Abstract
p63 is a crucial regulator of epidermal development, but its transcriptional control has remained elusive. Here, we report the identification of a long-range enhancer (p63LRE) that is composed of two evolutionary conserved modules (C38 and C40), acting in concert to control tissue- and layer-specific expression of the p63 gene. Both modules are in an open and active chromatin state in human and mouse keratinocytes and in embryonic epidermis, and are strongly bound by p63. p63LRE activity is dependent on p63 expression in embryonic skin, and also in the commitment of human induced pluripotent stem cells toward an epithelial cell fate. A search for other transcription factors involved in p63LRE regulation revealed that the CAAT enhancer binding proteins Cebpa and Cebpb and the POU domain-containing protein Pou3f1 repress p63 expression during keratinocyte differentiation by binding the p63LRE enhancer. Collectively, our data indicate that p63LRE is composed of additive and partly redundant enhancer modules that act to direct robust p63 expression selectively in the basal layer of the epidermis.
INTRODUCTION
The transcription factor (TF) p63, a p53 family member, is a master regulator of epidermal morphogenesis as demonstrated by the absence of surface epithelium in neonatal mice lacking p63 (1,2). p63, and more specifically the more abundant ΔNp63α isoform, has been associated with several crucial functions in epidermal cells including cell proliferation and stem cell renewal (2–8), commitment to an epidermal cell fate and to cell differentiation (1,7,9–14) and cell adhesion (15–18).
During mouse embryonic development, p63 mRNA is one of the earliest epidermal-specific markers to be detected in the surface epithelium at embryonic day 8.5 (E8.5) (1–2,10,19). At E10.5, ectodermal cells begin to express keratin K5 and K14 mRNA in a highly region-specific pattern similar to that of p63 (20). Suprabasal keratins K1 and K10 are expressed just before stratification occurs at E14.5 (20), a process that is completed at E17.5 when the epidermis acquires its barrier function. During stratification, p63 expression becomes progressively confined to the basal undifferentiated layer of the epidermis, and is weakly expressed or absent in the suprabasal differentiated layers (21,22). A similar regulation of p63 expression is observed in human and mouse keratinocytes in culture, in which p63 is highly expressed under proliferative conditions and is downregulated in differentiated cells (12,22–24).
The mouse p63 gene (Trp63) comprises a large genomic portion (about 208 kb) on chromosome 16 (band 16qB1), and contains 16 exons and two independent promoters (P1 and P2) that drive the expression of two classes of transcripts: TAp63 containing an N-terminal transactivation domain and ΔNp63 lacking the TA domain and acting as either transcriptional activator or repressor (25–28). Alternative splicing at the 3′ gives rise to three different carboxyl-terminal isoforms designated α, β and γ. The ΔNp63α protein containing the longest C-terminal domain is the most abundant isoform in mature proliferating stratified epithelia such as those of the skin (21,22).
In spite of the essential role of p63 in development of the epidermis and other stratified epithelia, little is known about the regulation of p63 expression during epidermal development and in keratinocyte differentiation. Studies in zebrafish and xenopus indicate that, during epidermal development, p63 expression is induced by bone morphogenetic protein (BMP) (29,30), consistent with the ability of BMP signaling to act as an epidermal inducer and a suppressor of neural cell fate during development (31,32). Similarly, in human and mouse embryonic stem cells, and induced pluripotent stem cells (iPSCs), active BMP signaling is required for epidermal or corneal epithelial lineage commitment and for p63 expression (33–35). In contrast to BMP signaling, Notch is active prior to the onset of p63 expression in ectodermal progenitor cells and negatively regulates p63 expression in both developing mouse embryos and in human embryonic stem cells (35). At later developmental stages, Notch signaling is a crucial positive regulator of keratinocyte terminal differentiation (36–39), and negatively regulates p63 mRNA levels at least in part by inhibiting interferon responsive factors IRF3 and IRF7 (12). In addition, IRF6, another IRF family member with a specific role in epidermal development and differentiation (40,41), has been shown to downregulate p63 protein levels in a proteasome-dependent manner (42).
In spite of these observations, the mechanisms regulating p63 expression are still not fully understood. We previously reported that neither the TAp63 (2 kb) nor the ΔNp63 (4 kb) promoters are sufficient to confer keratinocyte-specific expression to a luciferase reporter gene (43), although a number of TFs including Sp1/Sp3 and NF-Y bind and regulate the ΔNp63 promoter (44). Similarly, a 10-Kb human genomic fragment upstream of the ΔNp63 transcription start site (TSS) is insufficient to mimic endogenous ΔNp63 expression in mammary epithelial cells (45). Thus, other regulatory elements are likely required to regulate tissue-specific expression of the p63 transcripts.
Developmental control genes, such as p63, often comprise a large genomic portion with numerous cis-regulatory elements, and tend to have longer introns consistent with the fact that introns frequently harbor enhancer regions (reviewed in (46)). We previously identified an evolutionary conserved regulatory element (C40) located in p63 intron 5, the largest intron downstream of ΔNp63 promoter (P2), that drives weak expression in the epidermis during embryogenesis (43). p63 binds C40 thereby sustaining its own expression in cooperation with AP-2, a family of crucial regulators of epidermal genes in embryonic development (47–49). However, the C40 element drives expression in both basal and suprabasal compartments in vivo, and is unaffected by calcium-induced differentiation in cultured keratinocytes, suggesting that other regulatory elements are required for basal cell-specific expression.
Here, we report the identification of a novel evolutionarily conserved cis-regulatory element (C38) that acts synergistically with the previously characterized C40 enhancer to activate keratinocyte-specific transcription. A 12-kb mouse genomic region encompassing the two regulatory elements is sufficient to drive robust basal specific expression in embryonic and newborn skin. Layer-specific regulation is likely caused by direct binding of Cebpa and Cebpb and Pou3f1, known regulators of epidermal differentiation, to both C38 and C40. Accordingly, depletion of these transcriptional regulators relieves differentiation-dependent p63 downregulation.
MATERIALS AND METHODS
Cell cultures, plasmids, transfection and reporter assays
Mouse primary keratinocytes were isolated from C57BL/6 newborn mice and cultured under low calcium conditions (0.05 mM) in the presence of 4% calcium-chelated Fetal Bovine Serum (FBS) (Invitrogen, Carlsbad, CA, USA) and Epidermal Growth Factor (EGF, Invitrogen) as previously described (43). Terminal differentiation was induced by addition of calcium chloride to the medium (0.6 mM or 2 mM for 12, or 24 h as specified). Human primary keratinocytes were kindly provided by Dr. G.P. Dotto, and cultured in Keratinocyte-Serum Free Medium supplemented with bovine pituitary extracts (30 μg/ml) and EGF (0.2 ng/ml) (Invitrogen). Mouse dermal fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS. Mouse C38 and C40 genomic regions were cloned in the TK-pGL3 luciferase reporter plasmid (43) (see Supplementary Data). p63LRE-TK-luc, ΔC38, ΔC40 and ΔC38ΔC40 were obtained by recombineering (see Supplementary Data). Cells were transfected using Lipofectamine 2000 (Invitrogen) with plasmids pCMV-Klf4 and pCMV-Klf5 (provided by Dr. Tommaso Russo; (50)), pCMV-FoxN1 and pCMV-NIC (activated Notch1) (provided by Dr. GP Dotto; (36)), pBabe-Cebpa (provided by Dr. Gökhan S. Hotamisligil; (51)), pBabe-Cebpb (provided by Dr. Roberto Mantovani), pXCX-Pou2f3 and pXCX-Pou3f1 (provided by Dr. Bogi Andersen; (52)). Luciferase activity was determined 48 h after transfection with the dual-luciferase reporter assay kit (Promega). Renilla activity was used to normalize transfection efficiency. p63, Irf6, Cebpa, Cebpb, Pou2f3, Pou3f1 knockdown were obtained by transient transfection of 100 nM small interfering RNA (siRNA) (see Supplementary Data for specific sequences). Human iPSCs were obtained by lentiviral infection of primary dermal fibroblasts derived from a healthy individual or an individual carrying the R304W mutation in the p63 DNA binding domain causative of EEC syndrome (ectrodactyly, ectodermal dysplasia and cleft lip/palate syndrome). Human iPSCs were differentiated in vitro in the presence of corneal fibroblasts conditioned medium and BMP-4 as previously described (53). C38C40-luc was transfected using Fugene HD (Promega) and luciferase was detected at days 4 and 8 by Luciferase assay kit (Promega).
