Abstract
SET and RING-finger-associated (SRA) domain is involved in establishment and maintenance of DNA methylation in eukaryotes. Proteins containing SRA domains exist in mammals, plants, even microorganisms. It has been established that mammalian SRA domain recognizes 5-methylcytosine (5mC) through a base-flipping mechanism. Here, we identified and characterized two SRA domain-containing proteins with the common domain architecture of N-terminal SRA domain and C-terminal HNH nuclease domain, Sco5333 from Streptomyces coelicolor and Tbis1 from Thermobispora bispora. Both sco5333 and tbis1 cannot establish in methylated Escherichia coli hosts (dcm+), and this in vivo toxicity requires both SRA and HNH domain. Purified Sco5333 and Tbis1 displayed weak DNA cleavage activity in the presence of Mg2+, Mn2+ and Co2+ and the cleavage activity was suppressed by Zn2+. Both Sco5333 and Tbis1 bind to 5mC-containing DNA in all sequence contexts and have at least a preference of 100 folds in binding affinity for methylated DNA over non-methylated one. We suggest that linkage of methyl-specific SRA domain and weakly active HNH domain may represent a universal mechanism in competing alien methylated DNA but to maximum extent minimizing damage to its own chromosome.
INTRODUCTION
DNA methylation occurs at the C-5 position of cytosine in most eukaryotic organisms, resulting in 5-methylcytosine (5mC) (1,2). 5mC is found predominantly in the symmetric CpG context in mammals and other vertebrates (3,4), as well as CHG (H = A, T or C) and asymmetric CHH contexts in plants (5). They constitute important epigenetic marks that are implicated in repressed chromatin state, inhibition of transcription and genome stability (1,2). Faithful inheritance of these epigenetic marks is essential to cell functions (6) while aberrant DNA methylation is associated with various diseases and disorders (7,8). Major players in the maintenance of DNA methylation in mammals include DNA methyltransferase (Dnmt1) and UHRF1 (ubiquitin-like, containing PHD and RING finger domains) (9–12). UHRF1 contains a SET and RING-associated (SRA) domain that preferentially binds to hemi-methylated DNA relative to the fully methylated or unmodified DNA (10,11). In the structure of the DNA–SRA complex, the 5mC base is flipped out from the DNA duplex and is accommodated in a binding pocket of the SRA domain, potentially preventing the protein from sliding along the DNA strands (13–15). Similarly, the dimeric SRA domain from the Arabidopsis thaliana SUVH5 binds to 5mC-containing DNA either at fully-methylated or hemi-methylated CpG, CHG or methylated CHH sites. It flips out both 5mC and its partner base in the complementary strand from the DNA duplex. Each of the extruded bases is positioned in one binding pocket of an individual SRA domain (16).
The eukaryotic SRA-like domains are also found in bacteria, although they are not associated with the SET and RING domains in the genome. Oftentimes they are fused or associated with restriction endonucleases (REases) (17). For example, the latest structurally characterized type IV restriction endonucleases in bacteria, such as the MspJI family (16,18) and the PvuRts1I/AbaSI family (19,20), possess DNA binding domains that are structurally similar to the eukaryotic SRA domains and adopt similar base flipping mechanism to recognise 5mC. Interestingly, they do not share any sequence similarity with the eukaryotic SRA counterparts. On the other hand, by using the eukaryotic SRA sequence as query, one can readily pull out more than 100 genes from bacterial genomes in GenBank, most of which having an annotation of ‘SRA-YDG protein’. Beyond the computational prediction of the SRA presence in these genes, little is known about their functional roles. Sequence alignment reveals that these bacterial SRA domains share significant similarity to the eukaryotic counterparts, such as UHRF1 or SUVH5 SRA domains, suggesting a likely common ancestor. Conserved domain analysis suggests that most of these SRA-YDG genes consist of the N-terminal SRA domain and a C-terminal HNH-type nuclease domain. Given the binding preference to modified DNA for the eukaryotic SRA domains (13–16), it is likely that these genes may encode another class of the DNA modification-dependent restriction endonucleases.
Within this family of the SRA-HNH nucleases, we chose to study two close SRA homologs in Streptomyces coelicolor and Thermobispora bispora DSM 43833. In this study, we report the in vitro binding and cleavage of 5mC-containing DNA by the two proteins, as well as their in vivo toxicity in dcm+ Escherichia coli cells. Structural modeling for two bacterial SRA domains and superposition to eukaryotic SRA domains are extensively studied and discussed. To our knowledge, this represents the first study on the activities of bacterial-sourced SRA-HNH protein.
MATERIALS AND METHODS
Bacterial strains and plasmid constructs used in this study were shown in Supplementary Table S1. Primers used in this study are listed in Supplementary Table S2. Oligos with different methylation pattern used in this study were summarised in Supplementary Table S3. Growth of E. coli strains and DNA manipulation were carried out according to Sambrook et al. (21).
Construction of vectors expressing Sco5333 and its derivative
The coding sequence of Sco5333 except for stop codon was amplified from total DNA of S. coelicolor A (3) 2 using KOD DNA polymerase (TOYOBO) and primers 5333EX-F& 5333EX-R. The PCR fragment was digested with NdeI and XhoI, and was inserted into pET44b as pJTU4356 to express C-terminal His6-tagged Sco5333. N-terminal His-tagged Tbis1 was synthesized as overlapping gBLOCKs and cloned into pTXB1 (22,23) using Gibson assembly.
Mutant proteins Sco5333G32A, Sco5333Y50A of the SRA domain and Sco5333H228A, Sco5333H253A of the HNH motif, and pJTU4356M expressing Sco5333 triple amino acid changes of His228Ala, Asn244Ala and His253Ala of the HNH domain were individually constructed using the KOD-Plus Mutagenesis Kit (TOYOBO). Primer pairs G32A-F & R, Y50A-F & R, H228A-F & R, H253A-F & R, 4356M-F & R were respectively used for PCR with pJTU4356 DNA as template. PCR products were treated with DpnI prior to ligation in Solution I (TOYOBO) mixed with T4 Polynucleotide Kinase (NEB). The ligation products were introduced into dcm-deficient E. coli JTU006 (24) to generate pJTU4381, pJTU4382, pJTU4383, pJTU4384 and pJTU4356M (Supplementary Table S1).
Measuring transformation efficiency
For transformation efficiency assay, 0.1 μg of pJTU4356, pJTU4381, pJTU4382, pJTU4383 and pJTU4384 were individually introduced into E. coli DH10B and JTU006, respectively, and 0.1 μg pET44b was also performed as positive control. For measuring the transformation efficiency of pTbis1, 0.1 μg of pTXB1_Tbis1 was introduced into E. coli ER2566 (dcm−) and E. coli ER2984 (dcm+), and 0.1 μg pTXB1 was performed as positive control. Transformation of each plasmid DNA was performed in three replicates.
Over-expression and purification of Sco5333, Tbis1 and Dcm methyltransferase
Dcm coding sequence was amplified by colony-PCR from E. coli DH10B using KOD DNA polymerase (TOYOBO) and primers dcmEX-F & R, the PCR fragment was treated with NdeI and EcoRI and ligated into the expression vector pET15b, generating pJTU4357 for producing N-terminally His6-tagged Dcm.
