Abstract
Tick-borne spotted fever group (SFG) Rickettsia species are obligate intracellular bacteria capable of infecting both vertebrate and invertebrate host cells, an essential process for subsequent bacterial survival in distinct hosts. The host cell signaling molecules involved in the uptake of Rickettsia into mammalian and Drosophila cells have been identified; however, invasion into tick cells is understudied. Considering the movement of SFG Rickettsia between vertebrate and invertebrate hosts, the hypothesis is that conserved mechanisms are utilized for host cell invasion. The current study employed biochemical inhibition assays to determine the tick proteins involved in Rickettsia montanensis infection of tick-derived cells from a natural host, Dermacentor variabilis. The results revealed several tick proteins important for rickettsial invasion, including actin filaments, actin-related protein 2/3 complex, phosphatidylinositol-3′-kinase, protein tyrosine kinases (PTKs), Src family PTK, focal adhesion kinase, Rho GTPase Rac1, and neural Wiskott-Aldrich syndrome protein. Delineating the molecular mechanisms of rickettsial infection is critical to a thorough understanding of rickettsial transmission in tick populations and the ecology of tick-borne rickettsial diseases.
INTRODUCTION
Tick-borne rickettsial diseases caused by pathogenic bacteria belonging to the genera Rickettsia, Anaplasma, and Ehrlichia are steadily increasing in the United States (1). Infection associated with these obligate intracellular bacteria ranges from mild, self-limiting to severe, including death (2). In the typical transmission cycle, spotted fever group (SFG) rickettsiae transmit vertically during the tick life cycle stages and between vectors and vertebrate hosts during tick acquisition of a blood meal. In vertebrate hosts, mechanisms of SFG Rickettsia infection, including invasion, escape from the phagosome, intracellular growth, intracellular movement, and cell-to-cell spread, have been studied (3). Tick acquisition of SFG Rickettsia from rickettsemic hosts is poorly described, and despite the essential role for tick hosts in maintenance and transmission of SFG Rickettsia to vertebrate hosts, the biology of the tick cell-SFG Rickettsia interaction is understudied.
The obligate intracellular nature of Rickettsia requires bacteria to invade both vertebrate and invertebrate host cells, and this process, host cell invasion by SFG Rickettsia, has been studied in mammalian and Drosophila cell lines. Mammalian ligands Ku70 and α2β1 integrin interact with rickettsial outer membrane proteins B and A (OmpB and OmpA), respectively, and are involved in Rickettsia conorii invasion of mammalian host cells (4, 5). After binding to rickettsial OmpB, R. conorii induces signaling cascades in which Ku70 is initially ubiquitinated by c-Cbl ubiquitin ligase. Subsequently, signaling molecules, including Cdc42, protein tyrosine kinases (PTKs), phosphatidylinositol-3′-kinase (PI3-kinase), Src family tyrosine kinases (Src), focal adhesion kinase (FAK), and cortactin, coordinately activate the actin-related protein 2/3 (Arp2/3) complex. Activation of the Arp2/3 complex leads to actin polymerization and recruitment of the components of endocytic pathway, including clathrin and caveolin-2, at the bacterial entry site. The rearrangement of the actin cytoskeleton results in membrane extrusion and subsequent bacterial internalization into host cells (4, 6–8).
Differences between host factors central for rickettsial uptake in invertebrate- versus vertebrate-derived cells are present for Rickettsia parkeri. Assessed by an RNA interference (RNAi) screening approach, the GTPases Rac1 and Rac2, the WASP (Wiskott-Aldrich syndrome protein) family verprolin homologous protein (WAVE) nucleation-promoting factor complex, and the Arp2/3 complex contribute to the invasion process in Drosophila S2R+ cells. The Arp2/3 complex is also essential to rickettsial invasion of mammalian cells, yet the requirement of WAVE2 and Rho GTPases occurs in a cell-type-specific manner (9). These findings indicate a role for the Arp2/3 complex, WAVE, and Rho GTPases, with a degree of host-dependent variation, in SFG Rickettsia invasion of vertebrate and invertebrate cells.
The molecular mechanism of rickettsial invasion of tick cells has been examined in cell culture models and in tick tissues. In a tick-derived cell line, histone H2B facilitates the uptake of Rickettsia felis into host cells in vitro (10), consistent with a DNA-binding molecule serving as a receptor in mammalian cells (4). A homolog to mammalian vacuolar-ATPase (V-ATPase) was identified in Dermacentor variabilis, and a V-ATPase inhibition assay was used to demonstrate a role for V-ATPase in rickettsial internalization in a tick-derived cell line (11). Similarly, a recent molecular and functional characterization of the D. variabilis Arp2/3 complex identified a relatively conserved molecule that is involved in rickettsial entry. The Arp2/3 complex contributes to host cell invasion by Rickettsia montanensis in the natural arthropod vector, as determined using an ex vivo biochemical inhibition assay (12). Interestingly, downregulation of transcripts of α-catenin, a molecule associated with actin organization, in ticks upon rickettsial exposure suggests that ticks may actively respond to rickettsial infection by limiting the utilization of α-catenin (13). The presence of conserved molecules in multiple host backgrounds allows for further characterization in vector backgrounds.
The movement of SFG Rickettsia between vertebrate and invertebrate host cells during the transmission cycle suggests that conserved mechanisms are utilized for cell invasion in both hosts. To further delineate the process of SFG Rickettsia infection in natural arthropod vectors, it is necessary to identify the tick molecules involved in SFG Rickettsia invasion of host cells. A panel of inhibitors which impact components of the signal cascade, including actin polymerization, Arp2/3 complex, PTKs, and PI3-kinase, and impair rickettsial infection in both mammalian and insect cell lines (6, 9) was incorporated into biochemical inhibition assays to identify host proteins involved in the uptake of rickettsiae into tick cells. Because ticks play an important role as vectors and reservoirs in the ecology of rickettsiae, identification of tick-derived molecules associated with rickettsial invasion is essential to a thorough understanding of rickettsiosis.
(This work was conducted by N. Petchampai in partial fulfillment of the requirements for a Ph.D. from Louisiana State University, Baton Rouge, LA, 2013.)
MATERIALS AND METHODS
Cell culture and Rickettsia preparation.
Vero cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Invitrogen) and containing 5% heat-inactivated fetal bovine serum (FBS; HyClone) in a humidified 5% CO2 incubator at 34°C. Embryonically derived D. variabilis cells (DVE1) were grown in L15C medium (14) supplemented with 10% FBS, 5% tryptose phosphate broth (Difco), 0.1% lipoprotein-cholesterol concentrate (LPC; MP Biomedicals), 6 mM HEPES buffer (Sigma-Aldrich), and 0.06% sodium bicarbonate (Sigma-Aldrich) without CO2 at 32°C, as previously described (11).