Generation of transgenic mice and ß-galactosidase staining
A 12-kb genomic region spanning C38 and C40 enhancers (p63LRE) was cloned in the modified β-globin–lacZ vector p1229 (43) by recombineering (see Supplementary Data). The p63LRE-LacZ construct was linearized, purified and injected into fertilized oocytes of DBA X C57BL/6 mice at the CBRC Transgenic Facility (Massachusetts General Hospital, Boston, MA) and at the Transgenic Facility (Department of Molecular Biotechnology and Health Sciences, University of Turin). Nineteen pups were generated, six of which were positive by polymerase chain reaction (PCR) and three also expressed the LacZ transgene in a similar tissue-specific pattern. β-Galactosidase staining was previously described (43). Founders were backcrossed with C57BL/6 mice to establish mouse lines. p63−/−; p63LRE-LacZ mice were obtained crossing p63+/− mice with p63LRE-LacZ transgenic mice. All mouse experiments were approved by the Italian Ministry of Health.
Bioinformatics analysis of TF binding site and mutagenesis
Analysis of TF binding sites was performed using MatInspector Matrix Family Library Version 9.0 (August 2012) (GenomatixSuite v3.0) (54). Alignment of conserved non-coding element C38 in different species was performed with the ClustalW program (http://www.ebi.ac.uk/clustalw/). Mutagenesis reactions were performed using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) with the indicated oligonucleotides (see Supplementary Data).
Real-time RT-PCR, ChIP and immunoblotting analyses
RNA was extracted in TRIzol reagent (Invitrogen) and treated with DNase I (Roche). Complementary DNA (cDNA) synthesis was obtained using SuperScript Vilo (Invitrogen). Expression of target genes was normalized for the β-actin gene expression. ChIP analysis was performed as previously described (43) using rabbit polyclonal antibody anti-p63 (H137, Santa Cruz Biotechnology), anti-histone H3K4me3 (Upstate 07–473), anti-H3K27Ac (Diagenode), anti-Cebpa (14AA, Santa Cruz Biotechnology), anti-Cebpb (C-19, Santa Cruz Biotechnology), anti- Pou3f1 (kindly provided by Dr. Michael Wegner, Universität Erlangen-Nürnberg, Germany) (55) or rabbit anti-mouse IgG (Santa Cruz Biotechnology). Real-time RT-PCR was performed using the SYBR Green PCR master mix (Applied Biosystems) in an ABI PRISM 7500 (Applied Biosystems). Oligonucleotide sequences are given in the Supplementary Data. For immunoblotting, cells were lysed in sample buffer and immunoblotted as previously described (43). The following primary antibodies were used: anti-p63 (4A4, Santa Cruz Biotechnology), anti-β-actin (Santa Cruz Biotechnology). For immunoblotting, secondary antibodies were sheep anti-mouse IgG conjugated to horseradish peroxidase (GE Healthcare) and visualized using Enhanced chemiluminescence (ECL) western blotting system (GE Healthcare).
Statistics
All quantitative results are presented as mean ± SD of independently repeated experiments as indicated in the figure legends. Statistical significance was assessed by Student's t test using Microsoft Excel. P-value (P) is indicated in each single experiment.
RESULTS
Identification of a novel enhancer region in the p63 gene
To identify regulatory elements that may contribute to tissue-specific expression of p63, we previously performed a comparative sequence analysis of the p63 genomic region in vertebrates and characterized the highly conserved intronic C40 enhancer (43). In this analysis, another mouse genomic element (C38), located 7.6 kb upstream of the C40 and 33 kb downstream of the ΔNp63 TSS, was identified with a similarly high degree of conservation in even the most distant vertebrates (43,56) (Figure 1A). Given the high degree of evolutionary conservation restricted to the exons and the C38 and C40 element only, we hypothesized that C38 could be an important regulatory element for p63 expression in spite of the low enhancer activity when tested in transactivation assays (43).
Figure 1.
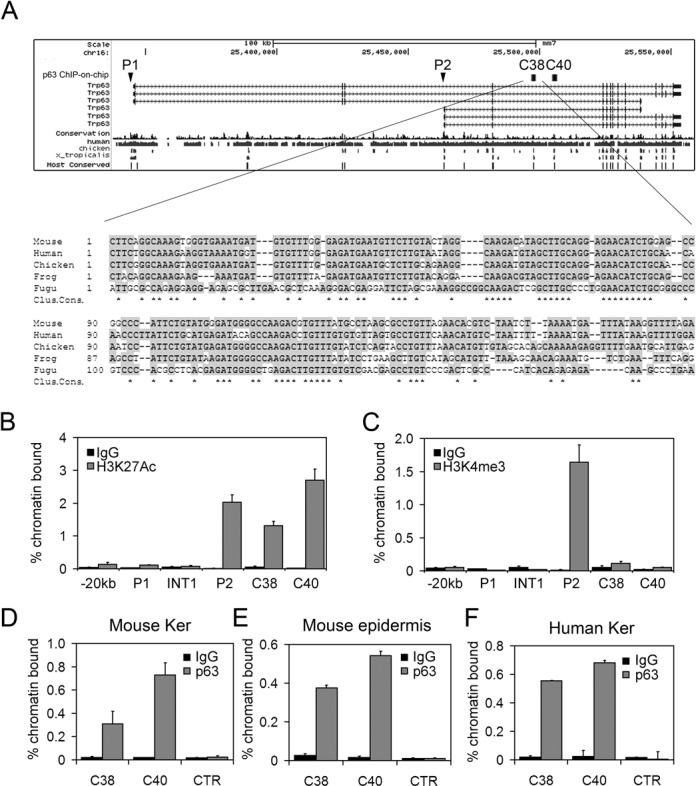
(A) Upper panel: mouse p63 gene structure. The P1 (TA) and P2 (ΔN) promoters are indicated. C38 and C40 correspond to highly conserved sequences in evolution as determined by the 17-way alignment predicted by the phastCons program (56). Genomic elements identified by ChIP-on-chip experiment using p63 antibodies are indicated with black squares (25). Lower panel: evolutionary conservation of the C38 element in multiple vertebrate species. Conserved nucleotides are shown in gray, and identical nucleotides in all species are indicated by stars (Clustal consensus). (B,C) ChIP-qPCR was performed on mouse primary keratinocyte using antibodies anti-H3K27Ac (B), anti-H3K4me3 (C) (gray bars) or rabbit IgG (black bars) as negative control. (D,E,F) ChIP-qPCR was performed on primary mouse keratinocytes (D), mouse newborn epidemis (E) and human keratinocytes (F) using p63 polyclonal antibodies (H-137) (gray bars), or rabbit IgG (black bars) as negative control. Oligonucleotide sequences are listed in Supplementary Data. Error bars denote SD.
