Abstract
Legionella pneumophila, the primary agent of Legionnaires' disease, flourishes in both natural and man-made environments by growing in a wide variety of aquatic amoebae. Recently, we determined that the Cas2 protein of L. pneumophila promotes intracellular infection of Acanthamoeba castellanii and Hartmannella vermiformis, the two amoebae most commonly linked to cases of disease. The Cas2 family of proteins is best known for its role in the bacterial and archeal clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein (Cas) system that constitutes a form of adaptive immunity against phage and plasmid. However, the infection event mediated by L. pneumophila Cas2 appeared to be distinct from this function, because cas2 mutants exhibited infectivity defects in the absence of added phage or plasmid and since mutants lacking the CRISPR array or any one of the other cas genes were not impaired in infection ability. We now report that the Cas2 protein of L. pneumophila has both RNase and DNase activities, with the RNase activity being more pronounced. By characterizing a catalytically deficient version of Cas2, we determined that nuclease activity is critical for promoting infection of amoebae. Also, introduction of Cas2, but not its catalytic mutant form, into a strain of L. pneumophila that naturally lacks a CRISPR-Cas locus caused that strain to be 40- to 80-fold more infective for amoebae, unequivocally demonstrating that Cas2 facilitates the infection process independently of any other component encoded within the CRISPR-Cas locus. Finally, a cas2 mutant was impaired for infection of Willaertia magna but not Naegleria lovaniensis, suggesting that Cas2 promotes infection of most but not all amoebal hosts.
INTRODUCTION
Legionella pneumophila is a Gram-negative, aquatic bacterium and the agent of a Legionnaires' disease pneumonia (1). The number of reported cases of Legionnaires' disease in the United States has more than tripled since 2001, with similar trends occurring in Canada and Europe (2). Humans acquire L. pneumophila mainly by inhaling contaminated water droplets from aerosol-generating devices (3). In the lung, the bacterium infects resident macrophages (4). Amoebae are the major replicative niche for L. pneumophila in natural and man-made water systems (5–7). Indeed, L. pneumophila infects ≥20 species of amoebae encompassing the genera Acanthamoeba, Echinamoeba, Hartmannella, Naegleria, Vahlkampfia, and Willaertia (8). L. pneumophila bacteria in amoebae remain viable for long periods of time and are resistant to biocides (9, 10), and internalization by amoebae resuscitates viable-but-not-culturable legionellae (11, 12). Also, amoebae may be part of the inoculum that initiates lung infection (3, 13, 14). Recently, we found that the Cas2 protein of L. pneumophila promotes infection of Acanthamoeba castellanii and Hartmannella vermiformis; i.e., although cas2 mutants grow normally in broth and macrophages, they exhibit a 1,000-fold reduced recovery from A. castellanii and a 20-fold defect in H. vermiformis (15). Because a complemented cas2 mutant behaves like the wild type does, these defects are due to the loss of Cas2. Compatible with these data, the levels of cas2 mRNA are higher during intracellular growth than during growth in broth (15). The significance of these findings is heightened by the fact that acanthamoebae and hartmannellae are the most common amoebae in waters linked to Legionnaires' disease (6, 7, 16, 17) and cas2 occurs in endemic strains linked to outbreaks (18).
The Cas2 family of proteins is best known for being part of the bacterial and archeal clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein (Cas) system, a recently described system that confers immunity against phage and plasmid (19–21). Present in the genomes of nearly all archaea and the genomes of approximately one-half of bacteria, the CRISPR-Cas locus consists of the CRISPR array, which is composed of a variable number of palindromic repeats separated by unique spacer sequences, and a set of cas genes which encodes a variable number of Cas proteins. On the basis of the nature of the cas genes and their operon organization, CRISPR-Cas loci are classified into types I, II, and III, which are further classified into subtypes A, B, etc. (22). In L. pneumophila, there is a type II-B locus that has a CRISPR array with 60 repeats and 58 unique spacers and a cas operon consisting of cas9, cas1, cas2, and cas4 (15). CRISPR-Cas-mediated immunity proceeds in three steps: first, a new spacer derived from an invading genetic element is incorporated into the CRISPR array via the action of Cas1 and Cas2; second, the array is transcribed, and small CRISPR RNAs (crRNAs) are generated by Cas6 homologs or RNase III; and third, the crRNAs direct other Cas nucleases to cleave newly invading nucleic acid (19, 20, 23, 24).
Our finding that Cas2 mutants of L. pneumophila have an impaired ability to infect host cells was part of an initial shift in thinking about other ways in which Cas proteins work. Because the cas2 mutants exhibit their defect in the absence of added phage, plasmid, or nucleic acid and because L. pneumophila mutants lacking cas9, cas1, cas4, or the CRISPR array are not impaired (15), the infection event mediated by L. pneumophila Cas2 must be distinct from the functions that have been traditionally ascribed to Cas proteins and CRISPR-Cas systems. There are now a variety of studies demonstrating that there are roles for other Cas proteins that are unrelated to phage/plasmid immunity (21, 25–29); e.g., Cas9 of Francisella novicida promotes virulence in a murine model of disease (30), and Campylobacter jejuni Cas9 enhances invasion of epithelial cells (31).
In beginning to consider how L. pneumophila Cas2 facilitates infection of amoebae, we hypothesized that the protein has nuclease activity (15). However, it was more difficult to posit the target of the activity, given the various results that had been reported for other Cas2 proteins. Initially, purified Cas2 proteins from the bacteria Nitrosomonas europaea and Thermotoga maritima and the archaea Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, and Sulfolobus solfataricus were found to have activity against single-stranded RNA (ssRNA) but not against single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA) (32), yet no RNase activity was detected when the Cas2 proteins from the bacteria Desulfovibrio vulgaris and Bacillus halodurans were examined, and only weak RNase activity was reported for Cas2 from Thermus thermophilus (33, 34). Moreover, further testing revealed that the Cas2 proteins of B. halodurans, Streptococcus pyogenes, T. thermophilus, and Xanthomonas oryzae have activity against dsDNA (34, 35). Recent work done with Escherichia coli has suggested that the nuclease activity of Cas2 proteins is not required for spacer acquisition during CRISPR-Cas immunity but might be involved in CRISPR-independent processes (19, 23). We now report that the Cas2 protein of L. pneumophila has both RNase and DNase activity and that this nuclease activity is critical for infection of amoebae. Also, we found, among other things, that introduction of cas2 into a strain of L. pneumophila that naturally lacks the CRISPR-Cas locus renders that strain more infective for amoebae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
L. pneumophila serogroup 1 strain 130b (ATCC BAA-74) served as our wild-type, parental strain (36). Mutants of 130b used in this study were cas1 mutant NU410, cas2 mutant NU411, cas4 mutant NU412, cas9 mutant NU413, and CRISPR array mutant NU414 (15). Another clinical isolate of L. pneumophila serogroup 1 that was examined was Philadelphia-1 (ATCC 33152) (37). Legionellae were routinely grown at 37°C on buffered charcoal yeast extract (BCYE) agar, which, when appropriate, contained chloramphenicol at 3 μg/ml or kanamycin at 25 μg/ml (15). E. coli strains DH5α (Invitrogen, Carlsbad, CA) and XL10-Gold (Agilent Technologies, La Jolla, CA) were hosts for recombinant plasmids and were grown in Luria-Bertani (LB) medium. The plasmid pCas2 represents the vector pMMB2002 into which the cas2 gene of strain 130b was cloned (15). Unless otherwise noted, chemicals were from Sigma-Aldrich (St. Louis, MO).
Cloning and purification of L. pneumophila Cas2.