Expression constructs pJTU4356, pJTU4357, pJTU4381, pJTU4382, pJTU4383 and pJTU4384 were introduced into BL21(DE3)/pLysS respectively. 10 ml overnight culture of each above strains was inoculated into 1-l LB medium supplied with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol, and was grown at 37°C to OD600 0.6, cooled to room temperature and isopropyl thiogalactoside (IPTG) was added to a final concentration of 0.4 mM, followed by another 5 h at 30°C. The cells were harvested and resuspended in 20 ml binding buffer (20 mM Tris–Cl and 150 mM NaCl pH 8.0) and lysed by sonication in an ice bath. After centrifugation (16 000 g for 30 min at 4°C), the supernatant was applied to a HisTrap HP column (GE Healthcare) and purified with an ÄKTA FPLC (GE Healthcare) by eluting with imidazole linear gradient 20–500 mM. The product was desalted by a HiTrap Desalting column (GE healthcare) and stored in 20mM Tris–Cl buffer pH 8.0 containing 100 mM NaCl and 50% glycerol at −30°C. Purified Sco5333 and Dcm were visualized by Coomassie-stained 12% sodium dodecyl sulfate-polyacrylamide gelelectrophoresis (SDS-PAGE) analysis. Protein concentration was determined using a Bradford Protein Assay Kit (Bio-Rad).
For Tbis1, expression construct pTXB1_Tbis1 was introduced into T7 Express (NEB). Ten milliliters overnight culture was inoculated 2 × 1 l Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin. Bacteria culture was grown at 37°C to OD600 0.6, cooled to room temperature and IPTG was added to a final concentration of 0.3 mM. The culture was grown for additional 16 h at 16°C. Cells were harvested and re-suspended in 50 ml sonication buffer (50 mM Tris–HCl, 50 mM NaCl, 1 mM ethylene diamine tetraacetic acid (EDTA), 1 mM dithiothreitol (DTT) and 2% glycerol at pH 8.0) and lysed by sonication in an ice bath. After centrifugation (23 000 g for 45 min at 4°C), the supernatant was applied to a HisTrap HP column (GE Healthcare) and purified with ÄKTA FPLC (GE Healthcare) by eluting with imidazole linear gradient 20–500mM. Those fractions of elutes were analysed by SDS-PAGE and nuclease activity on pBR322 (dcm+) (25) and active fractions were pooled and further purified with a HiTrap Heparin HP column (GE Healthcare) by eluting with NaCl linear gradient 20 mM–1 M. Fractions containing purified protein were pooled, dialysed in storage buffer (20 mM Tris–HCl, 300 mM NaCl, 1 mM EDTA, 1 mM DTT at pH 7.4), concentrated with VivaSpin concentrator (MWCO at 30 kDa) (VIVASCIENCE) and stored in 50% glycerol at −20°C. Purified Tbis1 was visualized by SimplyBlue™ Safe Stain (LifeTechnologies) stained 10–20% SDS-PAGE. Protein concentration was determined using Bradford Protein Assay Kit (Bio-Rad).
DNA cleavage assay by purified Sco5333 and Tbis1
For DNA cleavage assay, 0.25 μg pUC18 (26) plasmid DNA isolated from DH10B (dcm+) and JTU006(dcm−) strains was incubated with varied Sco5333 (0.0175–3.5 μM) in hydroxyethyl piperazine ethanesulfonic acid (HEPES) buffer (40 mM HEPES, pH 7.0, 50 mM NaCl). For divalent metal ion requirement, 2 mM divalent ion of Zn2+, Mg2+, Mn2+, Co2+, Ni2+, Cu2+ and Ca2+ (Sigma) was individually added into the reaction. Then the total volume of 20 μl reaction mixture was incubated at 37°C for 1 h, followed by digestion with 5 units of proteinase K (Roche), and then examined by electrophoresis in 0.75% or 1.5% agarose gel.
In the assay to compare the DNA cleavage activity of Sco5333 and its mutant protein Sco5333M (Supplementary Table S1), conditions used were same as above except for concentration range of varied Sco5333 and Sco5333M (0–8 μM).
For cleavage of plasmid DNA pBR322 by Tbis1, buffer used was 20 mM Tris–acetate pH 7.9, 50 mM potassium acetate, 1 mM DTT supplemented with 2 mM Zn2+, Mg2+, Mn2+, Co2+ or Ni2+.
0.2 μg pBR322 plasmid DNA isolated from ER2984 was incubated with Tbis1 of 0.01–3 pmol in 20 μl reaction system at 37°C for 1 h, followed by digestion with proteinase K at 55°C to remove the bound Tbis1, and examined by electrophoresis in 1.2% agarose gel.
Amplification, methylation of 219bp DNA fragments as the EMSA substrate for Sco5333
The 219 bp fragment from NT885–1103 of pUC18 (accession no.: L08752) that contains two Dcm-methylation sites (NT954–958; NT967–971) was PCR amplified using primers 219 bp-F & R and purified, and then treated with 2 μM purified Dcm methyltransferase in buffer (50 mM Tris–HCl pH 7.5, 5 mM β-mercaptoethanol, 10 mM EDTA and 160 μM SAM) in a volume of 30 μl at 37°C for 1 h followed by heat inactivation at 85°C for 10. In addition, the 219 bp DNA fragment was methylated by commercially available methylases M.AluI, M.HhaI, M.HpaII, M.MspI, GpC(M.CviPI) and CpG(M.SssI) (NEB) respectively. For EMSA, Sco5333 of varied concentrations (0–0.2 μM) was incubated with methylated 219 bp DNA at 37°C for 5 min in 20 μl binding buffer (20 mM Tris–Cl pH 8.0, 100 mM KCl), the reaction mixture was mixed with 4 μl 6 × loading buffer (30 mM EDTA, 36% glycerol, 0.035% xylene cyanol, 0.05% bromophenol blue (TAKARA) and then resolved by 6% native-PAGE (80:1, acrylamide/bis-acrylamide) in 0.5× Tris-borate EDTA (TBE) at 10 mA at room temperature.
EMSA conditions of 55nt fragments for Sco5333
The 55 nt oligos 55ntDcm1 (NT929–983) and 55ntDcm2 (NT942–996) of pUC18, were used to study the binding affinity and the sequence specificity for Sco5333. To generate fully-, hemi-, and non-methylated 55ntDcm1 duplexes, four single-strand oligos were synthesized as below (Genebioseq Ltd):
55nt-1: 5′-FAM-gaaacccgacaggactataaagatacmCaggcgtttccccctggaagctccctcgt-3′;
55nt-2: 5′-FAM-gaaacccgacaggactataaagataccaggcgtttccccctggaagctccctcgt-3′;
55nt-3: 5′-acgagggagcttccagggggaaacgcmCtggtatctttatagtcctgtcgggtttc-3′;
55nt-4: 5′-acgagggagcttccagggggaaacgcctggtatctttatagtcctgtcgggtttc-3′.
Pairs of 55nt-1&3, 1&4, 2&3 and 2&4 were mixed in 1:1 molar ratio and annealed ramping down from 100°C to room temperature in a water bath. The same strategy was used to generate four 55ntDcm2 oligos with different methylation pattern (55nt-5 to 8), as well as to 55ntDcm1 oligos harbouring systematic mutation of the base flanking the modified cytosine. All single-strand oligos were synthesized, and listed in Supplementary Table S3. In the EMSA of the 5-FAM labeled 55nt duplexes: 0.25 μM DNA oligos was incubated with 1 μM Sco5333 in 20 mM Tris–Cl pH 8.0, 100 mM KCl in a volume of 20 μl at 37°C for 5 min. The reaction was then mixed with 4 μl 6× loading buffer (30 mM EDTA, 36% glycerol, 0.035% xylene cyanol, 0.05% bromophenol blue. TAKARA) and examined by 6% native-PAGE (80:1, acrylamide/bis-acrylamide) in 0.5× TBE and 10 mA at room temperature. The gel was visualized by FUJIFILM FLA-3000 fluorescent image analyser (excitation wavelength 473 nm).