Histone modifications have proven effective in determining the location of cis-regulatory elements (reviewed in (57)). To test whether C38 was a putative enhancer, ChIP-qPCR was performed in mouse keratinocytes and embryonic skin using antibodies specific for acetylated H3K27 (H3K27ac), known to identify active regulatory regions including promoters and enhancers (58). As expected a strong signal was detected in keratinocyte for the ΔNp63 promoter (P2), consistent with high expression of ΔNp63 in these cells, whereas no signal was observed in the TAp63 promoter (P1), in a control region located 20kb upstream P1 or in p63 intron 1 (Figure 1B). Interestingly, a strong signal was detected in C38 and C40 both in mouse keratinocytes and in embryonic skin at E14.5 (Figure 1B and Supplementary Figure S1A), demonstrating that these elements are active regulatory regions. Conversely, H3K4me3, that marks promoter regions (59), bound specifically to the active ΔNp63 promoter (P2), but did not bind to either C38 or C40 (Figure 1C). In addition, using ENCODE data (60) a strong H3K27ac signal was detected on the C38 and C40 elements in human keratinocytes and in human mammary epithelial cells, but not in other cell types that do not express p63, consistent with the presence of active and specific enhancers in those genomic regions (Supplementary Figure S2). DNAse hypersensitive sites were detected in both C38 and C40 elements in human cells (Supplementary Figure S2), indicating that they are in an open chromatin state.
In addition ChIP-on-chip experiments in mouse keratinocytes revealed that in a genomic region spanning the ΔNp63 transcript plus 20 kb upstream of TSS, the p63 protein bound strongly and uniquely to the C38 and C40 elements (25) (Figure 1A). Supporting and extending these data, two independent genome-wide ChIP-seq analyses performed in human primary keratinocytes (61,62) revealed that in a 1-Mb region comprising the p63 locus, p63 protein bound to the C38 and C40 genomic elements with the highest signal scores (Supplementary Figure S2). ChIP-qPCR analysis confirmed that p63 efficiently bound both C38 and C40 in mouse primary keratinocytes, in postnatal epidermis and in embryonic skin (Figure 1D and E; Supplementary Figure S1B). Similar results were obtained in human keratinocytes (Figure 1F), suggesting that the function of these genomic elements is conserved between human and mice.
C38 and C40 elements cooperate to sustain transcription in basal keratinocytes
Since both C38 and C40 are highly conserved in evolution, display an active chromatin state and are bound by p63, we tested the hypothesis that these elements may act synergistically to regulate p63 expression. For this purpose, we generated a reporter construct containing both genomic elements upstream of the thymidine kinase minimal promoter driving the luciferase reporter gene (C38C40-Luc), and we tested its enhancer activity. The C38C40 transactivation activity was stronger than the C40 enhancer alone in mouse keratinocytes consistent with a synergistic contribution of the two elements (Figure 2A), whereas C38 had a very low enhancer activity by itself as previously reported (43).
Figure 2.
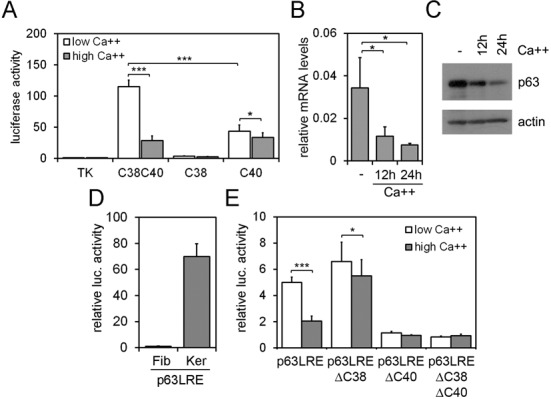
(A) C38-Luc, C40-Luc and C38C40-Luc enhancer activity was analyzed in primary mouse keratinocytes under proliferation (low calcium; white bars) and differentiation conditions (high calcium; gray bars) upon 24 h of calcium treatment (Ca++ 2 mM). Data are expressed relative to TK promoter activity used as control (*P ≤ 0.05; ***P ≤ 0.0005; n = 4). (B,C) p63 expression was detected at the mRNA (B) and protein (C) levels in primary mouse keratinocytes in proliferating and in calcium-induced differentiated keratinocytes at the indicated times (Ca++ 2 mM). (D) p63LRE luciferase activity was tested in mouse dermal fibroblasts (Fib) and primary keratinocytes (Ker). (E) TK-Luc construct carrying p63LRE, p63LREΔC38, p63LREΔC40 and p63LREΔC38ΔC40 were tested for luciferase activity in primary mouse keratinocytes under proliferation (white bars) and differentiation (gray bars) conditions (***P ≤ 0.0005; n = 6). Error bars denote SD.
In primary keratinocytes under basal conditions, p63 was expressed at high levels whereas its expression was reduced both at the RNA and protein levels in calcium-induced differentiation (Figure 2B and C) as previously reported (23,24). Regulatory elements conferring specificity to p63 expression in undifferentiating versus differentiating conditions have not been identified. C40 activity was poorly affected under differentiating conditions, whereas the C38C40 enhancer activity was strongly inhibited in mouse keratinocytes induced to differentiate by calcium treatment compared to the basal condition (Figure 2A).
To test the possibility that the two elements may have a similar synergistic response in undifferentiated conditions and may be responsive to calcium-induced differentiation even at their physiological distance, a 12 kb mouse genomic region comprising both elements was cloned in a luciferase reporter vector (p63LRE-Luc). Transactivation assays revealed a strong and specific enhancer activity in keratinocytes but not in fibroblasts (Figure 2D), indicating that p63LRE acts in a cell type-specific manner. In undifferentiating keratinocytes, deletion of the C40 element (p63LREΔC40) or concomitant deletion of both C38 and C40 elements (p63LREΔC38ΔC40) abolished p63LRE enhancer activity, whereas the absence of C38 element (p63LREΔC38) had little or no effect (Figure 2D). Similar to C38C40, p63LRE enhancer activity was inhibited in mouse keratinocytes induced to differentiate by calcium treatment compared to undifferentiating conditions, whereas p63LREΔC38 maintained a strong enhancer activity under both basal and differentiating conditions, demonstrating that the C38 element is required to confer responsiveness to differentiation (Figure 2E). The contribution of C40 in differentiating conditions could not be evaluated since p63LREΔC40 lost transactivation activity under both basal and differentiating conditions (Figure 2E).
LRE enhancer activity recapitulates p63 expression in a mouse model
To test the significance of these findings in vivo, we generated transgenic mice carrying the p63LRE upstream of the β-galactosidase gene (p63LRE-LacZ). The p63LRE enhancer activity drove strong and specific expression in the developing and newborn epidermis. At E10.5 LacZ expression was observed in the branchial arches and in the apical ectodermal ridge, with a similar but much stronger pattern of expression compared to the previously characterized C40×4-LacZ transgenic mice (Figure 3A). In addition, at this stage p63LRE drove expression in cells of the facial processes, the dorsal epidermal sheet and the tail bud, that never scored positive in the C40×4-LacZ. At E13.5 ß-galactosidase staining was present at the tip of the developing limbs, in the mammary buds, in the snout epidermis and to a lesser extent in several other areas of the skin (Supplementary Figure S3A). At E15.5 LacZ expression was detected in the entire embryonic skin of p63LRE-LacZ mice, whereas under the same experimental conditions, it was much weaker in the C40×4-LacZ mice (Figure 3B). In p63LRE-LacZ newborn mice, a strong and basal-specific LacZ expression was observed in the epidermis and in the hair follicles, whereas a weaker and diffuse expression was detected in C40×4-LacZ skin (Figure 3C). Co-localization of the ß-galactosidase staining and p63 expression was confirmed by immunohistochemistry with anti-p63 antibodies in p63LRE-LacZ skin (Supplementary Figure S3B), demonstrating that the p63LRE enhancer drives LacZ expression with a pattern identical to the endogenous p63.