The cas2 gene was PCR amplified from 130b genomic DNA using primers Cas2 LIC-F and Cas2 LIC-R (Table 1), and then, using the ligation-independent cloning technique of Aslanidis and de Jong (38), the PCR product was cloned into the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible protein expression vector pMCSG28 (39). The resultant plasmid, pMCSG28-Cas2, carried Cas2 with a C-terminal 6× His tag. To begin purification of Cas2, E. coli BL21(DE3) harboring pMCSG28-Cas2 was grown in 500 ml of LB medium (containing 100 μg/ml ampicillin) with shaking (200 rpm) at 37°C. Once the culture reached an optical density at 600 nm of 0.8, it was chilled to 16°C and IPTG was added to a concentration of 0.3 mM in order to begin to induce the expression of recombinant Cas2. The culture was incubated for 14 h with shaking at 16°C, and then after centrifugation (11,325 × g, 4°C for 15 min), the bacterial pellet was frozen at −80°C. After thawing the pellet for 15 min on ice, bacteria were resuspended in lysis buffer (1.5 mM magnesium acetate, 1 mM calcium chloride, 250 mM NaCl, 100 mM ammonium sulfate, 40 mM sodium phosphate dibasic, 3.25 mM citric acid, 5% glycerol, 5 mM imidazole, 0.08% lauryl-β-d-maltoside), 5 mM β-mercaptoethanol, 1 mg/ml fresh lysozyme at 5 ml per gram (wet weight) and stored on ice for 30 min. The bacterial suspension was then subjected to sonication, which involved six 10-s bursts at 30% amplitude with 10-s cooling periods between each burst. We used a Branson digital sonifier 450 equipped with a 3-mm microtip (Branson Ultrasonics, Danbury, CT). After centrifugation (10,000 × g, 4°C for 30 min) in order to obtain supernatant containing soluble proteins, recombinant Cas2 was purified by immobilized metal affinity chromatography using Ni-nitrilotriacetic acid (Qiagen, Valencia, CA). To that end, impurities were removed by washing the protein-containing column with 20 mM imidazole in 50 mM sodium phosphate dibasic, 300 mM NaCl buffer. Cas2 was then eluted from the column with 250 mM imidazole in 50 mM sodium phosphate dibasic, 300 mM NaCl buffer. Imidazole was removed from the sample by buffer exchange using an Amicon Ultra-15 centrifugal filter device (Millipore, Billerica, MA). Purified Cas2 was eluted from the filter with 10 mM Tris-HCl, pH 8, 100 mM NaCl, 2 mM dithiothreitol buffer. The purity of Cas2 was evaluated by SDS-PAGE and immunoblot analysis using anti-His–horseradish peroxidase (HRP) antibody (Life Technologies, Carlsbad, CA). Purified protein was flash frozen and stored at −80°C. In order to obtain a mutant version of Cas2, site-directed mutagenesis of cas2 in pMCSG28-Cas2 was done using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). As confirmed by sequencing, pMCSG28-Cas2 Y13A carried a mutated version of Cas2 in which tyrosine-13 was changed to an alanine. Cas2 Y13A was purified as described above for wild-type Cas2. To judge the protein conformation, the circular dichroism (CD) spectra of Cas2 and Cas2 Y13A were recorded at 0.1 mg/ml with a 0.2-cm-path-length cuvette using a Jasco J-815 spectrometer (Jasco Analytical Instruments, Easton, MD).
TABLE 1.
Primersa used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| Cas2 LIC-F | GTCTCTCCCATGCAAATAAGATTATACAAGGAATACATAGTTTCCTAC |
| Cas2 LIC-R | GGTTCTCCCCAGCTACGATGATGTGTCCATCCGG |
| Cas26×HIS-F1 | ATGCAAATAAGATTATACAAGGAATACAT |
| Cas26×HIS-R1 | TTAATGATGATGATGATGATGTACGATGATGTGTCCATCCGGTTT |
| Cas2-1F | TTGAAAGCATCATTCAAAGACAA |
| Cas2-2F | TAACCCCCGTTCAAAAATCA |
| Cas2 Y13A-F1 | GGAATACATAGTTTCCGCCGACATCAGGGACAAC |
| Cas2 Y13A-R1 | GTTGTCCCTGATGTCGGCGGAAACTATGTATTCC |
Primers were obtained from Integrated DNA Technologies (Coralville, IA).
Nuclease assays.
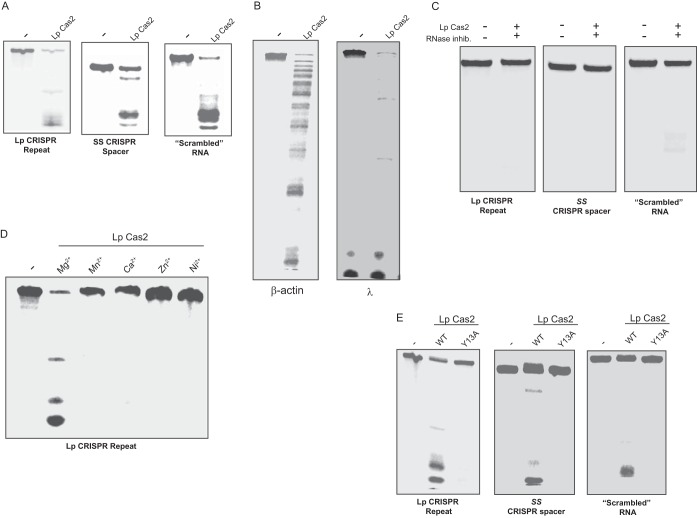
RNase assays were done as previously described for the study of other Cas2 proteins (32). The 10-μl reaction mixture contained 1.1 μM Cas2 or Cas2 Y13A (i.e.,1 μl taken from a 11 μM stock), 12 ng (i.e., 0.1 μM) of fluorescently labeled ssRNA, 50 mM Tris-HCl (pH 8.5), 100 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol. The ssRNA substrates tested were from the L. pneumophila 130b CRISPR repeat (5′-GGUUAUUAGGGAGUAGAUUUUUAGGUUGGUGACUUUG-3′), S. solfataricus CRISPR cluster 2, spacer 3 (5′-AAAUACGUUUUCUCCAUUGUCAUAUUGCGCAUAAGUUGA-3′), and scrambled RNA (5′-AUCAUUUUGUUAAGUACAGUCUUAAGUUUUCC-3′). The ssRNA substrates were synthesized with fluorescently labeled IR800CW dye on the 5′ end via a primary amine (Integrated DNA Technologies, Coralville, IA). The reaction mixture was incubated for 1 h at 37°C before being quenched with 10 μl of formamide loading buffer, and then the reaction products were resolved by electrophoresis using 15% polyacrylamide, 8 M urea gels (32). To detect the fluorescently labeled ssRNA, the gels were scanned using a LI-COR Odyssey Fc system (LI-COR Biosciences, Lincoln, NE). Control reactions utilizing RNase A (Life Technologies) confirmed that the substrates and assay conditions were working as expected (data not shown). In order to examine the effect of Cas2 on long ssRNA, pTRI-actin, encoding a 304-nucleotide (nt) portion of the mouse β-actin gene (MAXI script kit; Ambion, Austin, TX), and bacteriophage λ DNA, encoding a 523-nt portion of the phage genome (Jena Biosciences, Jena, Germany), were templates in in vitro transcription reactions using an Atto-680 RNA labeling kit (Jena Biosciences). The fluorescently labeled molecules were detected by the LI-COR system. To assay for RNase inhibition, the reaction mixtures were incubated in the presence or absence of 1 μl of RNaseOUT (Life Technologies). For DNase assays, 0.5 μg of either linear, double-stranded bacteriophage λ DNA (NEB, Ipswich, MA) or single-stranded M13mp18 DNA (NEB) was incubated with 1.1 μM Cas2 or Cas2 Y13A (1 μl of an 11 μM stock) in the presence of 25 mM HEPES-KOH (pH 7.5), 200 mM KCl, and 2.5 mM MgCl2 (34). The reaction mixtures were incubated for 2 h at 37°C, and the reaction products were separated by agarose (0.5%, wt/vol) gel electrophoresis and visualized by ethidium bromide staining. Control reactions utilizing DNase I (Life Technologies) confirmed that the substrates and the assay were working (data not shown). The effect of divalent cations was determined by replacing 5 mM MgCl2 in the RNase and DNase reaction mixtures with 5 mM MnCl2, 5 mM CaCl2, 5 mM ZnCl2, or 5 mM NiCl2, as well as by incorporating 0.05 mM EDTA into the reaction mixture.
Complementation analysis.