Measurement of the equilibrium dissociation constant (KD) via isothermal titration calorimetry (ITC)
MicroCal iTC200 system (GE healthcare) was used to measure the KD of Sco5333 to fully- and hemi-methylated 54nt fragment. 80 μl of 300 μM Sco5333 was injected, and 300 μl of 30 μM 54nt fragment was in the sample cell. The titration was followed with the default ITC procedure and the data was analysed using Origin 7.0.
RESULTS
Sequence features of SRA and HNH domain of Sco5333 and Tbis1
Two SRA-HNH genes, sco5333 and tbis1, were identified in the genome of S. coelicolor and T. bispora DSM 43833 with ∼ 40% identity to the well-studied eukaryotic SRAUHRF1 and SRASUVH5 (Figure 1B). Structural modelling of SRASco5333 and SRATbis1 was performed based on the crystal structure of SRAUHRF1 (PDB: 3CLZ) and SRASUVH5 (PDB: 3Q0C). The overall secondary structure of the four SRAs is very similar except for a short β-sheet (motif I), an α-helix (motif II) and an additional α-helix (motif III) encoded by SRAUHRF1 (Figure 1B). Superposition of SRASco5333 to SRATbis1 revealed that their overall structures are almost identical (Supplementary Figure S1), so we focus the following analysis on the SRASco5333 domain. SRASco5333 aligns well to the SRASUVH5 in the key folds (α-helix and β-sheets) as well as the functional motifs including the thumb, 5mC binding domain (Supplementary Figure S2A). In addition to the motif I and motif II (Supplementary Figure S2B), structural superposition revealed a much longer NKR finger loop in SRAUHRF1 which forms hydrogen bonds with the unpaired guanine and its adjacent cytosine via the side chain of N489 and R491 (Supplementary Figure S3). Despite that the structure of thumb finger in four domains aligned well, SRAUHRF1 uses V446 to replace the 5-methylcytosine, and form hydrogen bonds with R491, however, it does not directly pair with the orphaned guanine (>3.8 Å) that is anchored by the charged R491. By contrast, SRASco5333, SRATbis1 and SRASUVH5 all use glutamine (Q), which has a longer side chain compared to valine. It replaces the 5mC and directly forms stable hydrogen bonds with the orphaned guanine (Figure 1B, Supplementary Figures S3 and S4). The four SRA domains possess conserved 5mC binding elements either in the amino acids (marked with green dots above SUVH5, Figure 1B) or in the tertiary structures (Supplementary Figures S3 and S4).
Figure 1.
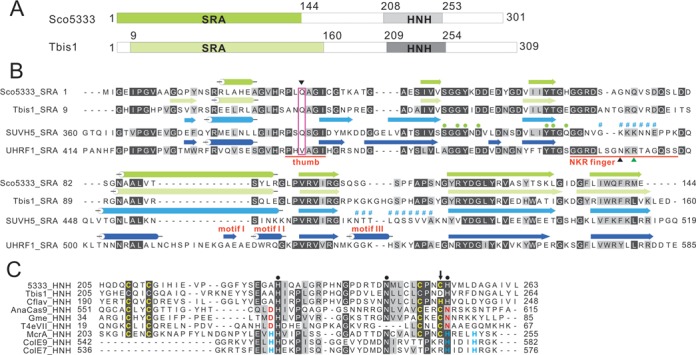
Sequence features of Sco5333 and Tbis1. (A) Domain architecture of Sco5333 and Tbis1, both of which contain a SRA domain (green) associated with a HNH motif (gray) at downstream. (B) Multiple sequence alignment and secondary structure comparison of SRA domains of Sco5333, Tbis1, SUVH5 and UHRF1. α-helices are in cylinders, and β-strands are in arrows. The thumb and NKR finger corresponding to UHRF1 are underlined in red. The inverted black triangle indicates the residue that inserts into the duplex and displaces the 5mC in the SUVH5 SRA. Filled green circles represent residues that interact with 5mC in the binding pocket of SUVH5 SRA and UHRF1 SRA, and the disordered region of SUVH5 SRA domain is represented by # #. Black and green upright triangles designate residues that replace the looped-out 5mC and mask the unmodified C in the UHRF1 SRA, respectively. (C) Multiple sequence alignment of HNH motifs of Sco5333, Tbis1, Cflav and the well studied HNH motif structure of AnaCas9, Gme, T4eVII, McrA, ColE9 and ColE7. Residues in golden designate the CCCC or CCCH type of zinc finger motifs. Filled black circles represent the conserved HNH motifs. The downward black arrow indicates that Tbis1 HNH displays a ruined CCCD zinc finger. The brown D and N are active sites of HNH motifs in AnaCas9, Gme and T4eVII that bind to divalent ions. The H in cyan designates the divalent ion-binding residues of HNH motifs in McrA, ColE9 and ColE7.
Sco5333 and Tbis1 both contain a HNH nuclease motif, which is found in more than 2000 proteins related to nucleic acid processing in GenBank. Within the HNH domain, Sco5333 harbours a CX2CX36CX2C motif (Figure 1C) that is characteristic of a zinc finger (ZF) structure, which is also present in other proteins containing HNH nuclease domains, such as AnaCas9, Gme, T4eVII and McrA (Figure 1C and Supplementary Figure S5). The four zinc finger proteins have a common ββα-fold in which an additional divalent metal ion is coordinated either by residues DN (red) for first three proteins or by H…H…H(cyan) for McrA, respectively (Figure 1C, Supplementary Figure S5). The zinc finger in AnaCas9 was explained to stabilize the adjacent ββα-Metal Fold (27). In some cases, the zinc finger could be CCHH, such as in Zif268 (28) or CCCH as in KpnI (29). As another example, the SRA-HNH protein Cflav (accession no.: ADG74130) contains a CCCH zinc finger (Figure 1C).
In vivo toxicity of Sco5333 and Tbis1 to E. coli with methylation
To purify Sco5333, its coding sequence was fused to 6×His-tag at C-terminus on pET44b and introduced to E. coli JTU006 (dam+, dcm−) and DH10B (dam+,dcm+), respectively. While JTU006 gave normal transformants with high efficiency, few transformants (<5) were obtained for DH10B, among which a spontaneous mutant Sco5333E42G was isolated. In order to evaluate the functional roles of the two domains for restriction of Dcm-methylated DNA, Sco5333 mutants, G32A and Y50A in the SRA domain, H228A and H253A at the HNH motif were individually introduced to E. coli DH10B or JTU006. Transformation efficiencies for all mutants into DH10B were restored to the level comparable to that for wild type sco5333 into JTU006 (Figure 2A), suggesting that the in vivo restriction of Dcm-methylated DNA requires cooperative function of the SRA domain and the HNH motif. Consistent with this result, the full-length tbis1 was restricted by ER2984 (dam+,dcm+) but not by ER2566 (dam+, dcm−). By comparison, transformation efficiencies of the SRATbis1 domain alone into ER2984 or ER2566 were not impaired and close to each other (Figure 2B).