Figure 3.
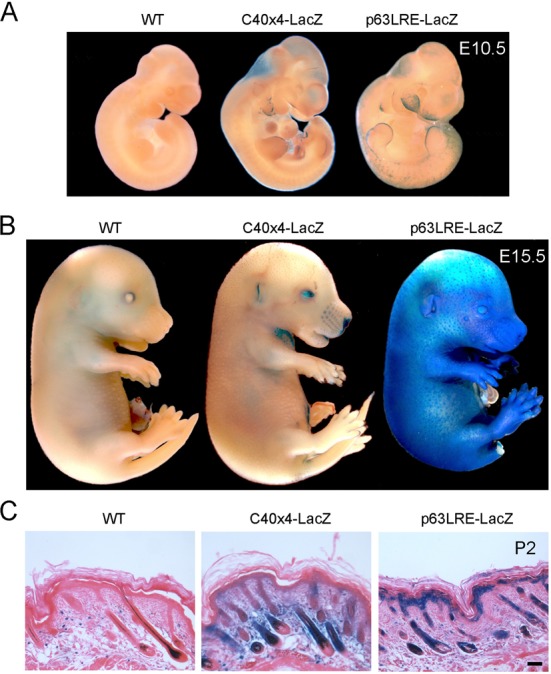
(A) E10.5 wild type (WT) and transgenic embryos containing four copies of C40 (C40×4-LacZ) or p63LRE-LacZ were tested for β-galactosidase activity. (B) Embryos at E15.5 were tested for β-galactosidase activity as in (A). (C) Histological sections of skin at P2 were processed for β-galactosidase staining and counterstained with eosin. Scale bar, 50 μm.
p63 is required for LRE enhancer activity
Since p63 protein binds both the C38 and C40 elements, we tested the possibility that p63 may be essential for p63LRE activity. In cultured p63-deficient keratinocytes, both C38C40 and p63LRE activities were markedly inhibited compared to p63 expressing cells (Supplementary Figure S4). To test whether the p63LRE enhancer was dependent on p63 expression also in vivo, we obtained mice carrying p63LRE enhancer driving LacZ expression in a p63−/− background (p63−/−; p63LRE-LacZ). Accordingly, LacZ expression was not detected in p63−/−; p63LRE-LacZ epidermis at E15.5 (Figure 4A and B), indicating that p63 is essential for p63LRE enhancer activity both in vitro and in vivo.
Figure 4.
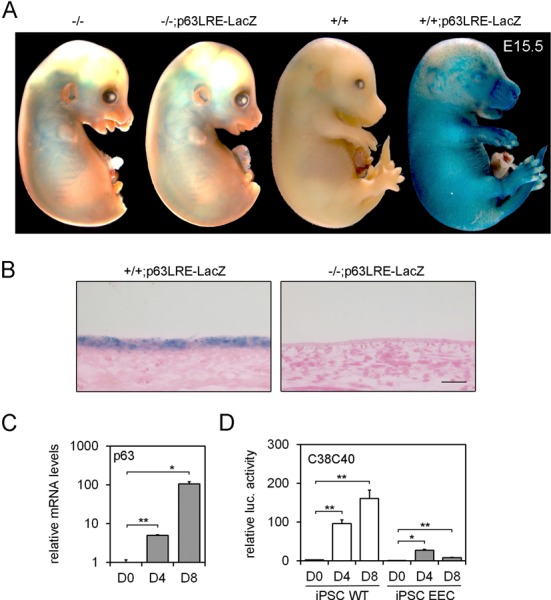
(A) E15.5 embryos carrying p63LRE in a wild type (+/+; p63LRE-LacZ) and in a p63-null background (-/-; p63LRE-LacZ) were tested for β-galactosidase activity. Wild-type (+/+) and p63-null (-/-) littermates were used as controls. (B) Histological sections of skin at E15.5 were obtained after β-galactosidase staining and were subsequently paraffin embedded and counterstained with eosin. Scale bar, 50 μm. (C) p63 expression was detected at the mRNA levels in human iPSCs at 0, 4 and 8 days (D0–D8) of corneal epithelial differentiation. Data are represented relative to p63 expression levels at day 0 (*P ≤ 0.05; **P ≤ 0.005; n = 3). (D) Luciferase activity of C38C40-Luc was tested in human iPSCs (WT and EEC) at D0–D8 of corneal epithelial differentiation. Data are represented relative to luciferase activity of TK control in D0 human iPSCs (*P ≤ 0.05; **P ≤ 0.005; n = 3).
To assess the requirement of p63 for the enhancer activity in epithelial commitment in vitro, we measured the C38C40 activity during epithelial differentiation of human iPSCs (Figure 4D). p63 is required for induction of an epidermal or corneal epithelial lineage commitment, two fates that are defective in cells derived from EEC patients (53). Using this system, we found that the enhancer activity was progressively activated in iPSC differentiation in parallel with p63 expression (Figure 4C and D). Importantly, p63 enhancer activity was impaired in iPSCs derived from an individual affected by EEC syndrome and carrying a deleterious missense mutation in p63, indicating that the p63 enhancer is dependent on p63 expression and is a useful tool to follow its activity.
A subset of TFs involved in keratinocyte differentiation regulates p63LRE activity
To identify the TFs regulating p63LRE activity, C38 and C40 sequences were analyzed for putative TF binding sites. Consistent with data shown above, bioinformatics analysis revealed putative evolutionary conserved p63 consensus sequences not only in C40 as previously reported (43) but also in C38 (Supplementary Figure S5 and Supplementary Table S1). Concomitant mutations of canonical p53/p63 binding site conserved in human and chicken identified by bioinformatics analysis in the C38 and C40 elements almost completely abolished C38C40 enhancer activity (Figure 5A). Since p63 expression is reduced during differentiation, we tested the hypothesis that the suppression of C38C40 enhancer activity in differentiating keratinocytes may be a consequence of reduced p63 protein levels. Surprisingly, C38C40 carrying deletions in the p63 binding sites was equally responsive to calcium induced differentiation even if its overall activity was reduced, indicating that reduced enhancer activity in differentiated cells was not merely a consequence of a progressive reduction in the p63 protein level (Figure 5B).
Figure 5.