As described by Sunyakumthorn et al. (13), R. montanensis was propagated in Vero cells and maintained in a humidified 5% CO2 incubator at 34°C. For each experiment, rickettsiae were purified using a modified protocol of Weiss et al. (15) as previously described (11). Briefly, Rickettsia-infected cells were detached using a sterile cell scraper (Sarstedt) and lysed by vortexing with sterile 3-mm borosilicate glass beads (Sigma-Aldrich) for 5 min. Cell lysate was then transferred aseptically to 15-ml centrifuge tubes and centrifuged at 4°C and 275 × g for 3 min to pellet cellular debris. The supernatant was transferred to a 10-ml syringe and filtered through a 2-μm-pore-size syringe filter. For all bioassays, rickettsiae were enumerated with a Live/Dead BacLight bacterial viability kit (Molecular Probes) in a Petroff-Hausser bacterial counting chamber (Hausser Scientific) and examined with a Leica microscope (16).
Database search for putative tick molecules.
The tBlastx algorithm was used to search publicly available D. variabilis transcriptome databases (GenBank accession numbers SRX018179, SRX001955, and SRX001954) (17) for partial transcripts with high similarity to molecules previously determined to be involved in the rickettsial invasion of mammalian or Drosophila cells. Nucleotide sequences from Ixodes scapularis, Drosophila melanogaster, or Homo sapiens for Src, N-WASP, Rac1, Cdc42, Rho1, PTK, PI3-kinase, and FAK were used for the search and analysis. Percent identity of resultant partial transcripts to the query sequences was determined using ClustalW across the aligned sequences.
Biochemical inhibition assays.
All inhibitors used in this study (Table 1) were purchased from EMD Chemicals, and dimethyl sulfoxide (DMSO) was obtained from Sigma-Aldrich. Effect of the inhibitors on tick cell viability was assessed by incubating tick cells with the highest concentration of each inhibitor used for 3 h prior to staining the cells with trypan blue (Gibco) and assessed by exclusion analysis. As previously described (11), DVE1 cells (1 × 105) were seeded onto 96-well plates and incubated at 32°C for 48 h. The cells were treated with three different concentrations of individual inhibitors or the control inhibitor vehicle (complete L15C medium or medium containing 1% DMSO) for 2 h. The experiments were performed in quadruplicate for each concentration of the inhibitor used, and the results were the combination of two independent experiments. The effect of DMSO on rickettsial infection of tick cells was determined by incubating cells with two concentrations of DMSO (0.1% and 1%) prior to performing biochemical inhibition assays. Treated cells were exposed to R. montanensis at a multiplicity of infection (MOI) of 10, and the plate was centrifuged at 700 × g for 2 min to facilitate rickettsial contact with host cells. After 1 h, unbound rickettsiae in the medium were removed, and host cells were harvested by pipetting 150 μl of phosphate-buffered saline (PBS) onto the cells several times; the suspension was centrifuged at 275 × g for 4 min. The cell pellet was washed with 1 ml of PBS and centrifuged at 275 × g for 4 min. The samples were stored at −20°C until used for genomic DNA (gDNA) isolation.
TABLE 1.
List of inhibitors, target molecule(s), and concentrations used in rickettsial invasion bioassays
| Inhibitor | Tick target molecule(s) | Concentrations used (μM) |
|---|---|---|
| Zygosporium mansonii cytochalasin Db | Actin polymerization | 1, 10, 100 |
| CK-666b | Arp2/3 complex | 5, 50, 500 |
| 187-1 | N-WASP | 1, 10, 100 |
| Clostridium difficile toxin Ba | Rho GTPases (Rho, Cdc42, and Rac) | 0.00001, 0.0001, 0.001 |
| Rac1 inhibitora | Rac1 | 10, 100, 1000 |
| Genisteinb | PTKs | 5, 50, 500 |
| PP2b | Src family PTKs | 2.5, 25, 250 |
| FAK inhibitor I | FAK | 5, 50, 500 |
| PI3-kinase inhibitor XI, HWT | PI3-kinase | 5, 50, 500 |
Dissolved in complete L15C medium, not 1% DMSO.
Known activity in Drosophila cell line (9).
To determine if rickettsial invasion of tick cells occurs through an active process, cells preincubated in medium supplemented with 1% DMSO were exposed, in triplicate, to either live or formalin-fixed Rickettsia (MOI of 10). Exposed cells were collected and washed as described above, and the cell pellet was stored for gDNA isolation. The results are a combination of two independent experiments.
For each sample, gDNA was extracted using a DNeasy blood and tissue kit (Qiagen) according to the manufacturer's instructions with the DNA eluted in 35 μl of DNase/RNase-free water. Numbers of rickettsiae and tick cells were then quantified by probe-based quantitative PCR (qPCR), measuring genomic equivalents of rickettsial OmpB and tick calreticulin genes, as previously described (11). Percent relative invasion was calculated by comparing the ratios of rickettsial genes to tick genes between inhibitor and inhibitor vehicle-treated groups.
Statistical analysis.
The SAS statistical package (version 9.3) was used for statistical analysis. For biochemical inhibition assays, two-way analysis of variance (ANOVA) was conducted using the general linear model (GLM) procedure. Pairwise t tests of least-squares means were used to examine the interaction effects of inhibitors, or inhibitor vehicle, on Rickettsia invasion of tick cells. The TTEST procedure was performed to compare percentages of relative invasion of live and formalin-fixed rickettsiae. P values of ≤0.05 were considered significantly different.
RESULTS AND DISCUSSION
Transcriptional evidence for tick molecules related to factors associated with rickettsial invasion in other cell types.
Ticks serve as both the transmission vectors and reservoirs for SFG Rickettsia; however, characterization of the molecular mechanisms by which SFG Rickettsia infects ticks is limited. With the paucity of genomic data available for D. variabilis, the presence of putative molecules can be inferred from analysis of published transcriptomes. Toward determining if molecules targeted by the inhibitors used in this study were present in D. variabilis, sequences for D. variabilis beta-actin (GenBank accession number EF488512.2) and the Arp2/3 complex (GenBank accession numbers KF780484.1, KF780485.1, KF780486.1, KF780487.1, KF780488.1, KF780489.1, and KF780490.1) were identified in GenBank. For the remaining molecules, partial transcripts for Src, Rac1, Cdc42, Rho1, PTK, PI3-kinase, and FAK, but not N-WASP, were identified in D. variabilis transcriptome databases (see Table S1 in the supplemental material). Although comprehensive analyses (11–13, 18–21) confirmed gene sequences for some of the molecules, the partial transcript sequence data for several D. variabilis molecules suggest a level of identity between ticks and other invertebrates relative to cytoskeletal molecules. The specificity of the inhibitors on tick proteins and undefined downstream effects influencing rickettsial invasion cannot be confirmed without detailed molecular and functional characterization of each target molecule; however, the combined characterization of inhibitors in mammalian and Drosophila systems and corresponding identification of putative factors in the current study suggest the potential for specific activity in ticks.