Figure 2.
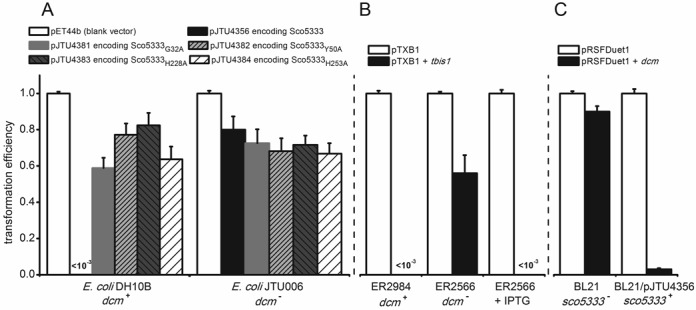
Transformations of plasmids encoding Tbis1 or Sco5333 and its mutants into dcm+, dcm− and sco5333+E. coli hosts. (A) The Sco5333 expression vector pJTU4356 and its mutants, pJTU4381 encoding Sco5333Gly32Ala and pJTU4382 encoding Sco5333Tyr50Ala in SRA domain, and pJTU4383 encoding Sco5333His228Ala and pJTU4384 encoding Sco5333His253Ala in HNH motif, were introduced into E. coli DH10B (dam+, dcm+) and its dcm knock-out mutant JTU006 (dam+, dcm−), respectively; All the amino acid changed proteins lost the lethal phenotype in dcm+E. coli hosts as pJTU4356 does, indicating that the in vivo restriction requires the SRA domain and HNH motif together. (B) pTbis1 encoding Tbis1 and was introduced into E. coli ER2566 (dam+, dcm−) and ER2984 (dam+, dcm+), respectively. Consistent with transformation of pJTU4356, pTbis1 leads cell death in ER2984 but not ER2566. However, over-expressing Tbis1 by IPTG induction in ER2566 leads cell lysis and death at high efficiency, implying promiscuous cleavage of non-Dcm methylated genome DNA was induced by high concentration of Tbis1. (C) pJTU4356 was introduced into E. coli BL21 previously, generating BL21/pJTU4356(dam+, dcm−, sco5333+). The dcm was cloned in pRSFDuet1, whose RSF origin is compatible with pBR322 origin of pJTU4356. pRSFDuet1 carrying dcm gene was efficiently introduced into BL21, but severely restricted by BL21 harboring sco5333.
We then measured the restriction of BL21(DE3) expressing Sco5333 to the dcm-methylated plasmid. As showed in Figure 2C, the plasmid pRSFDuet1+dcm was easily uptake by BL21(DE3), but its efficiency into BL21(DE3) expressing Sco5333 decreased by >90%. In contrast, transformation of strains with blank vector pRSFDuet1 showed little difference in transformation efficiency.
We then compared the growth curves for BL21(DE3)/plysS (dam+,dcm−) expressing Sco5333, Sco5333E42G and Sco5333M (triple amino acid changes of H228A,N244A,H253A) at 30°C with and without IPTG induction. Our results suggest that there are no differences in the growth rates among the wild type and the mutants (Supplementary Figure S6), implying that in vivo Sco5333 discriminates against Dcm-methylated DNA from non-methylated DNA at high specificity. In sharp contrast to this observation, over-expression of Tbis1 induced by IPTG led to in vivo toxicity to the Dcm-deficient strain ER2566, suggesting promiscuous cleavage of the non-methylated genome DNA at high level of Tbis1 (Figure 2B, third panel).
Effect of divalent metal ions to cleavage activity for both proteins
His-tagged Tbis1 and Sco5333 were over-expressed and purified (Figure 3A and B). In buffer without divalent metal ion, neither protein cleaved Dcm-methylated plasmid DNA, even with high protein:DNA ratio (120:1 for Sco5333, and 40:1 for Tbis1, Figure 3C and D). However, DNA cleavage activity were greatly stimulated in the presence of Ni2+, Co2+, Mn2+ and Mg2+ for Tbis1 (Figure 3D), and of Mn2+, Mg2+, Co2+ and Ni2+ for Sco5333 (Figure 3C). The remaining Ca2+, Cu2+ (data not shown) and Zn2+ did not stimulate DNA cleavage activity. On the contrary, the stimulated cleavage activity was suppressed by equal or excess molar of Zn2+ (Figure 4A and B). We then targeted the fourth cysteine by mutation in the zinc finger CX2CX36CX2C of Sco5333. Compared to the wild type, Sco5333C252D displayed higher DNA cleavage activity and required higher concentration of Zn2+ to suppress cleavage (Figure 4C), suggesting that a weaker zinc finger reduces the suppression of Zn2+. In addition, we compared the DNA binding affinity of Sco5333 by different metal ions. In the titration assay when the molar ratio of [protein]:[DNA binding site] is ∼7:1, excessive Zn2+ showed the most prominent effect on the DNA binding affinity to 5mC-DNA (Figure 5A). We then set the ratio of [Sco5333]:[DNA binding site] to ∼0.7:1 and increased Zn2+ concentration from 0.001 to 10 mM, and found increased shifting of 5mC-methylated DNA with the increasing concentration of Zn2+. (Figure 5B).
Figure 3.
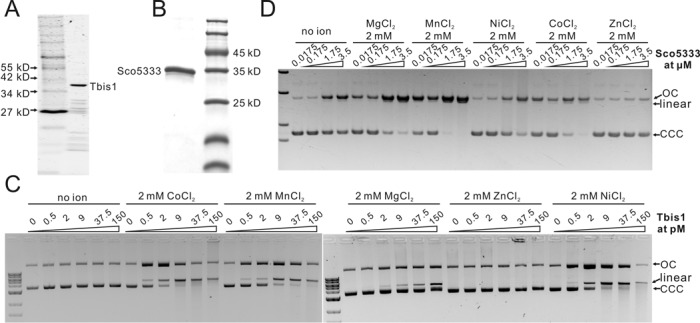
SDS-PAGE analysis and in vitro activity assay of Sco5333 and Tbis1 to methylated plasmid DNA in different divalent ions. (A) SDS-PAGE of Tbis1. (B) SDS-PAGE of Sco5333. (C) In vitro cleavage activity assays of Tbis1 in presence of 2 mM Co2+, Mg2+, Mn2+, Zn2+, Ni2+and no ion. pBR322 isolated from dcm+E. coli is used as the substrate. Cleavage activity is observed when supplied with 2 mM of Co2+, Mn2+, Ni2+ or Mg2+ in the reaction mixture, but not when supplied with Zn2+. (D) In vitro cleavage activity assays of Sco5333 in presence of 2 mM Mg2+, Mn2+, Ni2+, Co2+, Zn2+ and no ion. pUC18 isolated from dcm+E. coli host DH10B is used as the substrate. Cleavage activity is observed when supplied 2mM of Mn2+, Mg2+, Co2+ or Ni2+in the reaction mixture, and Zn2+could not stimulate cleavage activity of Sco5333.
Figure 4.