In order to assess complementation by a wild-type version and a mutant version of Cas2, cas2 was amplified from pCas2 DNA (15) using primers Cas2 6×HIS-F1 and Cas2 6×HIS-R1 (Table 1). The product was ligated into the pGEM-T Easy vector, and the resulting pCas2-His vector was sequenced using primers Cas2-1F and Cas2-2F (Table 1) to verify the insertion of the His tag at the C terminus of Cas2. Sequence data were analyzed using Lasergene software (DNASTAR, Madison, WI). pCas2-His was digested with SacI and SphI, and then the cas2-containing fragment was cloned into pMMB2002 (40), yielding pMMB2002-Cas2. Site-directed mutagenesis of cas2 in pMMB2002-Cas2 was done using the QuikChange XL kit, as noted above, using primers Cas2 Y13A-F1 and Cas2 Y13A-R1 (Table 1). As confirmed by DNA sequencing using primers Cas2-1F and Cas2-2F, pMMB2002-Cas2 Y13A carried a mutated version of Cas2 in which tyrosine-13 was changed to an alanine. pMMB2002-Cas2 and pMMB2002-Cas2 Y13A were separately electroporated into wild-type 130b, the 130b cas2 mutant NU411, and wild-type Philadelphia-1 using previously described protocols (40). To judge the relative levels of Cas2 and Cas2 Y13A in L. pneumophila, immunoblot analysis was done, along with assessment of cytosolic isocitrate dehydrogenase (ICDH) as a loading control (40). After lysates were obtained from 1-ml overnight cultures, proteins were separated by SDS-(12%) PAGE and then transferred onto a polyvinylidene fluoride membrane (Bio-Rad). Following incubation overnight at 4°C in Tris-buffered saline plus 0.1% Tween 20 and 5% nonfat dry milk (TBS-TM), the membrane was incubated overnight at 4°C in TBS-TM containing either murine anti-His antibody (catalog number 05-949; Millipore) at a 1:1,000 dilution or rabbit anti-ICDH antibody (40) at a 1:10,000 dilution. Following a wash step, the membrane containing the His-tagged Cas2 protein was incubated for 1 h with anti-mouse IgG HRP-linked antibody (Cell Signaling, Danvers, MA) at a 1:3,000 dilution in TBS-TM. The membrane containing ICDH was incubated for 1 h with anti-rabbit IgG HRP-linked antibody (Cell Signaling) at a 1:5,000 dilution in TBS-TM. After a final series of washes, the blots were developed with the Amersham ECL Prime Western blotting reagent (GE Healthcare, Buckinghamshire, United Kingdom).
Intracellular infection assays.
To examine the ability of L. pneumophila strains to grow within amoebae, A. castellanii (ATCC 30234), Naegleria lovaniensis (ATCC 30569), and Willaertia magna (ATCC 50036) were infected as previously described (8, 41). Briefly, bacteria were added to tissue culture wells containing 1 × 105 amoebae, and then at various times postinoculation, the numbers of L. pneumophila were counted by dilution plating aliquots from the wells' supernatants on BCYE agar. Since L. pneumophila does not grow in the medium, even after it is conditioned following infection of the amoebae, all increases in bacterial numbers result from intracellular infection and growth. Because infection of W. magna proceeds at a slower pace than does infection of A. castellanii or N. lovaniensis, assays using the willaertiae were done over a 96-h period, whereas experiments using acanthamoebae or naegleriae typically encompassed a 72-h incubation (8). The infection of U937 (ATCC CRL-1593.2) cells by L. pneumophila and the subsequent measurement of cytokine interleukin-6 (IL-6) levels produced by the infected macrophages were also done as previously described (42).
RESULTS
An L. pneumophila Cas2 mutant is impaired for infection of W. magna but not N. lovaniensis.
The fact that a cas2 mutant of L. pneumophila is impaired approximately 1,000-fold for infection of A. castellanii but 20-fold for infection of H. vermiformis indicates that the importance of Cas2 is dependent on the type of amoebal host being infected (15). Thus, we sought to determine whether the impaired infectivity of the cas2 mutant extended to other types of amoebal hosts. We compared wild-type strain 130b and its cas2 mutant for the ability to infect N. lovaniensis and W. magna, two other amoebae that are hosts in nature (8, 41). The cas2 mutant exhibited an impaired ability to infect W. magna, with ca. 5-fold and 10-fold-reduced recoveries being noted at 72 h and 96 h, respectively (Fig. 1A). The mutant was not defective for infection of N. lovaniensis (Fig. 1B). Thus, Cas2 appears to be required for infection of many but not all kinds of amoebae. That the mutant was not impaired in N. lovaniensis infection reinforces our previous conclusion that the loss of Cas2 does cause a generalized growth defect (15). We previously found that Cas2 is not needed for L. pneumophila growth in macrophages (15). After our report, a study of F. novicida found that a Cas protein can suppress the immune response of an infected macrophage; i.e., Cas9 can dampen IL-6 secretion (43). Thus, we repeated infection of U937 cell macrophages with wild-type 130b and our full set of CRISPR-Cas mutants, i.e., mutants lacking cas9, cas1, cas2, cas4, or the CRISPR array, and assayed for IL-6 production. None of the mutants triggered altered levels of cytokine (Fig. 2), indicating that, in the case of L. pneumophila, the CRISPR-Cas locus does not impact IL-6 production by infected macrophages. Given that L. pneumophila Cas2 had its greatest effect on infection of A. castellanii, we proceeded to focus on learning how Cas2 facilitates infection of the Acanthamoeba host and, in particular, if the Legionella Cas2 protein has a nuclease activity that is required for infection.
FIG 1.
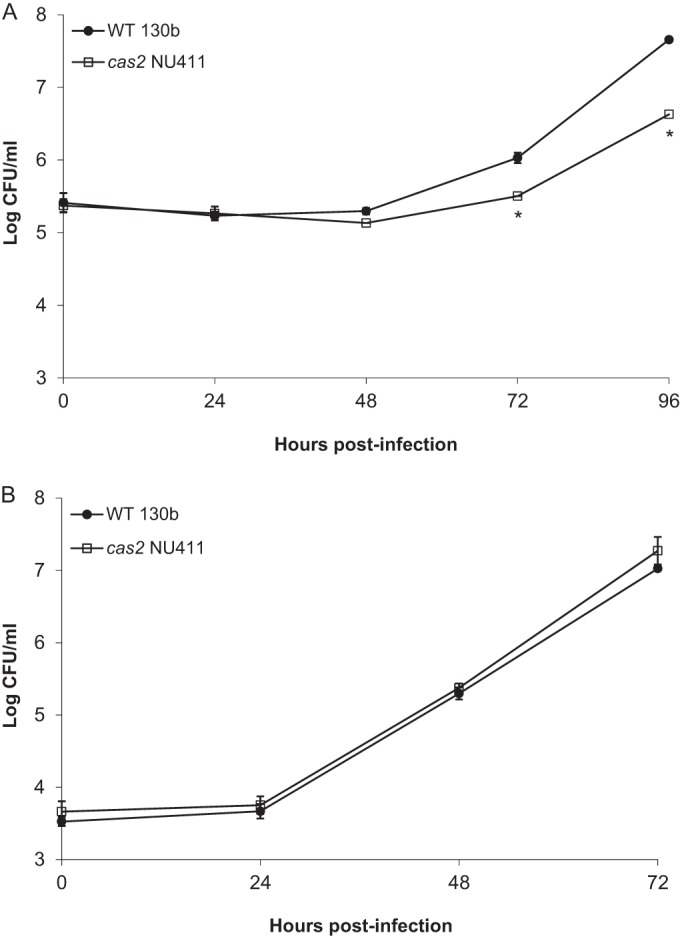
Intracellular infection of W. magna and N. lovaniensis by L. pneumophila wild-type (WT) strain 130b and its cas2 mutant. W. magna (A) and N. lovaniensis (B) were infected with strain 130b and cas2 mutant NU411, and then at the indicated times, the numbers of CFU from the infected monolayers were determined by plating. Data points are means and standard deviations for four infected wells. Asterisks in panel A indicate where the recovery of NU411 was significantly less than that of the wild-type strain (P < 0.001, Student's t test). The results in each panel are representative of those from three independent experiments. In one of the N. lovaniensis experiments, an additional data point taken at 96 h also did not reveal any difference between the wild type and the cas2 mutant.
