-score = 5.7, 3VZH), S. solfataricus Csa2 (Z-score 4.9, 3PS0). Notably, structural similarity of Cmr4 with these proteins is limited to the RAMP domain (Figure 4C).
The helical filament structure of Cmr4
Interestingly, the Cmr4 molecules in the crystal stack in a head-to-tail manner to form a helical filament along the 41 screw axis (Figure 5A). The filament consists of eight molecules per turn and exhibits a pitch length of 195 Å (which equals the length of the c axis in the unit cell). The two Cmr4 molecules in the ASU (hereafter referred to as molecules A and B) are crystallographically distinct and may form two types of interfaces along the filament: one between molecules A and B from an identical ASU and a second between molecule B and the symmetry-related molecule A’ from an adjacent ASU. Superposition of the entire A–B and B–A’ dimers results in an RMSD of 0.34 Å over 445 Cα atoms, indicating that the two interfaces are nearly identical. This result supports the physiologically relevance of the observed subunit interfaces, rather than simply resulting from the crystal packing.
Figure 5.
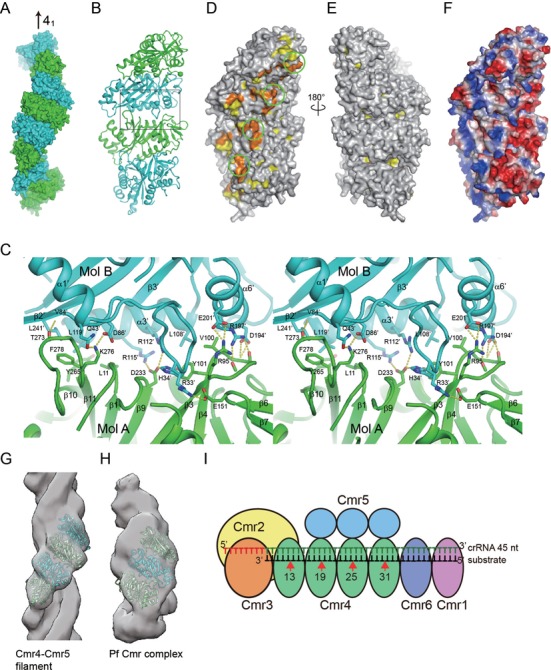
Filament structure of Cmr4. (A) Filament structure of Cmr4. Eight Cmr4 subunits constituting one helical turn are shown in surface representation and are alternately colored in blue and cyan. The crystallographic 41 axis is indicated. (B) Ribbon representation of four Cmr4 subunits viewed from the inner surface. The sphere (Cα of residue 28) indicates the approximate location of the putative catalytic D26 residue that is disordered in the crystal. The boxed region is shown in C. (C) Cross-eye stereoview of the interface between Cmr4 subunits. Residues involved in the interaction are shown as sticks. Dotted lines denote hydrogen bonds. Oxygen is red, nitrogen is blue and carbon is green for subunit A and cyan for subunit B. (D) Conservation surface of the Cmr4 filament. Residues with at least 95% and 80% conservation are colored orange and yellow, respectively. Same view as in B showing the inner surface. The putative active site is circled. (E) A 180 degree rotation of D showing the outer surface. (F) Electrostatic potential surface. The surface is colored blue to red from positively to negatively charged regions. Same view as in B. (G) The EM density of the Cmr4–Cmr5 filament is fit with the crystal filament structure of four Cmr4 subunits. (H) The EM density of the Pf Cmr complex is fit with the crystal filament structure of three Cmr4 subunits. (I) Cartoon model of the Pf Cmr complex containing four Cmr4 subunits and a 45 nt crRNA.
The intermolecular interface involves opposite faces of respective Cmr4 subunits (Figure 5B and C). The binding face in one subunit (molecule A in Figure 5C) is located at one end of the central β-sheet composed of the β10–β11 loop, the C-terminus of strand β1, the N-terminus of strand β9 and the β3–β4 loop. The binding face in the other subunit (molecule B in Figure 5C, denoted by primes) is composed of helices α3’ and α6’, the β2’–β3’ loop and a portion of the long loop linking strand β1’ and helix α1’. The interface is stabilized by a large number of electrostatic, hydrogen bonding and hydrophobic interactions and buries 1296 Å2 of solvent accessible surface area per subunit.