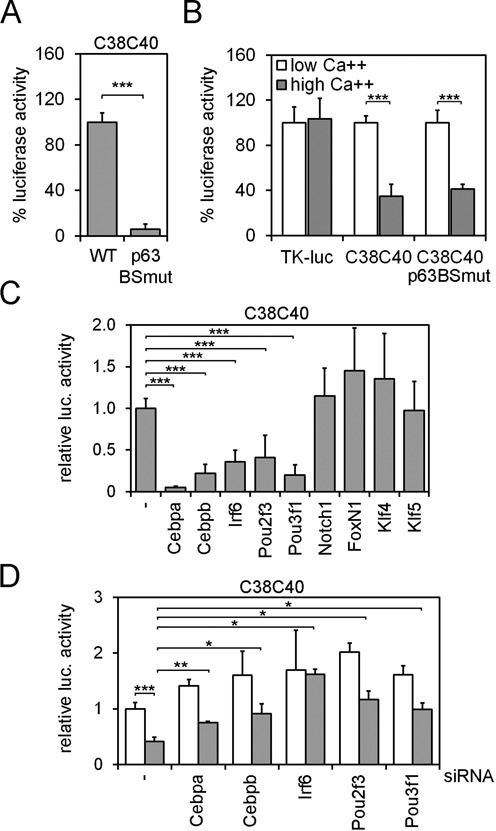
(A) Luciferase activity of the C38C40-Luc carrying either WT or mutant p63-binding sites (p63BSmut) was tested in mouse keratinocytes. Data are expressed as the% of wild type luciferase activity (***P ≤ 0.0005; n = 3). (B) The activity of wild type C38C40, C38C40p63BSmut and TK control was tested in proliferating (low Ca++; white bars) and differentiated keratinocytes (2 mM Ca++; gray bars), and plotted as% of each luciferase activity in low Ca++ conditions. (***P ≤ 0.0005; n = 3). (C) C38C40-Luc luciferase activity in the absence (−) or in the presence of expression vectors carrying the indicated TFs in mouse keratinocytes. (***P ≤ 0.0005; n = 7). (D) C38C40 luciferase activity was tested in differentiated (gray bars) and proliferating (white bars) mouse keratinocytes upon transfection of the indicated siRNAs. (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; n = 3). Error bars denote SD.
In addition to p63, we identified a number of putative sequence motifs for TFs known to be involved in keratinocyte differentiation including interferon regulatory factors (IRFs), Forkhead box family (FOXs or FKHDs), Krüppel-like family factors (KLFs), POU homeobox factors (POUs) and CCAAT/enhancer binding proteins (C/EBPs) (Supplementary Figure S5 and Supplementary Table S1). Despite its crucial role in epidermal differentiation (63), overexpression of the Krüppel-like factor 4 (Klf4) or of its family member Klf5 had no significant effect on the C38C40 activity (Figure 5C). Similarly, no effect was observed upon overexpression of the Forkhead box protein n1 (Foxn1), a regulator of epidermal and of hair follicle differentiation (64–67), or of a constitutively active Notch1 (Figure 5C), suggesting that Notch1 is likely to suppress p63 expression in a C38C40 independent manner.
In contrast, a significant repression of C38C40 enhancer activity was observed when Irf6, Cebpa, Cebpb, Pou2f3 (Skn-1a) and Pou3f1 (Tst-1) were overexpressed in mouse keratinocytes (Figure 5C). Expression levels of Cebpa, Cebpb, Irf6, Pou2f3 and Pou3f1 were all induced in differentiating keratinocytes as compared to undifferentiating conditions (Supplementary Figure S6A and SB), consistent with their functions in differentiated keratinocytes, and their crucial role in skin development and epidermal differentiation (40,68–71). To further explore the role of these TFs on C38 and C40 elements, we tested the effect of their depletion on C38C40 enhancer activity in keratinocytes using specific siRNA. Depletion of each TF induced C38C40 enhancer activity in differentiated keratinocytes (Figure 5D), indicating that these TFs have a significant inhibitor activity on the enhancer.
Direct regulation of LRE enhancer activity by a subset of TFs involved in keratinocyte differentiation
To determine whether any of the TFs that inhibit p63LRE function may be acting through physical interaction with the enhancer elements, ChIP were performed using specific antibodies. ChIP experiments in mouse and human keratinocytes indicated that Irf6 was unable to bind directly to either C38 or C40 (Supplementary Figure S7A and data not shown), consistent with previous ChIP-seq data (Antonio Costanzo, personal communication and (72)). In contrast, ChIP experiments performed in mouse keratinocytes using Cebpa- or Cebpb-specific antibodies revealed that these TFs bound both C38 and C40 elements to a similar extent (Figure 6A and B). Binding of Pou2f3 could not be assessed due to the lack of ChIP grade antibodies, however ChIP experiments using Pou3f1 antibodies indicated that Pou3f1 efficiently bound to both C38 and C40 (Figure 6C). In addition, ChIP experiments performed in differentiated keratinocytes revealed an increased Cebpa binding to the C38 element compared to proliferating conditions, whereas p63 binding was reduced upon calcium treatment (Supplementary Figure S7B). To further explore the putative role of each TF in regulating p63 expression, we analyzed p63 mRNA and protein levels in proliferating and differentiated keratinocytes using TF-specific siRNA. Interestingly, p63 expression levels were partially restored in differentiating conditions by depletion of Cebpa, Cebpb, Pou2f3 and Pou3f1 or by concomitant depletion of Cepba/Cebpb or Pou2f3/Pou3f1 (Figure 6D and E; Supplementary Figure S8A). Consistent with depletion studies, overexpression of Cebpa and Cebpb suppressed p63 expression levels (Supplementary Figure S8B). Taken together, these data indicate that these TFs contribute to p63 suppression during keratinocyte differentiation.
Figure 6.
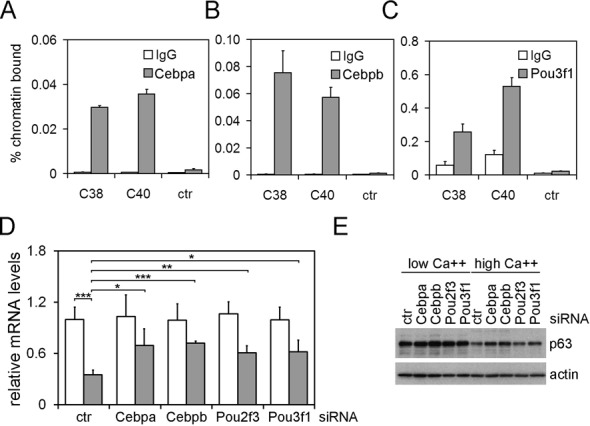
(A, B, C) ChIP-qPCR was performed on mouse primary keratinocytes using antibodies specific for Cebpa (A), Cebpb (B), Pou3f1 (C) (gray bars) or rabbit IgG as negative control (white bars). (D) p63 mRNA expression levels in differentiated (gray bars) and proliferating (white bars) keratinocytes upon transfection of the indicated siRNAs. (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005; n = 4). Error bars denote SD. (E) Immunoblotting of total cell extracts from primary mouse keratinocytes transfected as in (D) using antibodies against the indicated proteins. Keratinocytes were cultured in proliferation (low Ca++) or in differentiation conditions (0.6 mM Ca++ for 24 h).