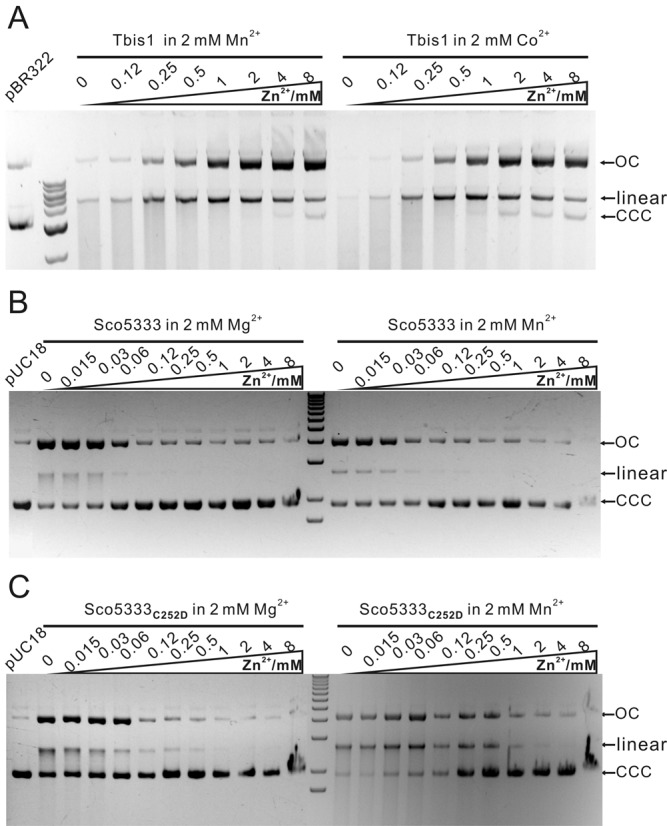
In vitro cleavage activity of Tbis1, Sco5333 and Sco5333C252D to methylated plasmid DNA is suppressed by Zn2+. (A) 0–8 mM Zn2+ is added into the in vitro cleavage reaction system of Tbis1 in presence of 2 mM Mn2+ or Co2+. (B) 0–8 mM Zn2+ is added into the in vitro cleavage reaction system of Sco5333 in presence of 2 mM Mg2+ or Mn2+. Cleavage efficacy of Sco5333 decreases along with the increasing Zn2+ concentration. (C) 0–8 mM Zn2+ is added into the in vitro cleavage reaction system of Sco5333C252D in presence of 2 mM Mg2+ or Mn2+. Cleavage efficacy of Sco5333C252D decreases with the increasing Zn2+ concentration.
Figure 5.
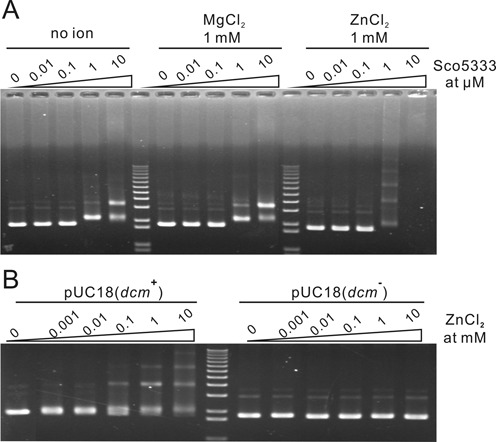
Zn2+ enhances DNA binding activity of Sco5333. (A) 0.01–10 μM of Sco5333 was added into the EMSA reaction system in presence of 1 mM Zn2+ respectively. EMSA system containing 1 mM Mg2+ or no ion was performed as control. Several shifted bands were observed when 1 μM Sco5333 or more was added into the EMSA reaction system, in which the [protein]:[DNA binding site] molar ratio ∼ 7:1. (B) 0.001–10 mM of Zn2+ was added into the EMSA reaction system in presence of 1 μM of Sco5333 respectively, in which the [protein]:[DNA binding site] molar ratio is ∼0.7:1. Several shifted bands were observed when 0.1 mM Zn2+ or more was added into the EMSA reaction system.
Run-off sequencing of the open circular and linear pUC18 revealed that the nicking and double strand breakage sites on pUC18 are randomly distributed, not sequence-specific and not associated with 5mC sites. Cleavage of 5mC-DNA and non-methylated DNA by Sco5333 in the presence of Mg2+ was compared. To our surprise, Mg2+ activated the DNA cleavage activity of Sco5333 to a similar extent on two type of DNA (Supplementary Figure S7). To determine if this weak non sequence- and non methylation-specific DNA cleavage is due to possible contamination of other DNA nuclease. Sco5333M, a Sco5333 mutant with all three catalytic residues (H228, N244 and H253) changed into alanine, was expressed and purified side-by-side with the wild-type. They were then assayed for their cleavage activity on pUC18 DNA with and without Dcm-methylation. Sco5333M completely lost the DNA cleavage activity even in high concentration (8 μM) while Sco5333 still displayed DNA cleavage activity (Supplementary Figure S8). Therefore, we ruled out possible contamination of nucleases and concluded that the weak and promiscuous DNA cleavage activity was indeed due to excessive amount of Sco5333.
Sco5333 and Tbis1 specifically bind to 5mC in all sequence contexts
When testing DNA cleavage activity of SRA-HNH proteins, strong mobility shift of pUC18 (dcm+) for Sco5333 was observed. In contrast there was no shift of non-methylated plasmid DNAs (Supplementary Figure S9). We then chose Sco5333 to test the binding specificity of 5mC-containing DNA. A pUC18-derived 219 bp DNA was PCR amplified and methylated by seven cytosine-5 DNA methylases with different specificities (Figure 6A). Binding of Sco5333 gave rise to discrete shifted-bands to all methylated DNA duplexes but not to the non-methylated PCR product. Shifting of methylated DNA by Sco5333 becomes stronger as the methylated sites increases (Figure 6A and C). Moreover, densely methylated substrates, such as CpG-methylated and GpC-methylated DNA fragments, resulted in multiple shifted bands (Figure 6C). These results demonstrated broad 5mC recognition specificity of Sco5333. No obvious DNA double strands breakage by Sco5333 to this linear fragment was observed (Supplementary Figure S10). In addition, hemi-methylated DNA at the Dcm site, either on top or bottom strand, is recognized by Sco5333 and shifted in the assay (Figures 6A and 7A). No shift was observed for the non-methylated 55nt duplexes (Figure 7A). Similarly, Tbis1 specifically binds to fully- and hemi-methylated DNA, but not to non-methylated DNA (Supplementary Figure S11).
Figure 6.
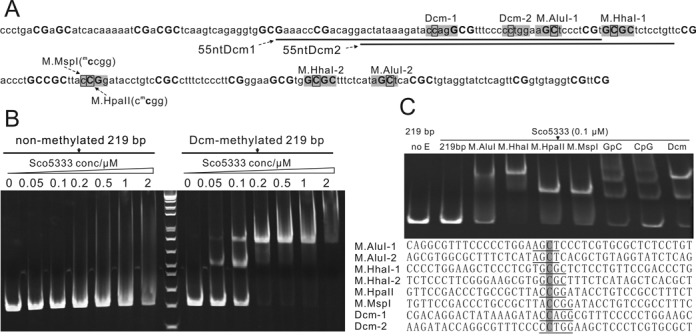
EMSA analysis of Sco5333 to methylated 219 bp derived from pUC18. (A) Sequence analysis of 219 bp which is PCR amplified from NT885–1103 of pUC18. Bases in shadow are modification sites of relative DNA MTases, and black rectangle designates the 5mC. C and G in bold represent the distribution of CpG and GpC modification sites. The underlined sequences are then synthesized and are named as 55ntDcm1 and 55ntDcm2. (B) Titration of Sco5333 on Dcm-modified and non-modified 219 bp fragment. Sco5333 shows high specificity to Dcm-modified 219 bp, and the two shifted bands in lane labelled 0.05 and 0.1 at right-half of the gel are due to the two Dcm modification sites which are centered at 219 bp. (C) EMSA of Sco5333 on 219 bp fragment modified by M.AluI, M.HhaI, M.HpaII, M.MspI, GpC, CpG or Dcm. Final concentration of Sco5333 is 0.1 μM. M.AluI, M.HhaI and Dcm have two modification sites on 219 bp, thus lanes 3, 4, and 9 show higher shift bands, whereas 219 bp modified by other MTases that have single modification site show lower shift bands. CpG and GpC have multiple modificaition sites (19 and 21 respectively), which result in multiple retarded bands. Statistics of Sco5333 binding sites on 219 bp shows poor sequence dependency.