FIG 2.
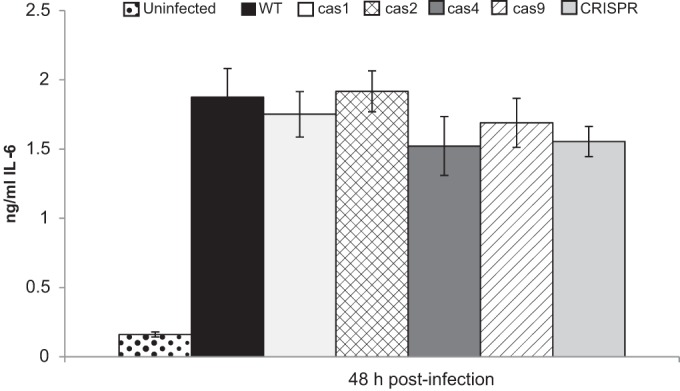
IL-6 output from macrophages infected with the wild type and CRISPR-Cas mutants. As indicated, U937 cells were either not infected or infected with wild-type (WT) strain 130b, the cas1 mutant NU410, the cas2 mutant NU411, the cas4 mutant NU413, the cas9 mutant NU409, or the CRISPR array mutant NU414, and then at 48 h postinoculation, the levels of IL-6 in culture supernatants were determined by enzyme-linked immunosorbent assay. Data are the means and standard deviations for triplicate wells and are representative of those from three independent experiments.
L. pneumophila Cas2 possesses RNase activity.
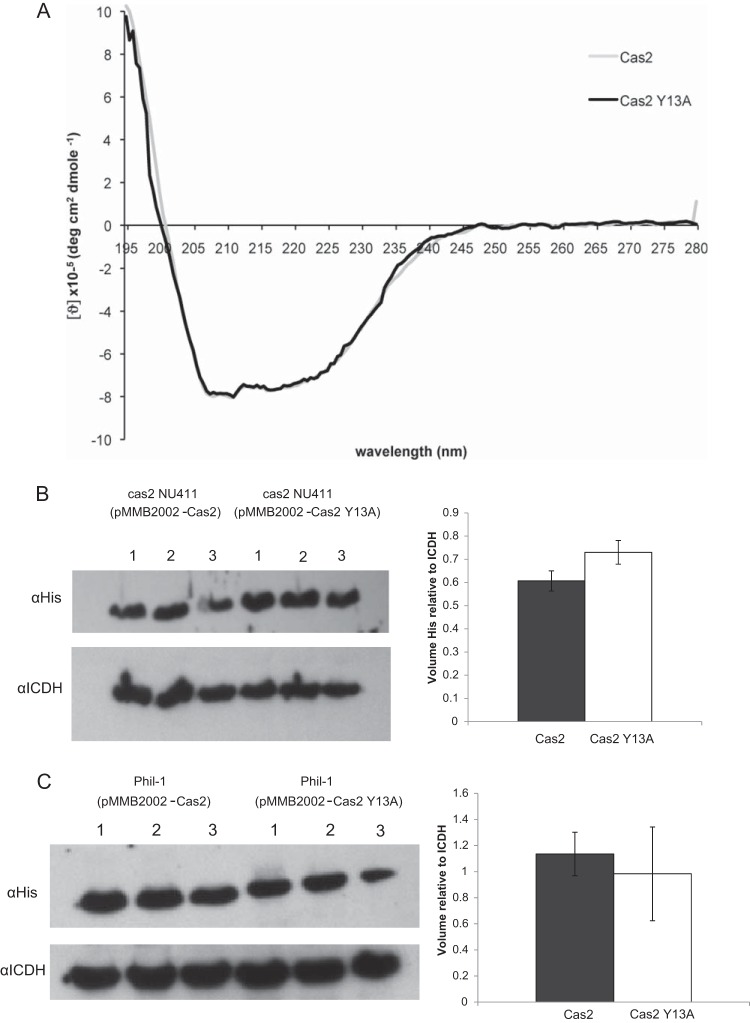
To determine whether the Cas2 protein of L. pneumophila has RNase activity, we cloned cas2 from strain 130b into E. coli, purified the recombinant protein, and examined its ability to cleave ssRNA substrates utilizing procedures that had been used to assay a variety of Cas2 proteins (32). The protein readily cleaved a 37-nt substrate that is identical to the sense strand of the L. pneumophila CRISPR array repeat (Fig. 3A). It also easily cleaved a 39-nt ssRNA that matches a CRISPR spacer from S. solfataricus (32) as well as a 32-nt scrambled RNA (Fig. 3A). No RNase activity was detected, when we processed, in parallel, material obtained from E. coli lacking the cloned cas2 gene (data not shown). To determine if L. pneumophila Cas2 can cleave longer RNAs, we generated a 304-nt molecule derived from actin mRNA and a 523-nt molecule originating from λ phage and repeated the assay. Once again, Legionella Cas2 exhibited strong RNA activity and had seemingly greater activity against the λ phage-derived ssRNA (Fig. 3B). The degradation of RNA substrates was blocked when an RNase inhibitor was added to the reaction mixture (Fig. 3C). Together, these data indicate that Cas2 of L. pneumophila exhibits nonspecific RNase activity. When we replaced the Mg2+ in the reaction mixture with Mn2+, Ca2+, Zn2+, or Ni2+, there was little to no evidence of RNase activity (Fig. 3D), indicating that L. pneumophila Cas2 has a strong preference for Mg2+. Members of the Cas2 family contain both a ferredoxin fold that is often found in RNA-binding proteins and an N-terminal β strand followed by a polar amino acid (32, 33, 44, 45). L. pneumophila Cas2 has both an N-terminal β strand followed by an aspartate and a ferredoxin fold that is analogous to what exists in the Cas2 proteins of B. halodurans, D. vulgaris, Pyrococcus furiosus, S. solfataricus, and T. thermophilus (15). The Legionella protein also has a tyrosine in position 13 that is analogous to a tyrosine residue that was defined as being important for RNase activity in S. solfataricus (32). Previously, Beloglazova et al. converted the tyrosine in position 9 of the Sulfolobus Cas2 protein to an alanine and then observed a loss of enzyme activity (32). Hence, we performed site-directed mutagenesis on the cloned L. pneumophila cas2 gene and purified a recombinant variant of Cas2 that has the residue at position 13 changed from tyrosine to alanine, i.e., Cas2 Y13A. CD spectrometry analysis determined that the variant protein assumed a conformation that was equivalent to that of the wild-type protein (Fig. 4A). When Cas2 Y13A was tested in the RNase assay, it lacked activity (Fig. 3E), indicating that the tyrosine residue is part of the catalytic site.
FIG 3.
Cleavage of RNA substrates by the L. pneumophila Cas2 protein. (A and B) Fluorescently labeled ssRNA corresponding to the CRISPR repeat of L. pneumophila (Lp) strain 130b (A, left), spacer 3 within the CRISPR cluster 2 of S. solfataricus (SS) (A, center), a scrambled RNA (A, right), β-actin RNA (β) (B, left), or λ phage RNA (λ) (B, right) was incubated with purified L. pneumophila Cas2, and then the cleavage products were separated by PAGE and visualized using a LI-COR system. As a negative control, the RNAs were incubated in the absence of any added protein (lanes marked −). (C) The ssRNA substrates used for the assay whose results are presented in panel A were incubated with purified L. pneumophila Cas2, but in the presence of an RNase inhibitor (inhib.), and the reaction products were analyzed as described above. RNA substrates incubated in the absence of added protein or inhibitor appear in the left lanes. (D) ssRNA corresponding to the L. pneumophila CRISPR spacer was incubated with purified L. pneumophila Cas2 either in buffer containing MgCl2, as was done for the assay whose results are presented in panels A to C, or in buffer in which the MgCl2 was replaced with an equimolar amount of MnCl2, CaCl2, ZnCl2, or NiCl2. Reaction products, along with untreated substrate (lane −), were visualized after PAGE. (E) The indicated ssRNA substrates were incubated with either wild-type (WT) Cas2, as was done in assays whose results are presented in panels A to D, or its Cas2 Y13A mutant form (Y13A) and then analyzed as described above. Lanes −, untreated substrate. The data presented in panel A are representative of those from at least four independent experiments, the data presented in panels D and E are typical of those from three independent experiments, and those presented in panels B and C are representative of those from two independent trials.