The β10–β11 loop mediates major interactions at one side of the interface. These interactions include a hydrogen bond between the backbone carbonyl oxygen of T273 and backbone amide nitrogen of K46’, and a hydrophobic cluster composed of residues Y265, F278, L11, L119’ and L241’. In addition, a salt bridge is formed between the highly conserved K276 and D86’. The D86A mutation reduced the cleavage activity of the Cmr complex probably by interfering with the filament structure. At the center of the interface, D233 of subunit A interacts with R112’, R115’ and H34’ from subunit B. At the other side of the interface, the β3–β4 loop makes extensive contacts with helix α6’, including four hydrogen bonds between the backbone atoms in the β3–β4 loop and the side chains of D194’ and R197’, a salt bridge between R95 and E201’ and hydrophobic interactions among Y101, V100 and L108’.
The helical filament structure of Cmr4 mostly likely corresponds to its organization in the Cmr complex. In the Cmr complex, the inner surface of the Cmr4 helix is supposed to bind the crRNA and substrate and to be involved substrate cleavage. The inner surface of the Cmr4 filament harbors most of the conserved surface residues, whereas the outer surface is composed of highly variable residues (Figure 5D and E). This bipolar distribution of conserved residue is consistent with the functional importance of the inner surface. In addition, residues 17–27, which harbor the putative catalytic residue D26, and residues 205–227 are highly conserved but are disordered in our structure (and hence are not displayed in the conservation surface). Both loops are located in the inner surface and likely play important role in RNA binding and catalysis. The inner surface of the Cmr4 filament is positively charged (Figure 5F), which would facilitate binding of the crRNA and substrate. In addition, Cmr4 and Cmr5 have been shown to assemble into a filament structure (rise = 24.4 Å and twist = 48°) (43), whose geometry closely matches that of the Cmr4 filament in the crystal (rise = 24.4 Å and twist = 45°). Four consecutive Cmr4 subunits from the crystal filament can be directly fit into the EM density map of the Cmr4–Cmr5 filament (Figure 5G). Three or four Cmr4 molecules also fit well into the EM density maps of the Pf or Tt Cmr complexes (Figure 5H) (43,44).
DISCUSSION
In this study, we show that the Pf Cmr complex cleaves target RNAs at multiple sites separated by 6 nt and that the cleavage positions are determined by their distances from the 5′-end of the crRNA. Our results clarify the mode of action of the Pf Cmr complex and demonstrate that the Pf and Tt Cmr complexes, which are of archaeal and bacterial origin, respectively, are homologous at both the structural and biochemical level.
We have shown that Cmr4 is the slicer and determined its helical filament structure, which most likely reflects its organization in the Cmr complex. We also identified D26 essential for the cleavage activity that may be involved in catalysis or binding target RNA. Interestingly, D26 is located in a disordered loop in the RNA-free structure. The active site is likely formed upon association of the crRNA and substrate.
As illustrated by the EM structures (43,44), the multiple-copy Cmr4 and Cmr5 subunits form a helical backbone. One end of the backbone is capped by the Cmr2–Cmr3 heterodimer and the other end by Cmr6 and Cmr1. UV-crosslinking studies have shown that the 5′-tag of the crRNA is anchored at the Cmr2–Cmr3 dimer and the 3′-end of the crRNA is placed at the Cmr1 side (43). Although the EM structures cannot resolve the crRNA, it most likely binds at the inner surface of the Cmr4 helical filament.
Our results provide new insights into the structural organization of the Pf Cmr complex (Figure 5I). The helical structure of Cmr4 corresponds well with the periodic pattern of Cmr-mediated RNA cleavage. It is most likely that each Cmr4 is responsible for cut at one periodic site. We detected a total of five cleavage sites using various crRNAs; however, not all possible products were observed for each crRNA. The number of actual cleavage sites is closely related to the length of the crRNA. The possible reason is two-fold. First, Cmr complexes contain a variable number of Cmr4 subunits and short crRNAs selectively associate with complexes containing fewer Cmr4 subunits, thereby limiting the number of cleavage sites. The heterogeneity of Cmr complexes has been experimentally demonstrated. The EM structures of Pf and Tt Cmr complexes contain three and four Cmr4 subunits, respectively (43,44). The 5′+5 (50 nt) and longer crRNAs should associate with at least five Cmr4 molecules to allow cleavage at site 37. Second, although crRNAs may assemble with various Cmr complexes in vitro, the length of crRNA determines how many sites are cleaved. Analysis of the minimal length of crRNA that allows detection of cleavage at a site shows that cleavage at sites 25, 31 and 41 require a crRNA length of at least 35 nt (3′-10), 41 nt (3′-4) and 50 nt (5′+5), respectively. Therefore, cleavage of the furthest site (relative to the 5′ end of the crRNA) requires the presence of at least 10 (sites 25 and 31) or 13 nt (site 37) of guide sequences downstream of the site. The 3′ region of the crRNA may contact with Cmr6, Cmr1 or inactive Cmr4 subunits that do not cut the substrate and is likely required to maintain the active conformation of the furthest cleavage site.