Figure 7.
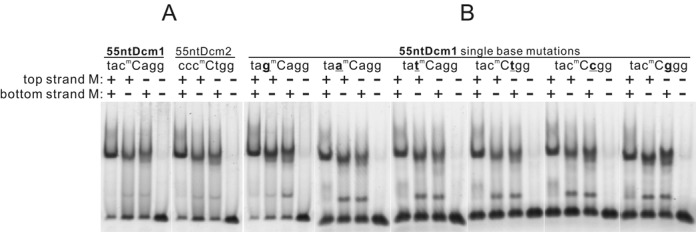
EMSA analysis of Sco5333 on 55ntDcm1, 55ntDcm2 and single-base substitutes of 55ntDcm1. (A) The 5-FAM labeled 55 nt oligos 55ntDcm1 (NT929–983) and 55ntDcm2 (NT942–996) of pUC18 are synthesized and annealed to generate fully-, top-, bottom-, and non-methylated 55nt oligos. Sco5333 shows high affinity to fully-, top-, bottom-, but not non-methylated 55nts. (B) The same strategy is performed to generate six groups of single-base substitutions of 55ntDcm1 in which the base flanking the 5mC is replaced by other six bases. Sco5333 shows the same activity as that for 55ntDcm1, 55ntDcm2 and 55ntDcm1 single-base mutants, indicating Sco5333 recognizes and binds to all the 5mC contexts for either fully- or hemi-methylation.
To further look into the sequence binding specificity of Sco5333, we systematically changed the base flanking the inner C of the Dcm site (CmCWGG) into other three bases, 24 hybrid duplexes were generated and tested (Figure 7B). In all sequence contexts, the fully- or hemi-methylated duplexes clearly gave shifted bands. These results support that Sco5333 binds specifically to 5-methylcytosine in all sequence contexts. Interestingly, binding of single stranded methylated DNA, but not non-methylated one, were also observed for either Sco5333 (Supplementary Figure S12A) or Tbis1 (Supplementary Figure S12B).
As some eukaryotic SRA domains can bind to both 5mC and 5hmC (30,31), Tbis1 was measured for its binding affinity to 5hmC substrates by EMSA. Results showed comparable affinity for 5mC and 5hmC for Tbis1 (Supplementary Figure S13).
In order to know the roles of SRA and HNH motif in the in vitro binding of 5mC sites, EMSA for wild-type Sco5333 and its mutants, G32A, Y50A in the SRA domain, and H228A, H253A at the HNH motif were performed and compared (Supplementary Figure S14). Results showed that amino acid changes in the SRA domain (G32A and Y50A) abolished the in vitro binding to 5mC, but amino acid changes at the HNH motif (H228A and H253A) did not, demonstrating that in vitro binding to 5mC is governed by the SRA domain alone (Supplementary Figure S14).
Binding properties of SRA-HNH proteins to methylated DNA
We then compared the binding properties of the bacterial SRA domain to the MBD (77–165) domain of mouse MeCP2 (32). EMSA of the 54nt DNA with different methylation patterns showed that MBD preferentially binds to fully-methylated DNA but not to hemi-methylated DNA (Supplementary Figure S14). Titration assay of Tbis1 and MBD of human MeCP2 (77–166, Cayman) demonstrated that Tbis1 had >100-fold preference on fully-methylated DNA over non-methylated DNA (Supplementary Figure S15) whereas MBD of human MeCP2 had 64-fold preference to fully-methylated DNA (Supplementary Figure S16), suggesting that the bacterial SRA domain is more specific in binding 5mC DNA.
To determine the affinity of Sco5333 binding to 5mC sites, the ITC approach was used to study the affinity parameters. The 54nt duplexes of fully-methylated, hemi-methylated on top or bottom strand are used as the DNA substrates. Sco5333 binds to fully-methylated 54nt duplex with an equilibrium dissociation constant (KD) of 4.1 μM (Figure 8A), slightly higher than 3.1 μM for the top strand hemi-methylated and also >1.5 μM for the bottom strand hemi-methylated DNA (Figure 8B&C). These dissociation constants are very close to that for SRASUVH5 (16).
Figure 8.
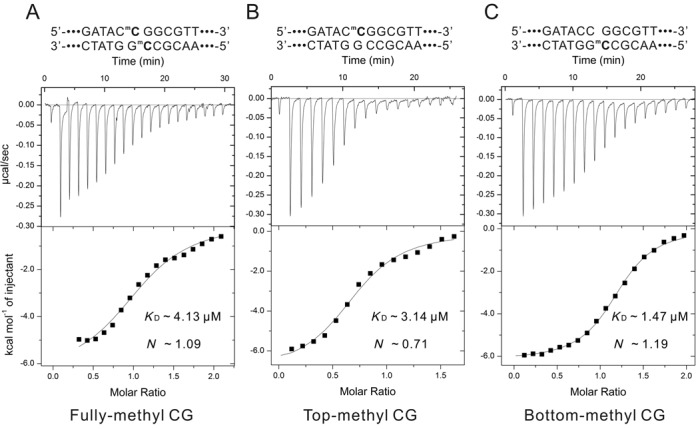
Equilibrium dissociation constant (KD) measurement via Isothermal titration calorimetry (ITC). The fully-, top- and bottom-methylated 54nt fragment with a central CmCGG pattern were performed as substrates for KD measurement via ITC. (A) KD for fully-methylated 54nt is 4.13 μM, and the binding molar ratio is ∼1.09. (B) KD for top-methylated 54nt is 3.14 μM, and the binding molar ratio is ∼0.71. (C) KD for bottom-methylated 54nt is 1.47 μM, and the binding molar ratio is ∼1.19.
According to tertiary structure prediction of Sco5333 and Tbis1 by the i-TASSER server (33,34), these two proteins adopt approximately globular shape. Gel-filtration analysis of Sco5333 and Tbis1 showed that Sco5333 forms as a homodimer in solution (Supplementary Figure S17), while Tbis1 is predominantly a monomer (Supplementary Figure S18). The ITC assay suggested that the ratio for Sco5333 to DNA was ∼1 (Figure 8A–C), which reflects that 1 DNA duplex, with a symmetrically methylated site, might be bound by two Sco5333, namely one dimer of Sco5333 per DNA duplex.