DISCUSSION
Here, we demonstrate that p63 transcription is regulated by an evolutionary conserved molecular mechanism involving a long-range enhancer, p63LRE. Two small genomic elements (C38 and C40) are essential for the p63LRE function and for basal keratinocyte-specific expression. These elements are unique in the p63 genomic region for the following reasons: (i) they represent the only two non-transcribed elements to be evolutionarily conserved in the most distant vertebrates; (ii) the p63 protein binds far more strongly to these two elements than to any other region in the p63 genomic locus. Evolutionary sequence conservation has been extensively used as filter to identify putative gene regulatory elements (73). However, coupling sequence conservation with ChIP studies using DNAseI sensitivity, histone acetyltransferase or H3K27ac as marks of active enhancers, more precisely detects enhancer regions (58,74–78). We used the H3K27ac mark and p63 binding, coupled to transgenic mouse assays to examine p63LRE function. Comparative genomic analysis has revealed that enhancers active during early embryogenesis are evolutionarily more conserved than those active after mid-gestation (75). C38 and C40 are characterized by strong binding of p63 and H3K27ac not only in mouse keratinocytes and in newborn epidermis but also in embryonic epidermis, consistent with the early activation of p63LRE in embryogenesis. In agreement with an evolutionarily conserved function of the enhancer in p63-expressing epithelial cells, H3K27ac is detected in human keratinocytes and in mammary epithelial cells, but not in other six human cell types that are known to be devoid of p63. Thus, our data suggest that p63LRE belongs to the enhancer category that is not stage- or species-specific, but rather plays a fundamental role in regulating tissue-specific expression of the endogenous p63 gene.
In some cases, enhancers can bypass a nearby gene in order to activate a more distal transcription unit. For instance, the enhancer regulating Sonic Hedgehog in the developing limbs is located 1 Mb away, within the intron of another gene (79). We have strong reasons to believe that p63LRE regulates p63 expression: (i) p63 is the only gene in a 2-Mb genomic region with a precocious and spatially restricted expression in embryonic skin that fully overlaps with p63LRE enhancer activity ((23) and our unpublished observations); (ii) distal transcriptional enhancers are often associated with genes controlling critical developmental processes (73,80–81) such as p63, the only gene in the locus to have such a crucial regulatory function in development.
Direct and indirect positive autoregulation by a DNA-binding TF is a widely used strategy to ensure the long-term maintenance of its own gene expression, after the initiating signals and factors are no longer present (82,83). p63 protein positively controls its own transcription in a direct manner by strongly binding to both C38 and C40 elements. p63-depletion causes loss of p63LRE enhancer activity both in vitro and in vivo suggesting that this autoregulation is central to the control of the p63 gene expression after other initiating signals, possibly including BMP (29–30,33,35). Whether BMP signaling controls p63 expression in the developing surface ectoderm in mammals remains unclear. We found that SMAD1 is unable to bind p63LRE or ΔNp63 promoter in BMP-stimulated keratinocytes (our unpublished observations), however the possibility that BMP/SMAD signaling may control p63LRE in early development cannot be excluded. Nevertheless, careful dissection of the enhancer activity at different stages of development revealed that p63LRE becomes active after p63 expression, indicating that the enhancer is unlikely to contain binding sites for upstream transcriptional activators required to turn on p63. This hypothesis is consistent with the notion that developmentally regulated genes, such as p63, often have a complex transcriptional regulation with many enhancer modules contributing in a cumulative manner to the overall spatial and temporal regulation of the gene (84,85).
In addition, interconnected functional gene networks, rather than individual TFs, control single enhancers. Accordingly, besides p63 we found that Cebpa, Cebpb and Pou3f1 bind both C38 and C40 directly. C/EBPs are members of a highly conserved family of leucine zipper TFs involved in the control of cellular proliferation and differentiation of a number of cell types (86). Concomitant epidermal ablation of Cebpa and Cebpb leads to increased basal keratinocyte proliferation and severe defects in keratinocyte commitment and differentiation coupled with suprabasal expression of p63 (71). Although it has been shown that the DNA-binding domain of Cebpa is required for suppression of p63 expression, no direct regulation could be demonstrated (71). Here, we show that the suppression of p63 expression is due to a direct binding of Cebpa and Cebpb to the p63LRE elements. Other C/EBP family members may be involved in p63 regulation, such as Cebpd that has been reported to negatively regulate p63 in epidermis by binding the ΔNp63 promoter (87).
The two POU homeobox proteins, Pou2f3 and Pou3f1, are both expressed in the epidermis and more abundantly in the suprabasal compartment (69). Depletion of both POU proteins in mice has no overt phenotype at birth while adult skin is hyperplastic and fails to suppress expression of basal layer markers such as keratin14 (69). Our results show that p63 is suppressed by both Pou2f3 and Pou3f1, and that Pou3f1 binds directly to both C38 and C40 elements suggesting a direct regulation. These data are consistent with previous reports showing that Pou2f3 can repress K14 expression and that p63 and Pou2f3 exert antagonistic effects on K14 and other keratin expression (52,88). Thus, increased expression of the Cebps and Pous and concomitant reduced p63 expression are likely to be additively responsible for the enhancer specificity and for p63 regulation in cycling cells versus differentiating cells.
Consistent with the modularity of the enhancers, the requirement of C38 and C40 in the p63LRE enhancer activity in both proliferation and differentiation conditions suggests that C38 and C40 may have a partially overlapping role in p63LRE enhancer to control p63 expression in the epidermis. This is in agreement with the findings that several TFs are capable of binding both C38 and C40. Dividing the enhancer in various functionally connected and partially redundant activities may confer flexibility and robustness, preventing mutations from destroying the enhancer function. C38 and C40 are also likely to have distinct functions as C40 depletion leads to a significant decrease in the p63LRE enhancer activity in all tested conditions, whereas C38 depletion does not, however, combined with C40, C38 confers higher responsiveness to differentiation, confining p63 expression in the basal keratinocytes of the epidermis.
Mutations in non-coding portions of the p63 gene have not been described so far in any of the p63-related syndromes. Future studies should be directed at exploring the possibility that mutations or even relatively frequent sequence variations in the p63LRE may affect the penetrance and the expressivity of p63-related syndromes.
Beside its relevance in p63 biology, the identification of p63LRE will provide a useful tool to drive exogenous expression during embryonic development and in mature epidermis mimicking p63 expression pattern. In addition, we demonstrate here that p63LRE activity follows p63 expression in the conversion of iPSCs toward the epithelial cell fate. Thus, p63LRE could be a potentially useful tool to improve the efficiency of current protocols for iPSC differentiation into corneal and most likely epidermal differentiation, or to identify candidate molecules that can revert defects in p63 function in patient-derived iPSCs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to the members of the Missero's laboratory for providing helpful suggestions and for critically reading the manuscript. We thank Drs Bogi Andersen, Gian Paolo Dotto, Gökhan Hotamisligil, Roberto Mantovani, Tommaso Russo, and Michael Wegner for providing useful reagents, and Drs Huiqing Zhou and Antonio Costanzo for sharing p63 and IRF6 ChIP-seq data respectively.