The in vivo assembly pathway of the Cmr complex may differ from the above described pathway for in vitro assembly. The 39 and 45 nt crRNAs may specifically associate in vivo with the Cmr complex that contains three and four Cmr4 subunits, respectively, as a result of their 3′-processing pathway. The Cmr-bound crRNAs exist in two major sizes: 39 and 45 nt in Pf or 40 and 46 nt in Tt (35,44). Interestingly, the size difference between these two classes of crRNAs exactly matches the period of cleavage sites or the distance spanned by Cmr4. This correlation suggests that the 3′-processing of crRNA occurs in the Cmr complex and that the length of the mature crRNA is related to the number of Cmr4 subunits in the Cmr complex. Specifically, after the pre-crRNA is cleaved at repeat regions by Cas6, the crRNA intermediate associates with Cmr complexes that primarily assemble three or four Cmr4 molecules in vivo. An exonuclease subsequently digests the 3′-tail of the crRNA intermediate that is not protected by protein. One Cmr4 molecule can provide protection over 6 nt. As a result, crRNAs are processed to 39 nt in complexes containing three Cmr subunits and to 45 nt in complexes containing 4 Cmr4 subunits.
For a 5′ ruler mechanism, the crRNA must be properly aligned from its 5′-end to the furthest cleavage site within the Cmr complex. The essential 5′-tag appears to be anchored at the Cmr2–Cmr3 dimer (43). By prediction, each Cmr4 in the helical assembly should align with a specific 6-nt region of the spacer around the corresponding cleavage sites. The alignment between the Cmr4 filament and the spacer may not need prior binding of the substrate. We found that crRNAs containing a 5′-extended spacer yielded an identical cleavage pattern (Figure 1D-E, 5′+1 to 5′+7). Because the inserted 5′ spacer sequences did not pair with the substrate in our system, the register of crRNA within the Cmr complex is therefore independent of whether the spacer, at least its 5′-region, adopts a single- or double-stranded conformation.
Given that the Cmr complex contains multiple active sites, the substrate could be cleaved multiple times. This notion is supported by the apparent greater abundance of short products than long products for both 5′- and 3′-labeled substrates and the time-dependent accumulation of short products (Figure 1). The Tt Cmr complex has been shown to cleave the substrate in the 5′ to 3′ direction relative to the crRNA (44). However, the Pf Cmr complex appears to randomly cleave the substrate among potential sites. If the cleavage proceeds in a 5′ to 3′ direction relative to the crRNA, only the shortest 3′-product from site 13 would be observed for 3′-labeled substrates. Conversely, if the cleavage follows a 3′ to 5′ direction, only the shortest 5′-product would be observed for 5′-labeled substrates. However, multiple products were detected for both 5′- and 3′-labeled substrates in our studies. Therefore, cleavage at multiple sites does not follow a specific direction for the Pf Cmr complex.
When this manuscript was under submission and review, three papers reported the identification of Cmr4 as the slicer, the filament structure of Cmr4 and the revised biochemical properties of the Cmr complex (53–55). Our results here are largely consistent with these reports.
ACCESSION NUMBERS
The coordinates and structural factors have been deposited in Protein Data Bank with accession code 4WNZ.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
Acknowledgments
We thank the staff at the Shanghai Synchrotron Radiation Facility beamline BL17U for assistance in data collection and Yan Guo for technical assistance.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010203]; National Natural Science Foundation of China [31325007]; Ministry of Science and Technology of China [National Basic Research Program of China, 2010CB835402]; Beijing Municipal Government. Funding for open access charge: National Natural Science Foundation of China [31325007].
Conflict of interest statement. None declared.