Invasion of tick cells occurs through a process dependent on live Rickettsia.
In previous studies, functional and transcriptional analyses of tick-derived molecules utilizing whole organs in ticks (22) and in an ex vivo bioassay (12) provided advantages regarding natural infection targets but were limited by the constraints of whole-organ culture and controlling the parameters of infection, including consistent rickettsial exposure to all cells. An alternate approach employed in the current study utilizes the D. variabilis-derived DVE1 embryonic cell line. It is recognized that tick-derived cell lines comprise multiple cell types (23); with subculturing, the diversity of cell types in the line decline, and one or two cell types become dominant (24). A morphologically distinct cell type described in other tick cell lines was noted to contain apparent phagocytic activity as a portion of nonviable rickettsiae were able to invade cells (25). To discern if invasion is active or a result of phagocytic activity, rickettsiae can be fixed in formalin, which may alter the conformation of some components on the rickettsial surface but still allows rickettsiae to adhere to host cells (26). In the current study, Rickettsia internalization assays were performed using either live or formalin-fixed, nonviable R. montanensis, similar to a previously described protocol for an R. parkeri invasion bioassay (9). Formalin-fixed rickettsiae were intracellular significantly less (75%; P < 0.0001) than live rickettsiae, suggesting that rickettsial entry into tick cells occurs through an active Rickettsia-specific process (Fig. 1).
FIG 1.
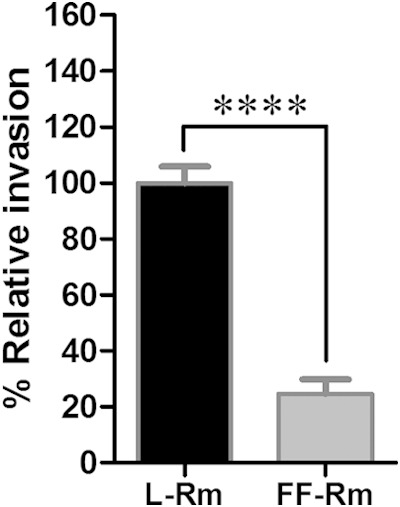
Invasion of tick cells occurs through a process dependent on live Rickettsia. Live (L-Rm) or formalin-fixed (FF-Rm) R. montanensis was used to infect DVE1 cells preincubated in medium supplemented with 1% DMSO. After 1 h, Rickettsia was removed, and the cells were washed twice with PBS. The samples were collected by low-speed centrifugation, and gDNA was extracted. Mean (± standard error of the mean) percentages of relative invasion, calculated as the ratio of rickettsial OmpB and tick calreticulin gene copy numbers, were compared between formalin-fixed and live R. montanensis samples. The experiments were performed in triplicate for each group, and the results are a combination of the results from two independent experiments. ****, P < 0.0001.
Tick-derived cells in biochemical inhibition assays.
The objective of this study was to use biochemical inhibition assays to elucidate the molecular mechanisms underlying rickettsial invasion of tick cells. The DVE1 cell line was used to study the factors influencing Rickettsia peacockii infectivity (16); in the current study, the R. montanensis and DVE1 cell model represents a natural SFG Rickettsia and tick pairing (27–31). The concentration of inhibitors used in each experiment was based on the literature, solubility, and the effect of the inhibitors on tick cell viability. At the highest concentration of each inhibitor, tick cell viability was not affected when assessed by a trypan blue exclusion assay. Because most inhibitors were reconstituted in DMSO, the concentration of DMSO was optimized to minimize the influence of DMSO in the assays. Similar to the chemical inhibitors, DMSO at concentrations up to 1% of culture medium did not affect cell viability. To assess the influence of DMSO on rickettsial invasion, tick cells were seeded, treated with 0.1% or 1% DMSO, infected with Rickettsia, and processed for gDNA extraction. Real-time qPCR (11) was used to enumerate the number of intracellular rickettsiae and tick cells. Compared to cells without DMSO, at DMSO concentrations of 0.1% or 1%, rickettsial infection did not vary by more than 12% (see Fig. S1 in the supplemental material). Combined, neither the vehicle nor the inhibitor had a direct effect on DVE1 cells, indicating that any decrease in cell infection was not due to cell death.
Host actin is required for R. montanensis invasion of tick cells.
Well-characterized pathogens such as Listeria, Shigella, and Yersinia utilize host cell actin to enter cells (32–35). Similarly, actin polymerization is important for rickettsial invasion of Drosophila and mammalian cells (6, 9); thus, the role of actin polymerization was examined for R. montanensis infection of tick cells. An inhibitor of actin polymerization, cytochalasin D, was used to treat DVE1 cells for 2 h prior to infection with R. montanensis for 1 h. The results showed that disruption of actin polymerization in tick cells significantly decreased percent relative rickettsial invasion to 40% (P < 0.0001), 51% (P < 0.0001), and 70% (P = 0.0001) at 100, 10, and 1 μM concentrations of the inhibitor used, respectively (Fig. 2). Consistently with rickettsial invasion of cells from other host backgrounds, R. montanensis invasion of tick cells occurs through a process that depends on tick actin.
FIG 2.
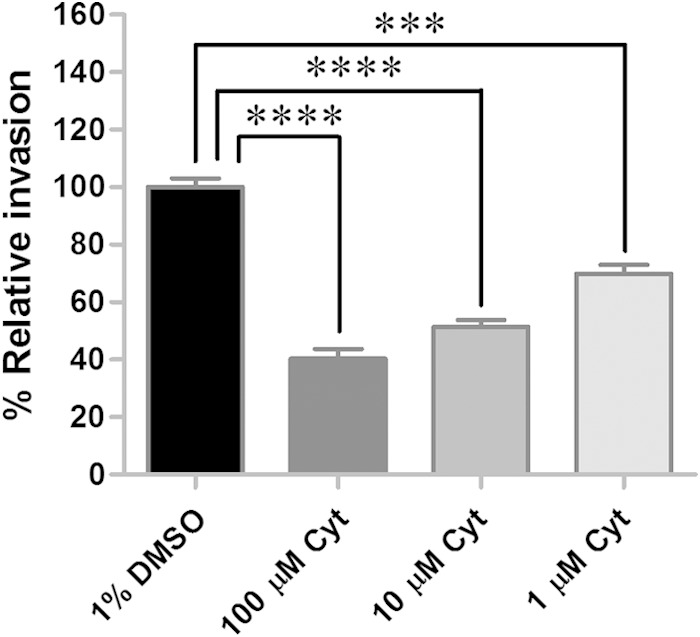
Actin polymerization is essential for R. montanensis invasion of tick cells. Tick cells were treated with an actin depolymerizing agent, cytochalasin D (Cyt). Numbers of intracellular rickettsiae and host cells were quantified by qPCR, and the mean (± standard error of the mean) percent relative invasion of each treatment was compared to that in the untreated control. All treatments were performed in quadruplicate for each experiment, and the results are a combination of the results from two independent experiments. ****, P < 0.0001; ***, P = 0.0001.