DISCUSSION
Combinatorial domain assortment of SRA and DNA cleavage domains in bacteria
SRA domains, originally identified in mammalian genes, play versatile roles in epigenetic regulation in eukaryotes. With its ability to bind to sites containing methylated cytosine, SRA domains may recruit associated proteins to interact with the DNA methylation machinery. In bacteria, eukaryotic SRA-like domains are often fused with nuclease domains containing HNH motif (17,35,36). However, the vast gene family of these SRA-HNH remain functionally elusive. Recently, a number of structural work revealed that the common SRA fold is widely adopted by many bacterial type IV DNA modification-dependent restriction endonucleases. Examples include the DNA binding domains of the MspJI family (18,37), which recognizes both 5-mC and 5hmC; and those of the PvuRts1I/AbaSI family (19,20), which recognizes 5hmC and glucosylated 5hmC. In all cases, there is no apparent sequence similarity among them, possibly suggesting convergent evolution in action.
Examination of the domain association in these bacterial SRA-containing genes reveals that bacterial SRA domains are often associated with different types of DNA cleavage domains. For examples, in the MspJI family, the N-terminal SRA DNA binding domain is associated with the C-terminal type IIP-like endonuclease domain with characteristic D..(E/Q)XK motif (37). In the PvuRts1I/AbaSI family, the C-terminal SRA DNA binding domain is associated with the N-terminal Vsr-like endonuclease domain (19,20). Here in this paper, the genes under study have N-terminal SRA domain and C-terminal HNH type endonuclease domain. In addition, in at least one case, an MspJI-like SRA domain is also fused with the HNH-type endonuclease domain (SghWI in REBASE). The assortment of the domain association suggests a combinatorial nature between the bacterial SRA domain and different types of cleavage domains. The domain association may also shed light on its possible biological roles. In all cases, it seems that the role of the SRA domains is to bring the DNA cleavage activity close to the modified DNA sites. Such activity may become useful when the bacterial cells are under attack from bacteriophages with modified DNA.
SRA domain of Sco5333 and Tbis1 is much more like SRASUVH5 than SRAUHRF1
Sco5333 and SRASUVH5 bind to 5mC either in fully- or hemi-methylated DNA, in sharp contrast to SRAUHRF1 which preferentially binds to hemi-methylated DNA over fully-methylated one. ITC studies revealed that one molecule of 5mC DNA duplex was bound by two SRA domains in the dimer form whereas the hemi-methylated CpG is bound by one SRAUHRF1. This stoichiometry is quite like SRASUVH5 and might be related to the length of the NKR finger. In SRAUHRF1, a steric clash may arise if two long NKR fingers intercalated from opposite directions into DNA groove containing 5-methylcytosine (13–15). SRASUVH5 and SRASco5333 have a much shortened and disordered loop that corresponds to long NKR finger of SRAUHRF1 (Supplementary Figure S2), this loop does not insert into the DNA groove, therefore leaving enough space for another domain to bind and flip out the base on the complementary strand. Moreover, the key residue on the thumb which takes place the flipped base and pairs with the orphaned guanine on the complementary strand is glutamine Q392, Q30 and Q28 in SRASUVH5, Sco5333 and Tbis1, respectively, however is valine in SRAUHRF1. Compared to the valine, glutamine skeleton has two additional carbons and an amide group that makes it much closer to the unpaired guanine (<3.1Å) and forms stable hydrogen bonds with it (16). As a result, the SRA domain of Sco5333 functions more likely as SRASUVH5 than as SRAUHRF1.
The contribution of dual metal ion binding motifs to its cleavage activity
Two ion binding motifs were identified in the HNH domain of Sco5333 and its orthologs, one is zinc finger binding motif that is composed of CX2CX36CX2C, the other one is the HNH motif. They overlap in the primary amino acid sequence. Sco5333 showed non-specific DNA cleavage in the presence of Mn2+ or Mg2+, but DNA cleavage activity in Mg2+ was suppressed by equal molar or higher concentration of Zn2+. We hypothesize that coordination of Zn2+ by Zinc finger is much more stronger than Mg2+ or Mn2+ (Figure 4B), similar to R.KpnI in which binding affinity of Zn2+ by ZF is much higher than to HNH motif (38). Binding of Zn2+ to the Zinc finger may induce a conformational change of the overall structure of Sco5333. Our result demonstrated that increasing Zn2+ can enhance the binding affinity of Sco5333 to 5mC DNA (Figure 5A and B), implying this conformational change may occur to the SRA domain of Sco5333. Binding of Zn2+ to ZF might exclude metal ions like Mn2+ and Mg2+ coordinated by the HNH motif, and therefore eliminate the cleavage activity that requires Mg2+ and Mn2+. Consistent with this, Sco5333C252D with the destructed ZF showed weak DNA cleavage activity. It is postulated that the defective Zinc finger may have a decreased binding affinity for Zn2+ as supported by comparative Zn2+ suppression experiments (Figure 4B and C), therefore allowed HNH motif to compete for coordination of Zn2+ and activated the DNA cleavage activity in the presence of Zn2+ alone.
As another note, the predicted structure of HNH motifs of Sco5333 and Tbis1both possess the ββα-Metal Fold, but compared to other three active ZF–HNH dual motif domains, such as Anacas9, Gme_HNH and T4eVII, their first β-sheet lack an asparagine residue (Figure 1C and Supplementary Figure S5) that is crucial to the coordination of metal ion other than Zn2+. This defect might in part explain in vitro weak cleavage activity of Tbis1 and Sco5333 that requires at least 40-fold excess of protein to DNA.
The difference between in vivo toxicity and in vitro cleavage activity by SRA-HNH proteins
Our results have shown that plasmids expressing Sco5333 and Tbis1 could not be established in E. coli hosts with Dcm-methylation whereas they can be maintained in the hosts without 5mC methylation. However, when induced by IPTG, the methylation deficient E. coli ER2566 expressing Tbis1 lysed (Figure 2B, third panel) while E. coli BL21 (DE3) expressing Sco5333 showed similar growth curves between the wild type and mutants (Supplementary Figure S6). This difference might imply that HNH motif of Tbis1 is more promiscuously active than that of Sco5333 toward non-methylated DNA. This speculation is supported by lower transformation efficiency of pTbis1 into ER2566 than pJTU4356 (encoding Sco5333) into BL21 (DE3) (Figure 2A and B). Consistent with this observation, a much lower concentration of Tbis1 can generate linear plasmid in the in vitro cleavage assay.
The in vivo toxicity of SRA-HNH protein to E. coli strains with the Dcm methylation requires both SRA domain and HNH motif (Figure 2A). But under in vitro conditions, purified enzymes cannot discriminate methylated or non-methylated DNA with respect to DNA cleavage. We demonstrated that the in vitro non-specific cleavage indeed stems from the enzyme rather than the contaminating nuclease. It also correlates with the observation that methylation-deficient cell expressing Tbis1 lysed upon induction by IPTG. Therefore, DNA cleavage activity by SRA-HNH, regardless of in vivo and in vitro, is very weak compared to typical restriction endonucleases. This speculation is not contradictory with the toxicity to host with 5mC modification. As the SRA domain can specifically bind to 5mC with high affinity, its cognate HNH domain may continuously exert its cleavage activity in the vicinity of target 5mC sites; Moreover, the tight binding of SRA protein to its target sequence might prevent the DNA repair systems to access the damaged sites. For the non-methylated host, the HNH domain can only transiently nick the chromosome DNA without the SRA-directed binding, so the damage of DNA might be repaired in a timely manner.