FIG 4.
Conformation and expression of the catalytic mutant protein Cas2 Y13A. (A) Conformational analysis of wild-type and mutant Cas2. The CD spectra of purified Cas2 and Cas2 Y13A were recorded at 0.1 mg/ml. Spectral data are plotted as the mean residue ellipticity (θ) × 104 (deg cm2 dmol−1 residue−1). The spectra from buffer alone were subtracted from each curve, and for comparison, the data were overlaid using the Excel program. (B) Levels of wild-type and mutant Cas2 within L. pneumophila strain 130b. Lysates (n = 3) were obtained from strain 130b containing either pMMB2002-Cas2 or pMMB2002-Cas2 Y13A, separated by PAGE, and subjected to immunoblotting. (Top) Image obtained when one portion of the blot was exposed to anti-His antibodies for detection of the His-tagged Cas2 proteins; (bottom) the other part of the blot that was reacted with anti-ICDH antibodies and served as a loading control; (right) the relative levels of wild-type and mutant Cas2 normalized to the level of ICDH. The data presented are representative of those from three independent trials. (C) Levels of wild-type and mutant Cas2 proteins in L. pneumophila strain Philadelphia-1. Lysates (n = 3) were obtained from strain Philadelphia-1 (Phil-1) containing either pMMB2002-Cas2 or pMMB2002-Cas2 Y13A and then analyzed in the manner outlined in the legend to panel B.
L. pneumophila Cas2 possesses DNase activity.
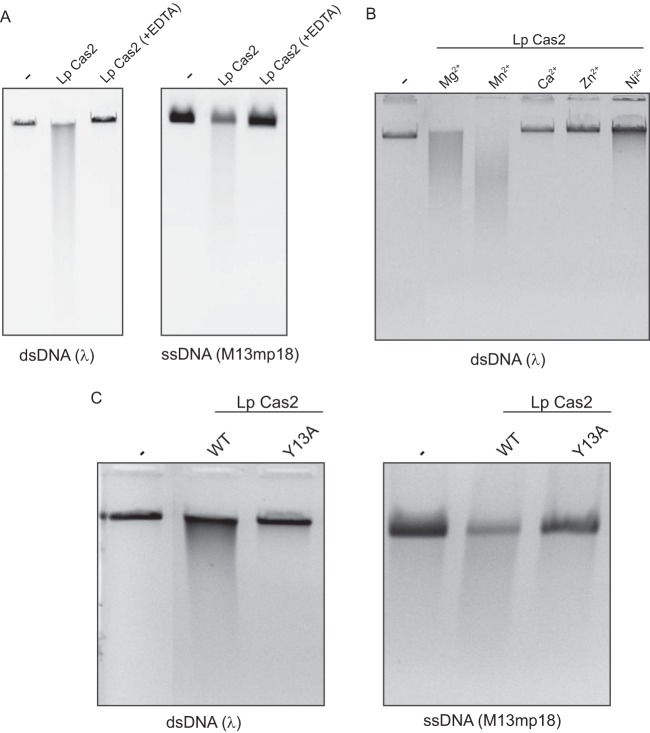
To ascertain whether the Cas2 protein of L. pneumophila also has DNase activity, we tested the ability of our purified Cas2 to cleave dsDNA and ssDNA following the protocol that had been used before to characterize the Cas2 proteins of B. halodurans and T. thermophilus (34). L. pneumophila Cas2 cleaved both the dsDNA of λ phage and the ssDNA of M13mp18 phage (Fig. 5A). No DNase activity was detected, when we assayed, in parallel, material obtained from E. coli lacking the cloned cas2 gene (data not shown). The activity of L. pneumophila Cas2 was abolished when EDTA was added to the reaction mixture (Fig. 5A), suggesting that a divalent cation is needed for DNase activity, as was the case for the RNase activity. Further assays revealed that Mg2+ and Mn2+, but not Ca2+, Zn2+, or Ni2+, facilitated DNase activity (Fig. 5B). Interestingly, there was more activity in the Mn2+-containing reactions than there was in the Mg2+-containing reactions, suggesting that Mn2+ is the preferred cation for DNase activity, whereas Mg2+ is the preferred cation for RNase activity (Fig. 3D). Further comparison of the RNase and DNase experiments suggested that L. pneumophila Cas2 is more effective as an RNase than as a DNase; e.g., substantial RNA degradation was achieved with 12.3 ng of protein during a 1-h incubation period, whereas DNA cleavage was observed with 0.5 μg of protein after a 2-h incubation. Mutant protein Cas Y13A lacked DNase activity (Fig. 5C), indicating that the tyrosine residue at position 13 is important for both DNase and RNase activity and suggesting that the catalytic site for the two activities is overlapping. In sum, L. pneumophila Cas2 possesses a DNase activity against both dsDNA and ssDNA.
FIG 5.
Cleavage of DNA substrates by L. pneumophila Cas2. (A) dsDNA of λ phage [dsDNA (λ)] and ssDNA of M13mp18 phage [ssDNA (M13mp18)] were incubated with L. pneumophila (Lp) Cas2 in the absence or presence of added EDTA, and then the reaction products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. As a negative control, DNA substrates were incubated in the absence of added protein (lanes marked −). (B) Phage λ DNA was incubated with L. pneumophila Cas2 either in buffer containing MgCl2, as described in the legend to panel A, or in buffer in which the MgCl2 was replaced with an equimolar amount of MnCl2, CaCl2, ZnCl2, or NiCl2. Reaction products, along with untreated substrate (lane −), were visualized after gel electrophoresis. (C) dsDNA of λ phage [dsDNA (λ)] and ssDNA of M13mp18 phage [ssDNA (M13mp18)] were incubated with either wild-type (WT) Cas2, as was done in the assays whose results are presented in panels A and B, or its catalytic mutant (Y13A) and then analyzed as described above. Lanes −, untreated substrate. The data presented in panels A to C are representative of those from at least three independent experiments.
The nuclease activity of Cas2 promotes amoebal infection by L. pneumophila.
To determine if the newly found nuclease activity of L. pneumophila Cas2 is required for intracellular infection of amoebae, we tested the ability of the Cas2 Y13A catalytic mutant to restore the growth of a cas2 mutant in A. castellanii (Fig. 6A). As we had observed before (15), the cas2 mutant NU411 exhibited a markedly reduced recovery at both 48 h and 72 h postinoculation, and the growth of a complemented mutant carrying an intact cas2 (i.e., pMMB2002-Cas2) was restored to the wild-type level of growth. Importantly, when a plasmid that carries the catalytic mutant of Cas2 (i.e., pMMB2002-pCas2 Y13A) was introduced into NU411, there was no improvement in the growth of the mutant. That pMMB2002-pCas2 Y13A, like pMMB2002-Cas2, did not alter the replication of parental, wild-type 130b (Fig. 6B) indicated that the inability of Cas2 Y13A to rescue the mutant was not because of a generalized, negative effect on bacterial replication. Immunoblot analysis confirmed that the wild-type and mutant forms of Cas2 were equally expressed in the L. pneumophila strains (Fig. 4B). Taken together, these data indicate that nuclease activity of Cas2 is essential for the protein's ability to promote L. pneumophila infection of amoebae.
FIG 6.