The Arp2/3 complex is important for R. montanensis invasion of DVE1 cells.
The Arp2/3 complex is a seven-subunit protein capable of nucleating actin filaments (36, 37); a homologous complex was recently identified in D. variabilis (12). For several bacterial pathogens, the Arp2/3 complex plays a role in invasion of host cells (38–45). Previous in vitro studies revealed the importance of the Arp2/3 complex in rickettsial internalization in Drosophila and mammalian cells (6, 9), as well as in whole tick organs ex vivo (12). To examine whether the molecule is essential for the entry of R. montanensis into tick cells, DVE1 cells were treated with CK-666, an Arp2/3 complex inhibitor, at 500, 50, and 5 μM for 2 h before being infected with R. montanensis for 1 h. Compared to the controls (no inhibitor, vehicle only), inhibition of the Arp2/3 complex significantly reduced (P < 0.0001) the percent relative invasion to 8% at the highest concentration of the inhibitor used (Fig. 3). Therefore, based on studies in varied host cell backgrounds, the Arp2/3 complex is important for rickettsial invasion of cells in both mammalian and vector hosts.
FIG 3.
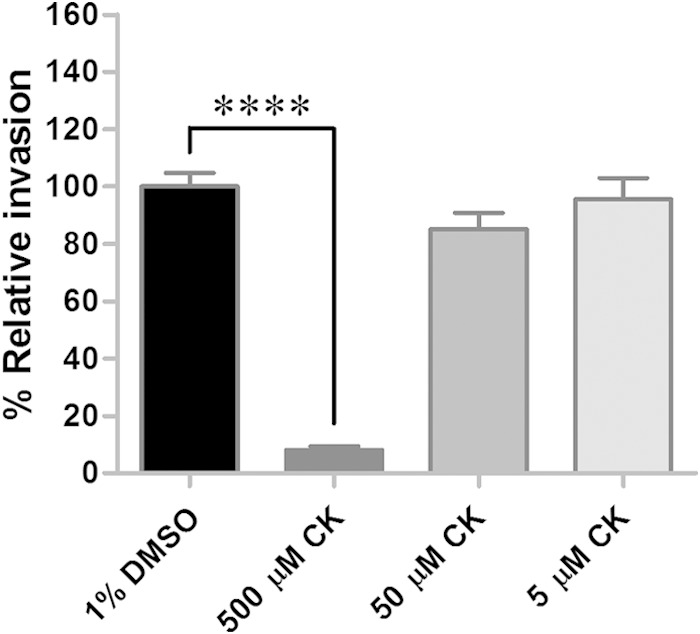
Arp2/3 complex is important for rickettsial internalization of DVE1 cells. Tick-derived DVE1 cells were treated with CK-666 (CK), an inhibitor of the Arp2/3 complex, at various concentrations, and the mean (± standard error of the mean) percent relative invasion of each treatment was compared to that of the untreated (vehicle only) control. All treatments were assessed in quadruplicate for each experiment, and the results are a combination of the results from two independent experiments. ****, P < 0.0001.
Inhibition of N-WASP has a moderate effect on R. montanensis invasion of DVE1 cells.
N-WASP is a cytoskeleton regulator that promotes actin nucleation by binding to and activating the Arp2/3 complex (46, 47). For some pathogens, such as Yersinia pseudotuberculosis (48) and Listeria monocytogenes (49), N-WASP facilitates internalization into host cells. Interestingly, it is not required for R. parkeri invasion of Drosophila and mammalian cells (9). To investigate the role of N-WASP in R. montanensis entry into tick cells, the N-WASP inhibitor 187-1 was used to treat tick cells at three different concentrations (100, 10, and 1 μM) prior to exposure to R. montanensis. At the highest concentration of 187-1, a moderate (20%), but significant (P = 0.0092), decrease in R. montanensis entry was observed for treated cells compared to that in the control (Fig. 4). The modest effect of the inhibitor on rickettsial invasion of tick cells may be due to multiple factors, including the inability for the inhibitor to affect tick N-WASP or poor solubility of the inhibitor in the medium, or, as with insect cells, the rickettsiae may utilize an alternate pathway (e.g., Rac-dependent) for infection.
FIG 4.
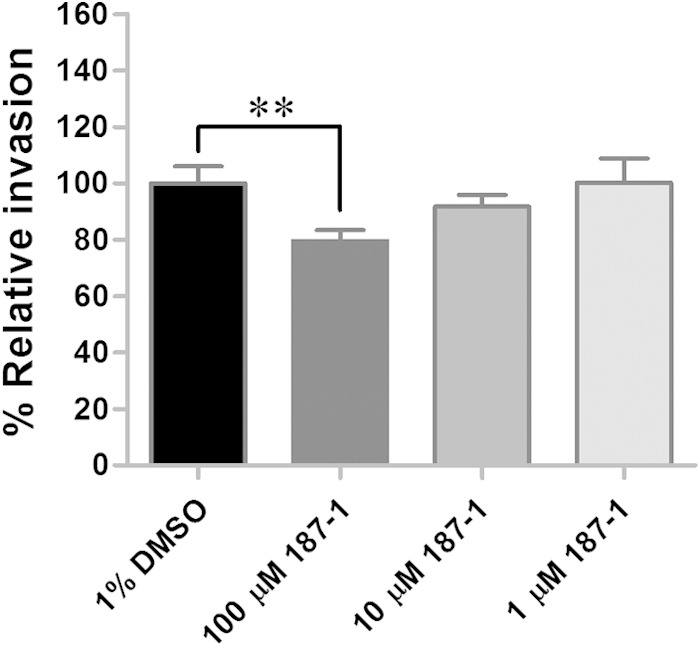
Inhibition of N-WASP slightly affects the ability of R. montanensis to invade tick cells. The mean (± standard error of the mean) percent relative invasion by R. montanensis was reduced when DVE1 cells were pretreated with the N-WASP inhibitor, 187-1. Inhibition by 187-1 was determined in quadruplicate for each experiment, and the results are a combination of the results from two independent experiments. **, P = 0.0092.