Possible roles for SRA-HNH protein in bacteria
Restriction endonucleases are often accompanied by DNA methylases (39,40). In the genome of S. coelicolor, sco5333 has an adjacent Type IIG restriction enzyme/N6-adenine DNA methyltransferase gene sco5331 located downstream. The neighbouring configuration of Sco5331 and Sco5333 were not conserved in other bacterial genomes, indicating that their association may be coincidental. For Tbis1, one of the immediate adjacent genes is a rRNA adenine methyltransferase.
Sco5333 was isolated from S. coelicolor, a model strain with stringent restriction of alien DNA bearing 5mC and 6mA methylation (41). We previously identified a type IV HNH endonuclease ScoMcrA which can restrict phosphorothioated DNA and Dcm-methylated DNA (42). But the scoMcrA knockout mutant still displayed strong restriction activity to 5mC DNA. Here we show that Sco5333 may contribute to the observed restriction.
Interestingly, sco5333 was located in a typical genomic island that is flanked by two almost identical copies of Arg-tRNA genes. Genomic islands are often associated with horizontal gene transfer (43). This finding is well fit to the notion that the restriction and methylation systems are located in the mobile element to respond to environmental threats such as phage attacks (44). Combination a typical SRA domain and an endonuclease characteristic HNH domain may represent a high efficient mechanism to counteract the threat of alien DNA bearing 5mC. The advantage by employing SRA-HNH is easy discrimination of 5mC DNA from its own DNA. More importantly, the extremely low DNA cleavage activity of the cognate zinc finger-HNH can to the maximum extent reduce the damage to the unmodified part of its chromosomal DNA. SRA-HNH might be an universal mechanism in restricting methylated DNA as most type IV restriction enzymes identified has the SRA structure and HNH motif.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to Zhang Yan and Dr Siu-Hong Chan (NEB) for their assistance in ITC measurement and data analyse. We thank Drs Bill Jack, Rich Roberts and Xiaodong Cheng (Emory University) for comments on an earlier version of the manuscript.
FUNDING
National Natural Science Foundation of China [31170083, 31130068, 31121064]; the Ministry of Science and Technology of China [2012CB721004]; the Ministry of Education of China [20110073130011; the Chen Xing Young Scholars Program of Shanghai Jiao Tong University awarded (to X.H.); New England Biolabs Inc. (to M.Y.-M., Y. Z.). Funding for open acess charge: National Natural Science Foundation of China [31170083, 31130068, 31121064]; the Ministry of Science and Technology of China [2012CB721004]; the Ministry of Education of China [20110073130011; the Chen Xing Young Scholars Program of Shanghai Jiao Tong University awarded (to X.H.); New England Biolabs Inc. (to M.Y.-M., Y. Z.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol. Biotechnol. 2010;44:71–81. doi: 10.1007/s12033-009-9216-2. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M., Gama-Sosa M.A., Huang L.H., Midgett R.M., Kuo K.C., McCune R.A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.-M. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson I.R., Jacobsen S.E. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 6.Probst A.V., Dunleavy E., Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 7.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M., Corn P.G., Baylin S.B., Herman J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 9.Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 10.Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 11.Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 12.Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet.: TIG. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 13.Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 14.Avvakumov G.V., Walker J.R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C.H., Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto H., Horton J.R., Zhang X., Bostick M., Jacobsen S.E., Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajakumara E., Law J.A., Simanshu D.K., Voigt P., Johnson L.M., Reinberg D., Patel D.J., Jacobsen S.E. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer L.M., Abhiman S., Aravind L. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 18.Horton J.R., Nugent R.L., Li A., Mabuchi M.Y., Fomenkov A., Cohen-Karni D., Griggs R.M., Zhang X., Wilson G.G., Zheng Y., et al. Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI. Sci. Rep. 2014;4:4246. doi: 10.1038/srep04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton J.R., Borgaro J.G., Griggs R.M., Quimby A., Guan S., Zhang X., Wilson G.G., Zheng Y., Zhu Z., Cheng X. Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA. Nucleic Acids Res. 2014;42:7947–7959. doi: 10.1093/nar/gku497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazrani A.A., Kowalska M., Czapinska H., Bochtler M. Crystal structure of the 5hmC specific endonuclease PvuRts1I. Nucleic Acids Res. 2014;42:5929–5936. doi: 10.1093/nar/gku186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T., Fritsch E.F., Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 22.Evans T.C., Jr, Benner J., Xu M.Q. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong S., Mersha F.B., Comb D.G., Scott M.E., Landry D., Vence L.M., Perler F.B., Benner J., Kucera R.B., Hirvonen C.A., et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H., Wang Y., Yu Y., Bai T., Chen L., Liu P., Guo H., Zhu C., Tao M., Deng Z. A non-restricting and non-methylating Escherichia coli strain for DNA cloning and high-throughput conjugation to Streptomyces coelicolor. Curr. Microbiol. 2012;64:185–190. doi: 10.1007/s00284-011-0048-5. [DOI] [PubMed] [Google Scholar]
- 25.Sutcliffe J.G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci.U.S.A. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Jinek M., Jiang F., Taylor D.W., Sternberg S.H., Kaya E., Ma E., Anders C., Hauer M., Zhou K., Lin S., et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavletich N.P., Pabo C.O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 29.Saravanan M., Bujnicki J.M., Cymerman I.A., Rao D.N., Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frauer C., Hoffmann T., Bultmann S., Casa V., Cardoso M.C., Antes I., Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou T., Xiong J., Wang M., Yang N., Wong J., Zhu B., Xu R.M. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol. Cell. 2014;54:879–886. doi: 10.1016/j.molcel.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Valinluck V., Tsai H.H., Rogstad D.K., Burdzy A., Bird A., Sowers L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumbusch L.O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R.B. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Citterio E., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Fiore P.P., Bonapace I.M. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horton J.R., Mabuchi M.Y., Cohen-Karni D., Zhang X., Griggs R.M., Samaranayake M., Roberts R.J., Zheng Y., Cheng X. Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease. Nucleic Acids Res. 2012;40:9763–9773. doi: 10.1093/nar/gks719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saravanan M., Vasu K., Ghosh S., Nagaraja V. Dual role for Zn2+ in maintaining structural integrity and inducing DNA sequence specificity in a promiscuous endonuclease. J. Biol. Chem. 2007;282:32320–32326. doi: 10.1074/jbc.M705927200. [DOI] [PubMed] [Google Scholar]
- 39.Jeltsch A., Kröger M., Pingoud A. Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene. 1995;160:7–16. doi: 10.1016/0378-1119(95)00181-5. [DOI] [PubMed] [Google Scholar]
- 40.Tock M.R., Dryden D.T. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez-Ceron G., Miranda-Olivares O.J., Servin-Gonzalez L. Characterization of the methyl-specific restriction system of Streptomyces coelicolor A3(2) and of the role played by laterally acquired nucleases. FEMS microbiology letters. 2009;301:35–43. doi: 10.1111/j.1574-6968.2009.01790.x. [DOI] [PubMed] [Google Scholar]
- 42.Liu G., Ou H.Y., Wang T., Li L., Tan H., Zhou X., Rajakumar K., Deng Z., He X. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juhas M., van der Meer J.R., Gaillard M., Harding R.M., Hood D.W., Crook D.W. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furuta Y., Abe K., Kobayashi I. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 2010;38:2428–2443. doi: 10.1093/nar/gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