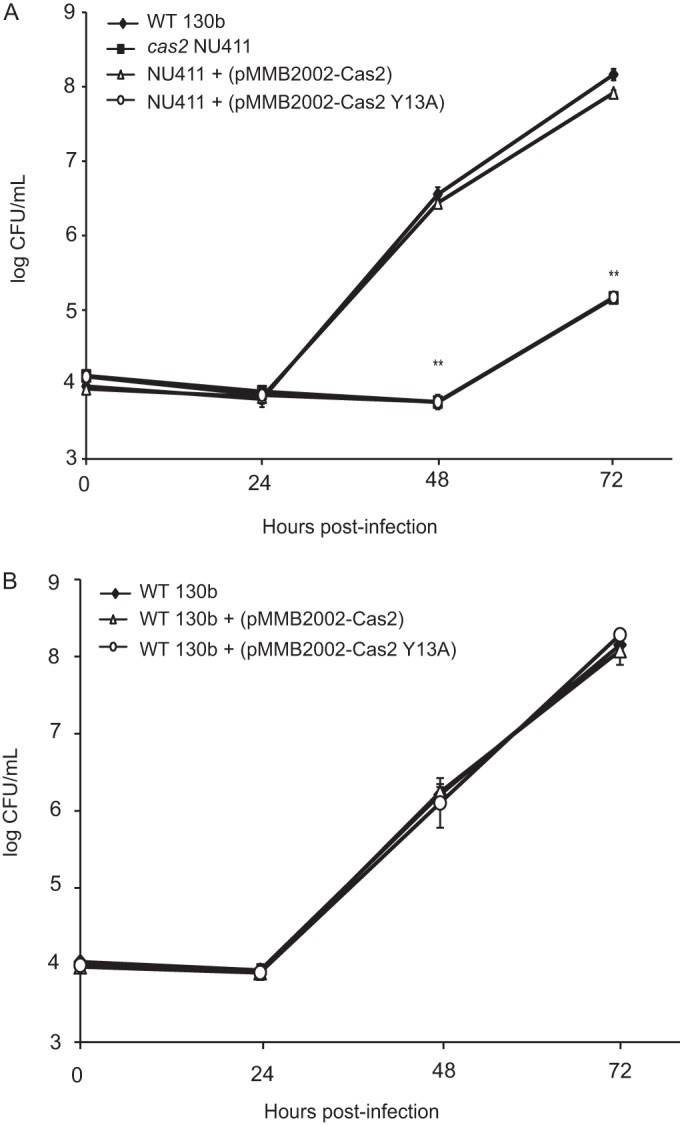
Intracellular infection of A. castellanii by L. pneumophila strain 130b and its cas2 mutant carrying plasmids that harbor wild-type Cas2 or catalytic mutant Cas2 Y13A. (A and B) Amoebae were infected with wild-type (WT) 130b, the cas2 mutant NU411, the wild-type strain carrying either a plasmid that harbors wild-type Cas2 (pMMB2002-Cas2) or a plasmid that harbors a catalytic mutant form of Cas2 (pMMB2002-Cas2 Y13A), or NU411 carrying either pMMB2002-Cas2 or pMMB2002-Cas2 Y13A, and at the indicated times, the numbers of CFU from the infected monolayers were determined. Data are means and standard deviations for four infected wells. Asterisks in panel A indicate when the recoveries of NU411 and NU411 (pCas2 Y13A) were significantly less than the recovery of the wild type (P = 0.002, Student's t test). The data in panel A are representative of those from three independent experiments, and the experiment whose results are presented in panel B was done twice, with comparable results obtained each time.
Cas2 can enhance the infectivity of an L. pneumophila strain that naturally lacks a CRISPR-Cas locus.
As has been previously noted (15, 18, 46), some strains of L. pneumophila do not possess a CRISPR-Cas locus. Therefore, we sought to determine if the introduction of cas2 into a strain that naturally lacks the CRISPR-Cas locus would render that strain more infective for an amoebal host. Among the L. pneumophila strains that do not have the locus is Philadelphia-1, a well-studied strain that, like 130b, is capable of infecting amoebal host cells. Thus, we introduced pMMB2002-Cas2 into strain Philadelphia-1 and then performed infections of A. castellanii. Remarkably, the introduction of Cas2 increased the intracellular growth of Philadelphia-1 by 40-fold at 48 h and 90-fold at 72 h (Fig. 7). In contrast, the introduction of a plasmid expressing the catalytic mutant of Cas2 did not alter the growth of Philadelphia-1 (Fig. 7). Immunoblot analysis confirmed that Cas2 and its mutant form are expressed in Philadelphia-1 and are expressed to a comparable degree (Fig. 4C). Together, these data confirm that the ability of Cas2 and its nuclease activity to enhance infection is not a peculiarity of strain 130b and is entirely independent of any other factor encoded by the CRISPR-Cas locus.
FIG 7.
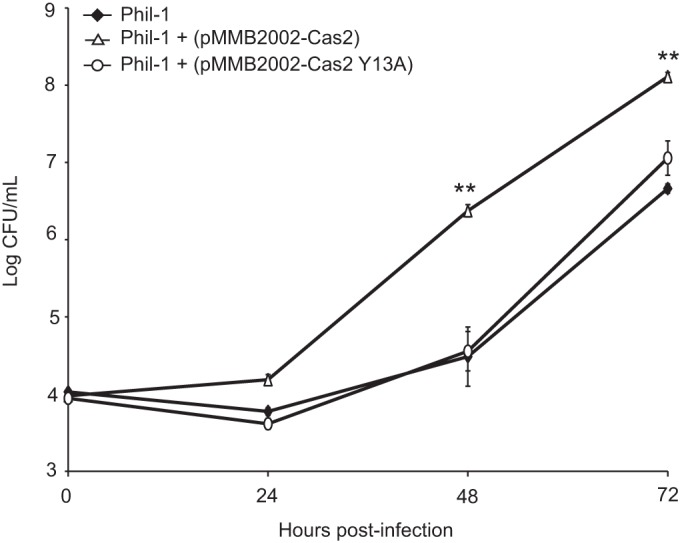
Infection of A. castellanii by L. pneumophila strain Philadelphia-1 (Phil-1) carrying the cloned cas2 gene. Amoebae were infected with wild-type strain Philadelphia-1 or Philadelphia-1 carrying a plasmid that harbors either a wild-type version of the 130b cas2 gene (pMMB2002-Cas2) or a catalytic mutant form of cas2 (pMMB2002-Cas2 Y13A), and at the indicated times, the numbers of CFU from the infected monolayers were determined. Data are means and standard deviations for four infected wells. Asterisks indicate points at which the recovery of Philadelphia-1(pMMB2002-Cas2) was significantly greater than that of Philadelphia-1 or Philadelphia-1(pMMB2002-Cas2 Y13A) (**, P = 0.006 at 48 h and P = 0.001 at 72 h, Student's t test). The data presented are representative of those from three independent experiments.
DISCUSSION
In defining the RNase and DNase activities of L. pneumophila Cas2, we have gained new insight into the types of activities that are possible with Cas2 proteins. That Cas2 of L. pneumophila exhibits activity against ssRNA is analogous to what is known for Cas2 originating from A. fulgidus, M. thermoautotrophicum, N. europaea, S. solfataricus, and T. maritima (32). On the other hand, that L. pneumophila Cas2 has activity against dsDNA is reminiscent of the activity of Cas2 from B. halodurans, S. pyogenes, and X. oryzae (34, 35). There is only one report of a Cas2 protein with activity against both ssRNA and dsDNA, and in that instance the protein from T. thermophilus was more active as a DNase (34). Thus, our demonstration that Cas2 of L. pneumophila is both an RNase and a DNase, with its greater activity being against ssRNA, represents a new type of enzymatic profile for a Cas2 protein. The ability of L. pneumophila Cas2 to cleave both ssDNA and dsDNA also denotes a new activity for a Cas2 protein. On the basis of a comparison of the crystal structures obtained for the Cas2 proteins of B. halodurans, D. vulgaris, S. pyogenes, S. solfataricus, and T. thermophilus, it has been suggested that differences in enzymatic activities might be explained by differences in certain loops within the proteins; i.e., a deeper and narrower binding cleft is linked to RNase activity, whereas a more shallow, wide groove favors DNA substrates (19, 34, 35). Hence, it will be instructive to obtain the three-dimensional structure of L. pneumophila Cas2, as it has the broadest enzyme activity among the characterized Cas2 proteins. It has also recently been suggested that the substrate preference of Cas2 enzymes might be CRISPR-Cas subtype specific (35). Our study is the first enzymatic characterization of a Cas2 protein from subtype II-B as well as from the type II group overall (47, 48). Interestingly, the type II group is the rarest bacterial CRISPR-Cas system, is absent from the archaea, and is overrepresented among commensals and pathogens, and subtype II-B is especially narrow in its distribution (22). In addition to L. pneumophila, genera that are in subtype II-B are Francisella, Parasutterella, Sutterella, Wolinella, and Leptospira.