Rho GTPase, Rac1, mediates R. montanensis uptake into tick cells.
Of the 22 Rho family GTPases, Cdc42 and Rac are the upstream signaling molecules that activate N-WASP and WAVE family proteins (50, 51). Cdc42 has been shown to trigger N-WASP, and Rac activates WAVE; interactions of these two pairs of molecules lead to Arp2/3 complex-mediated actin polymerization (52). In R. conorii, Cdc42 facilitates bacterial entry into mammalian cells (6); however, in R. parkeri, the Rho GTPases Rac1 and Cdc42 were proposed to cooperatively stimulate actin polymerization, leading to rickettsial internalization (9). The minimal effect that disruption of N-WASP had on rickettsial invasion of tick cells suggested that further examination was required of the parallel pathway, the Rac pathway, to N-WASP-mediated Arp2/3 complex activation. Using a broad-spectrum Rho family GTPase inhibitor, Clostridium difficile toxin B, which inhibits Rho, Rac, and Cdc42, demonstrated that inhibition of Rho GTPases with C. difficile toxin B had no significant effect on R. montanensis invasion of DVE1 cells (1 to 13% decrease in rickettsial invasion; P > 0.05) (Fig. 5A). To determine if ineffectiveness of the broad-range inhibitor was an issue of potency or specificity of the inhibitor, Rac1 was specifically targeted using the Rac1 inhibitor. As shown in Fig. 5B, inhibition of Rac1 significantly decreased the percent relative R. montanensis invasion of tick cells compared to that in the untreated control by 38% (P < 0.0001) at a 1 mM concentration of the inhibitor used. Consistently with the suggested role of Rac in R. parkeri, L. monocytogenes, and Candida albicans invasion of mammalian and Drosophila cells (9, 43, 53), Rac1 inhibition correlates with R. montanensis infection in tick cells. In contrast, experiments employing dominant negative Rac1 mutants suggested that Rac was not involved in R. conorii invasion of Vero cells (6). A possibility for the discrepancies between studies may relate to a severe phenotype that disrupts guanine nucleotide exchange factors for GTPase proteins in the Rac1 mutants, affecting alternate molecules also essential to rickettsial invasion (9). While additional methods to assess the role of Rac1 in different host backgrounds are needed, both Rac1 and N-WASP were identified to play a significant role in rickettsial invasion of tick cells.
FIG 5.
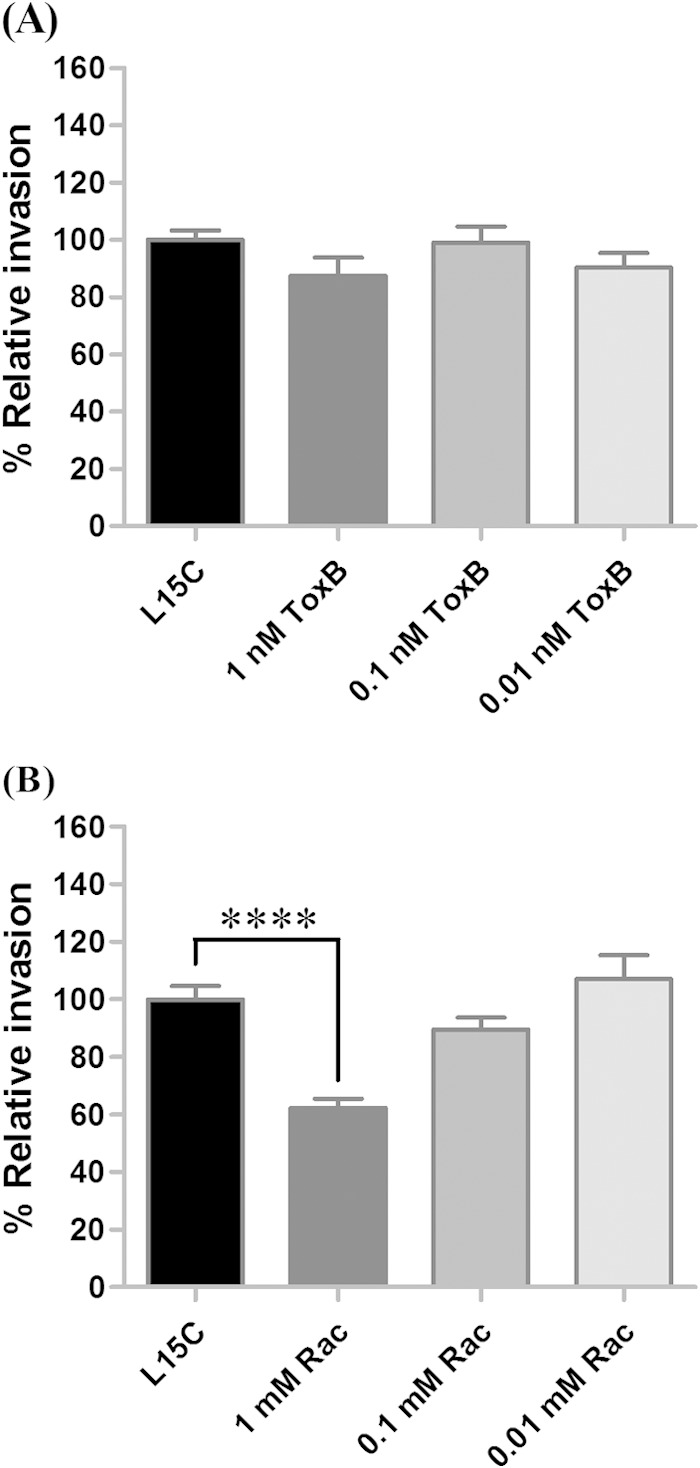
The Rho GTPase Rac1 facilitates R. montanensis entry into tick cells. DVE1 cells were treated with C. difficile toxin B (ToxB), a general inhibitor for Rho GTPases (A), or with the Rac1 inhibitor (Rac) (B), and the mean (± standard error of the mean) percent relative rickettsial invasion of each treatment was compared to that of the untreated (vehicle only) control. Each concentration of inhibitor was assessed in quadruplicate, and means represent a combination of results from two independent experiments. ****, P < 0.0001.
Protein tyrosine kinases play a role in R. montanensis invasion of DVE1 cells.
Studies utilizing SFG Rickettsia (6, 9) have illustrated that phosphorylation of proteins on tyrosine residues mediates internalization of bacteria into host cells. In the present study, three different concentrations (500, 50, and 5 μM) of genistein, a specific inhibitor of tyrosine-specific protein kinases, was used in our bioassay, and the inhibition of general tyrosine kinases reduced the ability of rickettsiae to invade tick cells compared to their ability to invade the untreated control (Fig. 6A). The significant reduction of invasion by 69% (P < 0.0001) and 23% (P = 0.0039) at 500 and 50 μM concentrations of the inhibitor used, respectively, indicated a role for PTKs in R. montanensis internalization of tick cells.