FUNDING
TELETHON, Italy [GGP09230 to C.M.]; Italian Association for Cancer Research (AIRC) [IG5348 to C.M.]; European ERA-Net Research Program on Rare Diseases (E-RARE-2) [Skin-Dev to C.M. and D.A.]; National Institutes of Health [AR045284 and AR055218 to J.L.B.]. Funding of Open Access funding: TELETHON, Italy (GGP09230).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 2.Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 3.Ferone G., Thomason H.A., Antonini D., De Rosa L., Hu B., Gemei M., Zhou H., Ambrosio R., Rice D.P., Acampora D., et al. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Mol. Med. 2012;4:192–205. doi: 10.1002/emmm.201100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 6.Antonini D., Russo M.T., De Rosa L., Gorrese M., Del Vecchio L., Missero C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J. Invest. Dermatol. 2010;130:1249–1257. doi: 10.1038/jid.2009.438. [DOI] [PubMed] [Google Scholar]
- 7.Truong A.B., Kretz M., Ridky T.W., Kimmel R., Khavari P.A. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H., Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- 9.De Rosa L., Antonini D., Ferone G., Russo M.T., Yu P.B., Han R., Missero C. p63 Suppresses non-epidermal lineage markers in a bone morphogenetic protein-dependent manner via repression of Smad7. J. Biol. Chem. 2009;284:30574–30582. doi: 10.1074/jbc.M109.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koster M.I., Kim S., Mills A.A., DeMayo F.J., Roop D.R. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perez C.A., Pietenpol J.A. Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle. 2007;6:246–254. doi: 10.4161/cc.6.3.3801. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen B.C., Lefort K., Mandinova A., Antonini D., Devgan V., Della Gatta G., Koster M.I., Zhang Z., Wang J., Tommasi di Vignano A., et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 2006;20:1028–1042. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shalom-Feuerstein R., Lena A.M., Zhou H., De La Forest Divonne S., Van Bokhoven H., Candi E., Melino G., Aberdam D. DeltaNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Diff. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fessing M.Y., Mardaryev A.N., Gdula M.R., Sharov A.A., Sharova T.Y., Rapisarda V., Gordon K.B., Smorodchenko A.D., Poterlowicz K., Ferone G., et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 2011;194:825–839. doi: 10.1083/jcb.201101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll D.K., Carroll J.S., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 16.Ferone G., Mollo M.R., Thomason H.A., Antonini D., Zhou H., Ambrosio R., De Rosa L., Salvatore D., Getsios S., van Bokhoven H., et al. p63 control of desmosome gene expression and adhesion is compromised in AEC syndrome. Hum. Mol. Genet. 2013;22:531–543. doi: 10.1093/hmg/dds464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihrie R.A., Marques M.R., Nguyen B.T., Horner J.S., Papazoglu C., Bronson R.T., Mills A.A., Attardi L.D. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Shimomura Y., Wajid M., Shapiro L., Christiano A.M. P-cadherin is a p63 target gene with a crucial role in the developing human limb bud and hair follicle. Development. 2008;135:743–753. doi: 10.1242/dev.006718. [DOI] [PubMed] [Google Scholar]
- 19.Laurikkala J., Mikkola M.L., James M., Tummers M., Mills A.A., Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- 20.Byrne C., Tainsky M., Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 21.Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dotsch V., Andrews N.C., Caput D., McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 22.Parsa R., Yang A., McKeon F., Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J. Invest. Dermatol. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 23.Antonini D., Dentice M., Mahtani P., De Rosa L., Della Gatta G., Mandinova A., Salvatore D., Stupka E., Missero C. Tprg, a gene predominantly expressed in skin, is a direct target of the transcription factor p63. J. Invest. Dermatol. 2008;128:1676–1685. doi: 10.1038/jid.2008.12. [DOI] [PubMed] [Google Scholar]
- 24.King K.E., Ponnamperuma R.M., Yamashita T., Tokino T., Lee L.A., Young M.F., Weinberg W.C. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 25.Della Gatta G., Bansal M., Ambesi-Impiombato A., Antonini D., Missero C., di Bernardo D. Direct targets of the TRP63 transcription factor revealed by a combination of gene expression profiling and reverse engineering. Genome Res. 2008;18:939–948. doi: 10.1101/gr.073601.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallant-Behm C.L., Ramsey M.R., Bensard C.L., Nojek I., Tran J., Liu M., Ellisen L.W., Espinosa J.M. DeltaNp63alpha represses anti-proliferative genes via H2AZ deposition. Genes Dev. 2012;26:2325–2336. doi: 10.1101/gad.198069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBoeuf M., Terrell A., Trivedi S., Sinha S., Epstein J.A., Olson E.N., Morrisey E.E., Millar S.E. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell. 2010;19:807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang A., Zhu Z., Kapranov P., McKeon F., Church G.M., Gingeras T.R., Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Bakkers J., Hild M., Kramer C., Furutani-Seiki M., Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- 30.Tribulo C., Guadalupe Barrionuevo M., Aguero T.H., Sanchez S.S., Calcaterra N.B., Aybar M.J. DeltaNp63 is regulated by BMP4 signaling and is required for early epidermal development in Xenopus. Dev. Dyn. 2012;241:257–269. doi: 10.1002/dvdy.23706. [DOI] [PubMed] [Google Scholar]
- 31.Stern C.D. Neural induction: 10 years on since the ‘default model’. Curr. Opin. Cell. Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Li L., Liu C., Biechele S., Zhu Q., Song L., Lanner F., Jing N., Rossant J. Location of transient ectodermal progenitor potential in mouse development. Development. 2013;140:4533–4543. doi: 10.1242/dev.092866. [DOI] [PubMed] [Google Scholar]
- 33.Aberdam D., Gambaro K., Rostagno P., Aberdam E., de la Forest Divonne S., Rouleau M. Key role of p63 in BMP-4-induced epidermal commitment of embryonic stem cells. Cell Cycle. 2007;6:291–294. doi: 10.4161/cc.6.3.3800. [DOI] [PubMed] [Google Scholar]
- 34.Shalom-Feuerstein R., Serror L., De La Forest Divonne S., Petit I., Aberdam E., Camargo L., Damour O., Vigouroux C., Solomon A., Gaggioli C., et al. Pluripotent stem cell model reveals essential roles for miR-450b-5p and miR-184 in embryonic corneal lineage specification. Stem Cells. 2012;30:898–909. doi: 10.1002/stem.1068. [DOI] [PubMed] [Google Scholar]
- 35.Tadeu A.M., Horsley V. Notch signaling represses p63 expression in the developing surface ectoderm. Development. 2013;140:3777–3786. doi: 10.1242/dev.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J.C., Krishna S., Metzger D., Chambon P., et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuyama R., Ogawa E., Nagoshi H., Yabuki M., Kurihara A., Terui T., Aiba S., Obinata M., Tagami H., Ikawa S. p53 homologue, p51/p63, maintains the immaturity of keratinocyte stem cells by inhibiting Notch1 activity. Oncogene. 2007;26:4478–4488. doi: 10.1038/sj.onc.1210235. [DOI] [PubMed] [Google Scholar]
- 38.Blanpain C., Lowry W.E., Pasolli H.A., Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriyama M., Durham A.D., Moriyama H., Hasegawa K., Nishikawa S., Radtke F., Osawa M. Multiple roles of Notch signaling in the regulation of epidermal development. Dev. Cell. 2008;14:594–604. doi: 10.1016/j.devcel.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Ingraham C.R., Kinoshita A., Kondo S., Yang B., Sajan S., Trout K.J., Malik M.I., Dunnwald M., Goudy S.L., Lovett M., et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat. Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richardson R.J., Dixon J., Malhotra S., Hardman M.J., Knowles L., Boot-Handford R.P., Shore P., Whitmarsh A., Dixon M.J. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat. Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- 42.Moretti F., Marinari B., Lo Iacono N., Botti E., Giunta A., Spallone G., Garaffo G., Vernersson-Lindahl E., Merlo G., Mills A.A., et al. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J. Clin. Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antonini D., Rossi B., Han R., Minichiello A., Di Palma T., Corrado M., Banfi S., Zannini M., Brissette J.L., Missero C. An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol. Cell. Biol. 2006;26:3308–3318. doi: 10.1128/MCB.26.8.3308-3318.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano R.A., Birkaya B., Sinha S. Defining the regulatory elements in the proximal promoter of DeltaNp63 in keratinocytes: Potential roles for Sp1/Sp3, NF-Y, and p63. J. Invest. Dermatol. 2006;126:1469–1479. doi: 10.1038/sj.jid.5700297. [DOI] [PubMed] [Google Scholar]