Besides having a broad spectrum of substrates, L. pneumophila Cas2 displayed a new type of metal dependency. For RNase activity, the protein had a strong preference for Mg2+ over Mn2+, Ca2+, Zn2+, and Ni2+, and this was akin to the metal dependency of S. solfataricus Cas2, the only other Cas2 examined in this way and which favored Mg2+ over Mn2+ (32). However, for targeting dsDNA, L. pneumophila Cas2 showed its greatest activity in the presence of Mn2+, and this was in contrast to the DNase activity of B. halodurans Cas2, which had a preference for Mg2+ over Mn2+ (34). The DNase activity of L. pneumophila Cas2 was more similar to that of T. thermophilus Cas2, which exhibited a preference for Mn2+ over Mg2+ (34). Since the effect of different metals on the RNase activity of T. thermophilus Cas2 had not been studied (34), it is not possible to say if there are other differences between the Legionella and Thermus proteins. Because L. pneumophila Cas2 has both RNase and DNase activities and the two activities have different metal dependencies, it is conceivable that switches in nuclease activity within the bacterial cell might be partly dictated by the concentration of metal cofactors.
On the basis of the inability of a catalytically inactive form of Cas2 to restore the cas2 mutant of strain 130b to full infectivity for H. vermiformis, the nuclease activity of Cas2 is required for optimal infection and might fully account for the importance of Cas2 in L. pneumophila infection of amoebae. Because introduction of wild-type Cas2, but not its catalytic mutant form, into Philadelphia-1 caused that strain to be more infective, we further conclude that Cas2 facilitates the infection process independently of any other Cas protein, CRISPR array sequence, or other RNA (trans-activating crRNA [tracrRNA] and small CRISPR/Cas-associated RNA [scaRNA]) or a sequence that is encoded within the CRISPR-Cas locus, although it is possible that there might be some CRISPR-Cas-related sequences in Philadelphia-1 that have gone undetected. Since the replacement of tyrosine-13 in the mutant Cas2 abolished both RNase and DNase activities, we do not know, at this time, if it is RNase activity, DNase activity, or both activities that are critical for infection. However, given that purified, wild-type Cas2 appeared to be more effective as an RNase than as a DNase, we would suspect that RNase activity is the more important activity during infection. Although Legionella Cas2, like all previously characterized Cas2 proteins (32, 34, 35), cleaved a range of nucleic acid sequences in vitro, we posit that some degree of specificity is achieved in the context of the bacterial cell. In considering how an RNase might promote infection, two basic scenarios can be envisioned. On the one hand, Cas2 might influence the amount, stability, or conformation of a Legionella mRNA(s) that encodes a factor(s) needed for infection and/or a regulatory RNA that modulates the expression of infectivity determinants. On the other hand, it is possible that Cas2 simply degrades L. pneumophila RNA as part of a nutritional pathway needed for intracellular replication. Cas2 DNase activity could potentially facilitate infection by mediating a DNA recombination/rearrangement event or by helping to repair damaged DNA within the bacterial cell, although we previously found that the L. pneumophila cas2 mutant is not hypersensitive to UV, mitomycin C, or nalidixic acid (15). Future work will strive to identify the molecular target(s) of the Cas2 nuclease as well as the stage(s) of infection that is promoted by Cas2.
L. pneumophila Cas2 is the only Cas2 protein thus far linked to an infectious process that is unrelated to phage/plasmid immunity. However, other types of bacterial cytoplasmic nucleases have been associated with virulence and/or regulatory events. These include the CvfA endoribonuclease of S. pyogenes, the RNase AS exoribonuclease of Mycobacterium tuberculosis, and the Cas9 nuclease of F. novicida, which, along with the tracrRNA and scaRNA from the Francisella CRISPR-Cas locus, degrade the mRNA for a lipoprotein that is recognized by the innate immune system (43, 49, 50). Perhaps most relevant to our current study are the VapD-like proteins, which exist in a range of bacteria and exhibit sequence similarity to Cas2 proteins (23, 32, 44, 51, 52). Some members of this group have RNase activity, and there is a suggestion that they might inhibit bacterial translation by mediating RNA cleavage (51, 52).
By including W. magna in our study, we have gained more evidence for the importance of Cas2 for L. pneumophila survival in the environment and, thus, disease transmission. Furthermore, since the loss of Cas2 impaired the ability of strain 130b to infect three out of the four types of amoebae tested (i.e., A. castellanii, H. vermiformis, and W. magna, but not N. lovaniensis) and because those three are very different kinds of amoebae, we strongly suspect that Cas2 helps L. pneumophila strains to infect yet additional host cells. To pursue this hypothesis, future work could involve Echinamoeba exudans and Vahlkampfia jugosa, two amoebae that coexist with and are known to be permissive for L. pneumophila, as well as species of Cochliopodium, Comandonia, Filoamoeba, Neoparamoeba, Paratetramitus, Platyamoebae, Saccamoeba, Thecamoeba, Vannella, and Vexillera, which are also present in Legionella-containing waters (8, 53, 54). That some L. pneumophila isolates lack cas2 (15, 18) means that Cas2 is not absolutely required by all strains in order to persist in the environment. However, because the introduction of cas2 into Philadelphia-1 caused that strain to be more infectious, there might be a selective advantage for strains to acquire cas2, perhaps via mobile genetic elements (21, 46). Thus, in the case of L. pneumophila, there appears to be an advantage for strains to acquire and retain cas2, independently of any role in canonical CRISPR-Cas phage/plasmid immunity. Compatible with this and moving beyond L. pneumophila, cas2, along with cas1, is both the most conserved gene in CRISPR-Cas loci and subject to relatively strong purifying selection (55).
ACKNOWLEDGMENTS
We thank members of the N. P. Cianciotto lab for helpful comments and advice. We also thank both Wayne Anderson and his lab for help with cloning L. pneumophila cas2 and the Center for Structural Biology at the University of Illinois at Chicago for use of their circular dichroism spectrometer.
This work was supported by NIH grant R21 AI103451, awarded to N.P.C.