FIG 6.
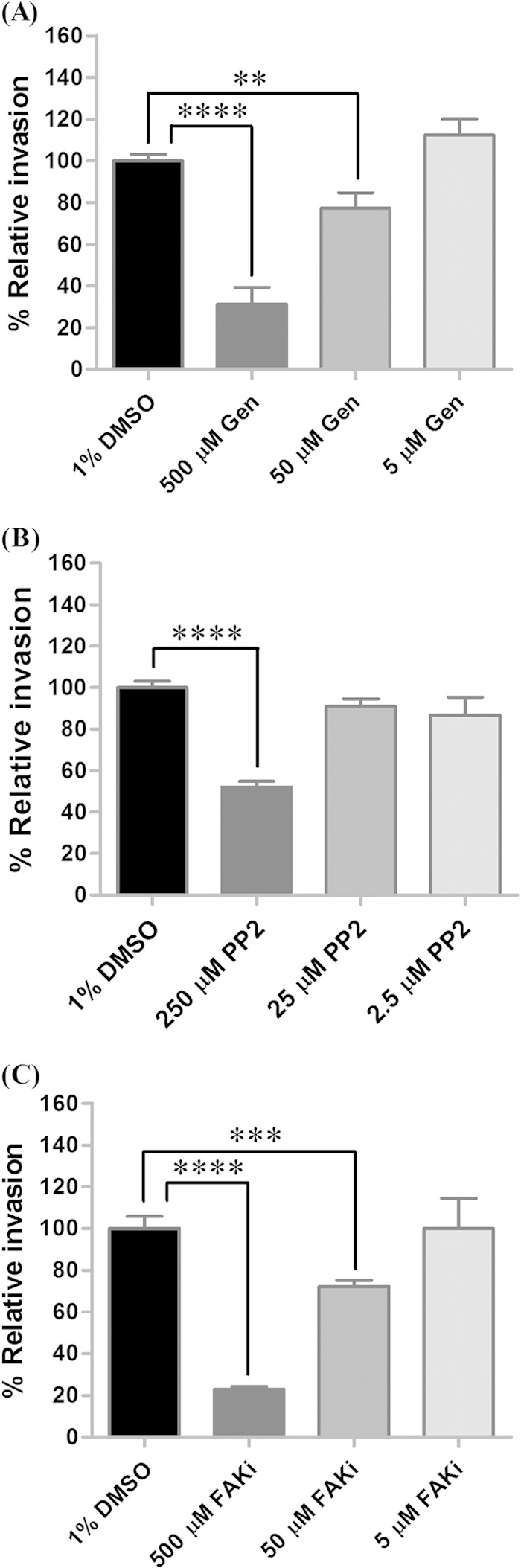
Protein tyrosine kinases play a role in R. montanensis invasion of DVE1 cells. Tick cells were treated with protein tyrosine kinase (PTK) inhibitors, including genistein (Gen), a general PTK inhibitor (A), PP2, an inhibitor of the Src family of PTKs (B), and the focal adhesion kinase inhibitor (FAKi) (C). The mean (± standard error of the mean) percent relative rickettsial invasion of each treatment group was compared to that of the untreated controls. Each treatment was assessed in quadruplicate for each experiment, and bars represent the combination of results from two independent experiments. ****, P < 0.0001; ***, P = 0.0004; **, P = 0.0039.
Invasion of mammalian cells by R. conorii revealed the involvement of Src and FAK in bacterial uptake, analogous to what occurs with Y. pseudotuberculosis, uropathogenic Escherichia coli (UPEC), or Shigella flexneri internalization into host cells (6, 54–57). The Src and FAK family kinases were shown to regulate actin cytoskeleton reorganization (58, 59); therefore, the importance of these molecules in R. montanensis invasion of tick cells was examined in this study. As shown in Fig. 6B, inhibition of Src family PTKs by PP2 significantly decreased (P < 0.0001) the percent relative rickettsial invasion to 52% at a 250 μM concentration of the inhibitor used. Similarly, disruption of FAK significantly reduced the ability of R. montanensis to invade tick cells to 23% (P < 0.0001) and 72% (P < 0.0001) at 500 and 50 μM concentrations of the inhibitor, respectively, compared to its ability to invade the untreated control (Fig. 6C). The results suggest that Src and FAK are important for R. montanensis internalization into DVE1 tick cells. However, these findings differ from the study of R. parkeri invasion of Drosophila and mammalian cells in which a specific tyrosine kinase, Src, was nonessential for the entry of the bacteria (9). The potential for either Rickettsia species or host-specific utilization of Src and FAK requires further study.
Inhibition of phosphatidylinositol-3′ kinase limits R. montanensis invasion of DVE1 cells.
PI3-kinase is involved in actin cytoskeleton remodeling (60), and R. conorii invasion of mammalian cells associates PI3-kinases with rickettsial uptake (6). To determine the role of the PI3-kinase in R. montanensis invasion of tick cells, three different concentrations (500, 50, and 5 μM) of PI3-kinase inhibitor XI, a wortmannin 17β-hydroxy analog (HWT), were assessed. The results showed a significant decrease (P < 0.0001) in R. montanensis invasion of DVE1 cells compared to that of the untreated control; the percent relative invasion was reduced to 40% at the highest concentration of the inhibitor used (Fig. 7). Although PI3-kinase is not required for R. parkeri invasion of Drosophila cells and some mammalian cell types (9), the results presented here indicate that PI3-kinase facilitates R. montanensis uptake into tick cells, consistent with R. conorii internalization into host cells. Further studies to demarcate the role of PTKs and PI3-kinase in other species of SFG Rickettsia invasion of tick cells are required.
FIG 7.