- 45.Harmes D.C., Bresnick E., Lubin E.A., Watson J.K., Heim K.E., Curtin J.C., Suskind A.M., Lamb J., DiRenzo J. Positive and negative regulation of deltaN-p63 promoter activity by p53 and deltaN-p63-alpha contributes to differential regulation of p53 target genes. Oncogene. 2003;22:7607–7616. doi: 10.1038/sj.onc.1207129. [DOI] [PubMed] [Google Scholar]
- 46.Zeitlinger J., Stark A. Developmental gene regulation in the era of genomics. Dev. Biol. 2010;339:230–239. doi: 10.1016/j.ydbio.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Bolotin D., Chu D.H., Polak L., Williams T., Fuchs E. AP-2alpha: a regulator of EGF receptor signaling and proliferation in skin epidermis. J. Cell Biol. 2006;172:409–421. doi: 10.1083/jcb.200510002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guttormsen J., Koster M.I., Stevens J.R., Roop D.R., Williams T., Winger Q.A. Disruption of epidermal specific gene expression and delayed skin development in AP-2 gamma mutant mice. Dev. Biol. 2008;317:187–195. doi: 10.1016/j.ydbio.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Pasolli H.A., Williams T., Fuchs E. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J. Cell Biol. 2008;183:37–48. doi: 10.1083/jcb.200804030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parisi S., Passaro F., Aloia L., Manabe I., Nagai R., Pastore L., Russo T. Klf5 is involved in self-renewal of mouse embryonic stem cells. J. Cell Sci. 2008;121:2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- 51.Tong Q., Tsai J., Tan G., Dalgin G., Hotamisligil G.S. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol. Cell. Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugihara T.M., Kudryavtseva E.I., Kumar V., Horridge J.J., Andersen B. The POU domain factor Skin-1a represses the keratin 14 promoter independent of DNA binding. A possible role for interactions between Skn-1a and CREB-binding protein/p300. J. Biol. Chem. 2001;276:33036–33044. doi: 10.1074/jbc.M103000200. [DOI] [PubMed] [Google Scholar]
- 53.Shalom-Feuerstein R., Serror L., Aberdam E., Muller F.J., van Bokhoven H., Wiman K.G., Zhou H., Aberdam D., Petit I. Impaired epithelial differentiation of induced pluripotent stem cells from ectodermal dysplasia-related patients is rescued by the small compound APR-246/PRIMA-1MET. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2152–2156. doi: 10.1073/pnas.1201753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 55.Renner K., Leger H., Wegner M. The POU domain protein Tst-1 and papovaviral large tumor antigen function synergistically to stimulate glia-specific gene expression of JC virus. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6433–6437. doi: 10.1073/pnas.91.14.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 58.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 60.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kouwenhoven E.N., van Heeringen S.J., Tena J.J., Oti M., Dutilh B.E., Alonso M.E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L., et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McDade S.S., Henry A.E., Pivato G.P., Kozarewa I., Mitsopoulos C., Fenwick K., Assiotis I., Hakas J., Zvelebil M., Orr N., et al. Genome-wide analysis of p63 binding sites identifies AP-2 factors as co-regulators of epidermal differentiation. Nucleic Acids Res. 2012;40:7190–7206. doi: 10.1093/nar/gks389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segre J.A., Bauer C., Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 64.Li J., Baxter R.M., Weiner L., Goetinck P.F., Calautti E., Brissette J.L. Foxn1 promotes keratinocyte differentiation by regulating the activity of protein kinase C. Differentiation. 2007;75:694–701. doi: 10.1111/j.1432-0436.2007.00176.x. [DOI] [PubMed] [Google Scholar]
- 65.Lee D., Prowse D.M., Brissette J.L. Association between mouse nude gene expression and the initiation of epithelial terminal differentiation. Dev. Biol. 1999;208:362–374. doi: 10.1006/dbio.1999.9221. [DOI] [PubMed] [Google Scholar]
- 66.Janes S.M., Ofstad T.A., Campbell D.H., Watt F.M., Prowse D.M. Transient activation of FOXN1 in keratinocytes induces a transcriptional programme that promotes terminal differentiation: contrasting roles of FOXN1 and Akt. J. Cell Sci. 2004;117:4157–4168. doi: 10.1242/jcs.01302. [DOI] [PubMed] [Google Scholar]
- 67.Brissette J.L., Li J., Kamimura J., Lee D., Dotto G.P. The product of the mouse nude locus, Whn, regulates the balance between epithelial cell growth and differentiation. Genes Dev. 1996;10:2212–2221. doi: 10.1101/gad.10.17.2212. [DOI] [PubMed] [Google Scholar]
- 68.Biggs L.C., Rhea L., Schutte B.C., Dunnwald M. Interferon regulatory factor 6 is necessary, but not sufficient, for keratinocyte differentiation. J. Invest. Dermatol. 2012;132:50–58. doi: 10.1038/jid.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andersen B., Weinberg W.C., Rennekampff O., McEvilly R.J., Bermingham J.R., Jr, Hooshmand F., Vasilyev V., Hansbrough J.F., Pittelkow M.R., Yuspa S.H., et al. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 1997;11:1873–1884. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- 70.Oh H.S., Smart R.C. Expression of CCAAT/enhancer binding proteins (C/EBP) is associated with squamous differentiation in epidermis and isolated primary keratinocytes and is altered in skin neoplasms. J. Invest. Dermatol. 1998;110:939–945. doi: 10.1046/j.1523-1747.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 71.Lopez R.G., Garcia-Silva S., Moore S.J., Bereshchenko O., Martinez-Cruz A.B., Ermakova O., Kurz E., Paramio J.M., Nerlov C. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat. Cell Biol. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 72.Botti E., Spallone G., Moretti F., Marinari B., Pinetti V., Galanti S., De Meo P.D., De Nicola F., Ganci F., Castrignano T., et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13710–13715. doi: 10.1073/pnas.1110931108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visel A., Rubin E.M., Pennacchio L.A. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nord A.S., Blow M.J., Attanasio C., Akiyama J.A., Holt A., Hosseini R., Phouanenavong S., Plajzer-Frick I., Shoukry M., Afzal V., et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- 80.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 81.Montavon T., Duboule D. Landscapes and archipelagos: spatial organization of gene regulation in vertebrates. Trends Cell Biol. 2012;22:347–354. doi: 10.1016/j.tcb.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 82.Crews S.T., Pearson J.C. Transcriptional autoregulation in development. Curr. Biol. 2009;19:R241–R246. doi: 10.1016/j.cub.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu F., Posakony J.W. An enhancer composed of interlocking submodules controls transcriptional autoregulation of suppressor of hairless. Dev. Cell. 2014;29:88–101. doi: 10.1016/j.devcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Laat W., Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 85.Harmston N., Lenhard B. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 2013;41:7185–7199. doi: 10.1093/nar/gkt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Borrelli S., Testoni B., Callari M., Alotto D., Castagnoli C., Romano R.A., Sinha S., Vigano A.M., Mantovani R. Reciprocal regulation of p63 by C/EBP delta in human keratinocytes. BMC Mol. Biol. 2007;8:85. doi: 10.1186/1471-2199-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lena A.M., Cipollone R., Amelio I., Catani M.V., Ramadan S., Browne G., Melino G., Candi E. Skn-1a/Oct-11 and DeltaNp63alpha exert antagonizing effects on human keratin expression. Biochem. Biophys. Res. Commun. 2010;401:568–573. doi: 10.1016/j.bbrc.2010.09.102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