REFERENCES
- 1.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscipl. Rev. RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 4.Terns R.M., Terns M.P. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet.: TIG. 2014;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 6.Mojica F.J., Diez-Villasenor C., Garcia-Martinez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 7.Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 8.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 10.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 13.Huo Y., Nam K.H., Ding F., Lee H., Wu L., Xiao Y., Farchione M.D., Jr, Zhou S., Rajashankar K., Kurinov I., et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat. Struct. Mol. Biol. 2014;21:771–777. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinkunas T., Gasiunas G., Waghmare S.P., Dickman M.J., Barrangou R., Horvath P., Siksnys V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32:385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulepati S., Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H., Sheng G., Wang J., Wang M., Bunkoczi G., Gong W., Wei Z., Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- 20.Mulepati S., Heroux A., Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson R.N., Golden S.M., van Erp P.B., Carter J., Westra E.R., Brouns S.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiedenheft B., Lander G.C., Zhou K., Jore M.M., Brouns S.J., van der Oost J., Doudna J.A., Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadan A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 29.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouillon C., Zhou M., Zhang J., Politis A., Beilsten-Edmands V., Cannone G., Graham S., Robinson C.V., Spagnolo L., White M.F. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R., Preamplume G., Terns M.P., Terns R.M., Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carte J., Wang R., Li H., Terns R.M., Terns M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hale C., Kleppe K., Terns R.M., Terns M.P. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hale C.R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A.M., Glover C.V., 3rd, Graveley B.R., Terns R.M., et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J., Jeon J.H., Shin M., Shin H.C., Oh B.H., Kim J.S. Crystal structure and CRISPR RNA-binding site of the Cmr1 subunit of the Cmr interference complex. Acta Crystallogr. D, Biol. Crystallogr. 2014;70:535–543. doi: 10.1107/S1399004713030290. [DOI] [PubMed] [Google Scholar]
- 37.Shao Y., Cocozaki A.I., Ramia N.F., Terns R.M., Terns M.P., Li H. Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.H., Sun J., Park S.Y., Hwang H.J., Park M.Y., Shin M., Kim J.S. Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications. FEBS Lett. 2013;587:562–568. doi: 10.1016/j.febslet.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 39.Zhu X., Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 40.Cocozaki A.I., Ramia N.F., Shao Y., Hale C.R., Terns R.M., Terns M.P., Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamoto K., Agari Y., Agari K., Yokoyama S., Kuramitsu S., Shinkai A. X-ray crystal structure of a CRISPR-associated RAMP superfamily protein, Cmr5, from Thermus thermophilus HB8. Proteins. 2009;75:528–532. doi: 10.1002/prot.22358. [DOI] [PubMed] [Google Scholar]
- 42.Osawa T., Inanaga H., Numata T. Crystal structure of the Cmr2-Cmr3 subcomplex in the CRISPR-Cas RNA silencing effector complex. J. Mol. Biol. 2013;425:3811–3823. doi: 10.1016/j.jmb.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 43.Spilman M., Cocozaki A., Hale C., Shao Y., Ramia N., Terns R., Terns M., Li H., Stagg S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol. Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staals R.H., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D.W., van Duijn E., Barendregt A., Vlot M., Koehorst J.J., Sakamoto K., et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J., Rouillon C., Kerou M., Reeks J., Brugger K., Graham S., Reimann J., Cannone G., Liu H., Albers S.V., et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price S.R., Ito N., Oubridge C., Avis J.M., Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- 47.Van Duyne G.D., Standaert R.F., Karplus P.A., Schreiber S.L., Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.DeLano W.L. The PyMOL User's Manual. San Carlos, CA, USA: Delano Scientific; 2002. [Google Scholar]
- 53.Benda C., Ebert J., Scheltema R.A., Schiller H.B., Baumgartner M., Bonneau F., Mann M., Conti E. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol. Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Hale C.R., Cocozaki A., Li H., Terns R.M., Terns M.P. Target RNA capture and cleavage by the Cmr type III-B CRISPR-Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramia N.F., Spilman M., Tang L., Shao Y., Elmore J., Hale C., Cocozaki A., Bhattacharya N., Terns R.M., Terns M.P., et al. Essential structural and functional roles of the Cmr4 Subunit in RNA cleavage by the Cmr CRISPR-Cas Complex. Cell Rep. 2014;9:1610–1617. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