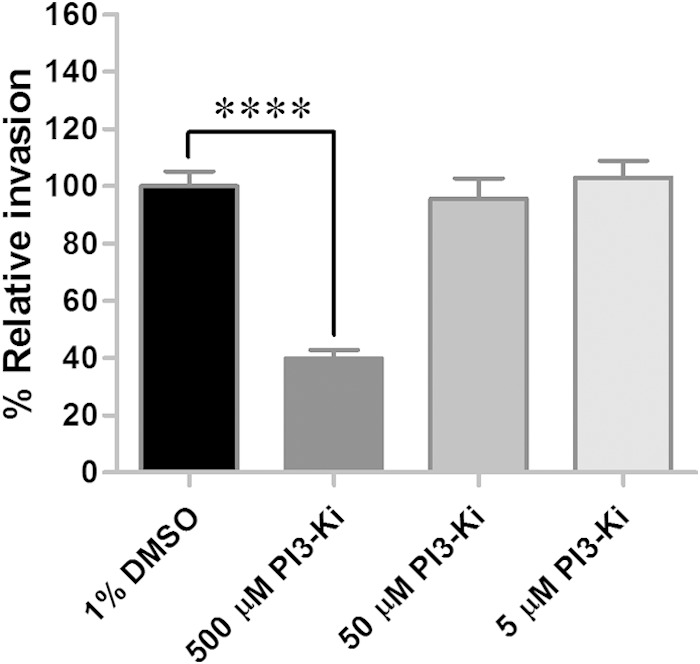
Inhibition of phosphatidylinositol-3′-kinase limits R. montanensis entry into tick cells. DVE1 cells were treated with PI3-kinase inhibitor XI, HWT (PI3-Ki), and the percent relative rickettsial invasion (mean ± standard error of the mean) for each treatment group was determined in quadruplicate for each experiment, and results are the combination of results from two independent experiments. ****, P < 0.0001.
In summary, tick signaling molecules associated with R. montanensis invasion were characterized using a tick and SFG Rickettsia pairing commonly observed in field-collected ticks (27–31). Utilization of the natural host cell background may illuminate unique pathways or identify novel rickettsial mechanisms for cell invasion. Consistently with the studies in mammalian and Drosophila cell lines and whole tick tissues (6, 9, 12), the Arp2/3 complex and actin are the central molecules activated during the entry of R. montanensis into tick cells. The upstream signaling proteins cooperating to regulate the Arp2/3 complex are similar but not identical between host cell backgrounds. Although differences in SFG Rickettsia species, molecules targeted, and techniques used in this and previous studies make comparing the means of invasion of mammalian, Drosophila, and tick cells by SFG Rickettsia difficult, it can be concluded that conserved mechanisms with some degree of variation are utilized in SFG Rickettsia invasion of vertebrate and invertebrate cells. The bioassay described here can be expanded to examine alternate tick-derived cell lines and rickettsial pathogens to ascertain if the differences are host background or rickettsia dependent. To expand upon the current findings using the in vitro model presented, further studies are required to investigate the mechanisms by which SFG Rickettsia manipulates tick proteins to facilitate rickettsial infection of tick hosts and transition between ticks and vertebrate hosts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Timothy Kurtti and Ulrike Munderloh for DVE1 cells. We also thank Jacqueline Macaluso for helpful comments.
This research was supported by National Institutes of Health grant AI077784 (K.R.M.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02888-14.
REFERENCES
- 1.Dumler JS., Jr 2010. Fitness and freezing: vector biology and human health. J Clin Invest 120:3087–3090. doi: 10.1172/JCI44402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker DH, Ismail N. 2008. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol 6:375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- 3.Welch MD, Reed SC, Haglund CM. 2012. Establishing intracellular infection: escape from the phagosome and intracellular colonization (Rickettsiaceae), p 154–174. In Palmer GH, Azad AF, Tan M (ed), Intracellular pathogens II: Rickettsiales. ASM Press, Washington, DC. [Google Scholar]
- 4.Martinez JJ, Seveau S, Veiga E, Matsuyama S, Cossart P. 2005. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell 123:1013–1023. doi: 10.1016/j.cell.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 5.Hillman RD Jr, Baktash YM, Martinez JJ. 2013. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with α2β1 integrin. Cell Microbiol 15:727–741. doi: 10.1111/cmi.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez JJ, Cossart P. 2004. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci 117:5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- 7.Chan YG, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol 11:629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan YG, Riley SP, Martinez JJ. 2010. Adherence to and invasion of host cells by spotted fever group Rickettsia species. Front Microbiol 1:139. doi: 10.3389/fmicb.2010.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed SC, Serio AW, Welch MD. 2012. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol 14:529–545. doi: 10.1111/j.1462-5822.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thepparit C, Bourchookarn A, Petchampai N, Barker SA, Macaluso KR. 2010. Interaction of Rickettsia felis with histone H2B facilitates the infection of a tick cell line. Microbiology 156:2855–2863. doi: 10.1099/mic.0.041400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petchampai N, Sunyakumthorn P, Guillotte ML, Thepparit C, Kearney MT, Mulenga A, Azad AF, Macaluso KR. 2014. Molecular and functional characterization of vacuolar-ATPase from the American dog tick Dermacentor variabilis. Insect Mol Biol 23:42–51. doi: 10.1111/imb.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petchampai N, Sunyakumthorn P, Guillotte ML, Verhoeve VI, Banajee KH, Kearney MT, Macaluso KR. 2014. Novel identification of Dermacentor variabilis Arp2/3 complex and its role in rickettsial infection of the arthropod vector. PLoS One 9:e93768. doi: 10.1371/journal.pone.0093768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunyakumthorn P, Petchampai N, Kearney MT, Sonenshine DE, Macaluso KR. 2012. Molecular characterization and tissue-specific gene expression of Dermacentor variabilis α-catenin in response to rickettsial infection. Insect Mol Biol 21:197–204. doi: 10.1111/j.1365-2583.2011.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver JD, Burkhardt NY, Felsheim RF, Kurtti TJ, Munderloh UG. 2014. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA. Appl Environ Microbiol 80:1170–1176. doi: 10.1128/AEM.03352-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss E. 1973. Growth and physiology of rickettsiae. Bacteriol Rev 37:259–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurtti TJ, Simser JA, Baldridge GD, Palmer AT, Munderloh UG. 2005. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae). J Invertebr Pathol 90:177–186. doi: 10.1016/j.jip.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaworski DC, Zou Z, Bowen CJ, Wasala NB, Madden R, Wang Y, Kocan KM, Jiang H, Dillwith JW. 2010. Pyrosequencing and characterization of immune response genes from the American dog tick, Dermacentor variabilis (L). Insect Mol Biol 19:617–630. doi: 10.1111/j.1365-2583.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulenga A, Macaluso KR, Simser JA, Azad AF. 2003. The American dog tick, Dermacentor variabilis, encodes a functional histamine release factor homolog. Insect Biochem Mol Biol 33:911–919. doi: 10.1016/S0965-1748(03)00097-3. [DOI] [PubMed] [Google Scholar]