REFERENCES
- 1.Cianciotto NP, Hilbi H, Buchrieser C. 2013. Legionnaires' disease, p 147–217. In Rosenberg E, DeLong EF, Stackebrandt E, Thompson F, Lory S (ed), The prokaryotes—human microbiology, 4th ed. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 2.Parr A, Whitney EA, Berkelman RL. 8 September 2014. Legionellosis on the rise: a review of guidelines for prevention in the United States. J Public Health Manag Pract. doi: 10.1097/PHH.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pagnier I, Merchat M, La Scola B. 2009. Potentially pathogenic amoeba-associated microorganisms in cooling towers and their control. Future Microbiol 4:615–629. doi: 10.2217/fmb.09.25. [DOI] [PubMed] [Google Scholar]
- 4.Newton HJ, Ang DK, van Driel IR, Hartland EL. 2010. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas V, McDonnell G, Denyer SP, Maillard JY. 2010. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev 34:231–259. doi: 10.1111/j.1574-6976.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 6.Taylor M, Ross K, Bentham R. 2009. Legionella, protozoa, and biofilms: interactions within complex microbial systems. Microb Ecol 58:538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- 7.Buse HY, Schoen ME, Ashbolt NJ. 2012. Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Res 46:921–933. doi: 10.1016/j.watres.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Tyson JY, Vargas P, Cianciotto NP. 2014. The novel Legionella pneumophila type II secretion substrate NttC contributes to infection of amoebae Hartmannella vermiformis and Willaertia magna. Microbiology 160(Pt 2):2732–2744. doi: 10.1099/mic.0.082750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouyer S, Imbert C, Rodier MH, Hechard Y. 2007. Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ Microbiol 9:1341–1344. doi: 10.1111/j.1462-2920.2006.01229.x. [DOI] [PubMed] [Google Scholar]
- 10.Barker J, Lambert PA, Brown MR. 1993. Influence of intra-amoebic and other growth conditions on the surface properties of Legionella pneumophila. Infect Immun 61:3503–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang MG, Katayama H, Ohgaki S. 2006. Effect of intracellular resuscitation of Legionella pneumophila in Acanthamoeba polyphage cells on the antimicrobial properties of silver and copper. Environ Sci Technol 40:7434–7439. doi: 10.1021/es060412t. [DOI] [PubMed] [Google Scholar]
- 12.Dusserre E, Ginevra C, Hallier-Soulier S, Vandenesch F, Festoc G, Etienne J, Jarraud S, Molmeret M. 2008. A PCR-based method for monitoring Legionella pneumophila in water samples detects viable but noncultivable legionellae that can recover their cultivability. Appl Environ Microbiol 74:4817–4824. doi: 10.1128/AEM.02899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brieland JK, Fantone JC, Remick DG, LeGendre M, McClain M, Engleberg NC. 1997. The role of Legionella pneumophila-infected Hartmannella vermiformis as an infectious particle in a murine model of Legionnaire's disease. Infect Immun 65:5330–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berk SG, Ting RS, Turner GW, Ashburn RJ. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol 64:279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunderson FF, Cianciotto NP. 2013. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio 4(2):e00074-13. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valster RM, Wullings BA, van den Berg R, van der Kooij D. 2011. Relationships between free-living protozoa, cultivable Legionella spp., and water quality characteristics in three drinking water supplies in the Caribbean. Appl Environ Microbiol 77:7321–7328. doi: 10.1128/AEM.05575-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu BM, Huang CC, Chen JS, Chen NH, Huang JT. 2011. Comparison of potentially pathogenic free-living amoeba hosts by Legionella spp. in substrate-associated biofilms and floating biofilms from spring environments. Water Res 45:5171–5183. doi: 10.1016/j.watres.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 18.Gomgnimbou MK, Ginevra C, Peron-Cane C, Versapuech M, Refregier G, Jacotin N, Sola C, Jarraud S. 2014. Validation of a microbead-based format for spoligotyping of Legionella pneumophila. J Clin Microbiol 52:2410–2415. doi: 10.1128/JCM.00219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol 12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heler R, Marraffini LA, Bikard D. 2014. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol 93:1–9. doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondy-Denomy J, Davidson AR. 2014. To acquire or resist: the complex biological effects of CRISPR-Cas systems. Trends Microbiol 22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U. 2014. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res 42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson TR, Weiss DS. 2013. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLoS Pathog 9:e1003621. doi: 10.1371/journal.ppat.1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatoum-Aslan A, Marraffini LA. 2014. Impact of CRISPR immunity on the emergence and virulence of bacterial pathogens. Curr Opin Microbiol 17:82–90. doi: 10.1016/j.mib.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louwen R, Staals RH, Endtz HP, van Baarlen P, van der Oost J. 2014. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev 78:74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veesenmeyer JL, Andersen AW, Lu X, Hussa EA, Murfin KE, Chaston JM, Dillman AR, Wassarman KM, Sternberg PW, Goodrich-Blair H. 2014. NilD CRISPR RNA contributes to Xenorhabdus nematophila colonization of symbiotic host nematodes. Mol Microbiol 93:1026–1042. doi: 10.1111/mmi.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace RA, Black WP, Yang X, Yang Z. 2014. A CRISPR with roles in Myxococcus xanthus development and exopolysaccharide production. J Bacteriol 196:4036–4043. doi: 10.1128/JB.02035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin CY, Ratner HK, Llewellyn AC, Jones CL, Laroui H, Merlin D, Zhou P, Endtz HP, Weiss DS. 2014. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci U S A 111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, Heikema AP, Timms AR, Jacobs BC, Wagenaar JA, Endtz HP, van der Oost J, Wells JM, Nieuwenhuis EE, van Vliet AH, Willemsen PT, van Baarlen P, van Belkum A. 2013. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis 32:207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- 32.Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, Koonin EV, Edwards AM, Savchenko A, Yakunin AF. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem 283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samai P, Smith P, Shuman S. 2010. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr 66:1552–1556. doi: 10.1107/S1744309110039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nam KH, Ding F, Haitjema C, Huang Q, Delisa MP, Ke A. 2012. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J Biol Chem 287:35943–35952. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ka D, Kim D, Baek G, Bae E. 2014. Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochem Biophys Res Commun 451:152–157. doi: 10.1016/j.bbrc.2014.07.087. [DOI] [PubMed] [Google Scholar]
- 36.Stewart CR, Burnside DM, Cianciotto NP. 2011. The surfactant of Legionella pneumophila is secreted in a TolC-dependent manner and is antagonistic toward other Legionella species. J Bacteriol 193:5971–5984. doi: 10.1128/JB.05405-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brenner DJ, Steigerwalt AG, McDade JE. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med 90:656–658. doi: 10.7326/0003-4819-90-4-656. [DOI] [PubMed] [Google Scholar]
- 38.Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res 18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eschenfeldt WH, Maltseva N, Stols L, Donnelly MI, Gu M, Nocek B, Tan K, Kim Y, Joachimiak A. 2010. Cleavable C-terminal His-tag vectors for structure determination. J Struct Funct Genomics 11:31–39. doi: 10.1007/s10969-010-9082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatfield CH, Mulhern BJ, Burnside DM, Cianciotto NP. 2011. Legionella pneumophila LbtU acts as a novel, TonB-independent receptor for the legiobactin siderophore. J Bacteriol 193:1563–1575. doi: 10.1128/JB.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyson JY, Pearce MM, Vargas P, Bagchi S, Mulhern BJ, Cianciotto NP. 2013. Multiple Legionella pneumophila type II secretion substrates, including a novel protein, contribute to differential infection of amoebae Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria lovaniensis. Infect Immun 81:1399–1410. doi: 10.1128/IAI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy-Simandle K, Stewart CR, Dao J, Debroy S, Rossier O, Bryce PJ, Cianciotto NP. 2011. Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect Immun 79:1984–1997. doi: 10.1128/IAI.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maris C, Dominguez C, Allain FH. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 46.D'Auria G, Jimenez-Hernandez N, Peris-Bondia F, Moya A, Latorre A. 2010. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11:181. doi: 10.1186/1471-2164-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makarova KS, Aravind L, Wolf YI, Koonin EV. 2011. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang SO, Caparon MG, Cho KH. 2010. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect Immun 78:2754–2767. doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romano M, van de Weerd R, Brouwer FC, Roviello GN, Lacroix R, Sparrius M, van den Brink-van Stempvoort G, Maaskant JJ, van der Sar AM, Appelmelk BJ, Geurtsen JJ, Berisio R. 2014. Structure and function of RNase AS, a polyadenylate-specific exoribonuclease affecting mycobacterial virulence in vivo. Structure 22:719–730. doi: 10.1016/j.str.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 51.Kwon AR, Kim JH, Park SJ, Lee KY, Min YH, Im H, Lee I, Lee BJ. 2012. Structural and biochemical characterization of HP0315 from Helicobacter pylori as a VapD protein with an endoribonuclease activity. Nucleic Acids Res 40:4216–4228. doi: 10.1093/nar/gkr1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makarova KS, Anantharaman V, Aravind L, Koonin EV. 2012. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct 7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fields BS, Sanden GN, Barbaree JM, Morrill WE, Wadowsky RM, White EH, Feeley JC. 1989. Intracellular multiplication of Legionella pneumophila in amoebae isolated from hospital hot water tanks. Curr Microbiol 18:131–137. doi: 10.1007/BF01570838. [DOI] [Google Scholar]
- 54.Rowbotham TJ. 1986. Current views on the relationships between amoebae, legionellae and man. Isr J Med Sci 22:678–689. [PubMed] [Google Scholar]
- 55.Takeuchi N, Wolf YI, Makarova KS, Koonin EV. 2012. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol 194:1216–1225. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]