- 19.Xu G, Fang QQ, Keirans JE, Durden LA. 2003. Cloning and sequencing of putative acetylcholinesterase cDNAs from the American dog tick, Dermacentor variabilis, and the brown dog tick, Rhipicephalus sanguineus (Acari: Ixodidae). J Med Entomol 40:890–896. doi: 10.1603/0022-2585-40.6.890. [DOI] [PubMed] [Google Scholar]
- 20.Xu G, Fang QQ, Keirans JE, Durden LA. 2004. Cloning and sequencing of putative calreticulin complementary DNAs from four hard tick species. J Parasitol 90:73–78. doi: 10.1645/GE-157R. [DOI] [PubMed] [Google Scholar]
- 21.Dreher-Lesnick SM, Mulenga A, Simser JA, Azad AF. 2006. Differential expression of two glutathione S-transferases identified from the American dog tick, Dermacentor variabilis. Insect Mol Biol 15:445–453. doi: 10.1111/j.1365-2583.2006.00657.x. [DOI] [PubMed] [Google Scholar]
- 22.Sunyakumthorn P, Petchampai N, Grasperge BJ, Kearney MT, Sonenshine DE, Macaluso KR. 2013. Gene expression of tissue-specific molecules in ex vivo Dermacentor variabilis (Acari: Ixodidae) during rickettsial exposure. J Med Entomol 50:1089–1096. doi: 10.1603/ME12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell-Sakyi L, Zweygarth E, Blouin EF, Gould EA, Jongejan F. 2007. Tick cell lines: tools for tick and tick-borne disease research. Trends Parasitol 23:450–457. doi: 10.1016/j.pt.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Munderloh UG, Liu Y, Wang M, Chen C, Kurtti TJ. 1994. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J Parasitol 80:533–543. doi: 10.2307/3283188. [DOI] [PubMed] [Google Scholar]
- 25.Mattila JT, Munderloh UG, Kurtti TJ. 2007. Phagocytosis of the Lyme disease spirochete, Borrelia burgdorferi, by cells from the ticks, Ixodes scapularis and Dermacentor andersoni, infected with an endosymbiont, Rickettsia peacockii. J Insect Sci 7:58. doi: 10.1673/031.007.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker TS, Brown JS, Hoover CS, Morgan DA. 1990. Endothelial prostaglandin secretion: effects of typhus rickettsiae. J Infect Dis 162:1136–1144. doi: 10.1093/infdis/162.5.1136. [DOI] [PubMed] [Google Scholar]
- 27.Ammerman NC, Swanson KI, Anderson JM, Schwartz TR, Seaberg EC, Glass GE, Norris DE. 2004. Spotted-fever group Rickettsia in Dermacentor variabilis, Maryland. Emerg Infect Dis 10:1478–1481. doi: 10.3201/eid1008.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henning TC, Orr JM, Smith JD, Arias JR, Norris DE. 2014. Spotted fever group rickettsiae in multiple hard tick species from Fairfax County, Virginia. Vector Borne Zoonotic Dis 14:482–485. doi: 10.1089/vbz.2013.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng WC, Murray ES, Burgdorfer W, Spielman JM, Rosenberg G, Dang K, Smith C, Spickert C, Waner JL. 1980. Spotted fever group rickettsiae in Dermacentor variabilis from Cape Cod, Massachusetts. Am J Trop Med Hyg 29:691–694. [DOI] [PubMed] [Google Scholar]
- 30.Anderson JF, Magnarelli LA, Philip RN, Burgdorfer W. 1986. Rickettsia rickettsii and Rickettsia montana from Ixodid ticks in Connecticut. Am J Trop Med Hyg 35:187–191. [DOI] [PubMed] [Google Scholar]
- 31.Pretzman C, Daugherty N, Poetter K, Ralph D. 1990. The distribution and dynamics of Rickettsia in the tick population of Ohio. Ann N Y Acad Sci 590:227–336. doi: 10.1111/j.1749-6632.1990.tb42224.x. [DOI] [PubMed] [Google Scholar]
- 32.Dramsi S, Cossart P. 1998. Intracellular pathogens and the actin cytoskeleton. Annu Rev Cell Dev Biol 14:137–166. doi: 10.1146/annurev.cellbio.14.1.137. [DOI] [PubMed] [Google Scholar]
- 33.Cossart P, Sansonetti PJ. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 34.Haglund CM, Welch MD. 2011. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol 195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva CV, Cruz L, Araújo Nda S, Angeloni MB, Fonseca BB, Gomes Ade O, Carvalho Fdos R, Gonçalves AL, Barbosa Bde F. 2012. A glance at Listeria and Salmonella cell invasion: different strategies to promote host actin polymerization. Int J Med Microbiol 302:19–32. doi: 10.1016/j.ijmm.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Mullins RD, Pollard TD. 1999. Structure and function of the Arp2/3 complex. Curr Opin Struct Biol 9:244–249. doi: 10.1016/S0959-440X(99)80034-7. [DOI] [PubMed] [Google Scholar]
- 37.Goley ED, Welch MD. 2006. The Arp2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 10:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 38.Alrutz MA, Srivastava A, Wong KW, D'Souza-Schorey C, Tang M, Ch'Ng LE, Snapper SB, Isberg RR. 2001. Efficient uptake of Yersinia pseudotuberculosis via integrin receptors involves a Rac1-Arp 2/3 pathway that bypasses N-WASP function. Mol Microbiol 42:689–703. doi: 10.1046/j.1365-2958.2001.02676.x. [DOI] [PubMed] [Google Scholar]
- 39.Unsworth KE, Way M, McNiven M, Machesky L, Holden DW. 2004. Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cell Microbiol 6:1041–1055. doi: 10.1111/j.1462-5822.2004.00417.x. [DOI] [PubMed] [Google Scholar]
- 40.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 41.Carabeo RA, Dooley CA, Grieshaber SS, Hackstadt T. 2007. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell Microbiol 9:2278–2288. doi: 10.1111/j.1462-5822.2007.00958.x. [DOI] [PubMed] [Google Scholar]
- 42.Hybiske K, Stephens RS. 2007. Mechanisms of Chlamydia trachomatis entry into nonphagocytic cells. Infect Immun 75:3925–3934. doi: 10.1128/IAI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sousa S, Cabanes D, Bougnères L, Lecuit M, Sansonetti P, Tran-Van-Nhieu G, Cossart P. 2007. Src, cortactin and Arp2/3 complex are required for E-cadherin-mediated internalization of Listeria into cells. Cell Microbiol 9:2629–2643. doi: 10.1111/j.1462-5822.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- 44.Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. 2008. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog 4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pielage JF, Powell KR, Kalman D, Engel JN. 2008. RNAi screen reveals an Abl kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog 4:e1000031. doi: 10.1371/journal.ppat.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takenawa T, Suetsugu S. 2007. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 47.Bershadsky A. 2004. Magic touch: how does cell-cell adhesion trigger actin assembly? Trends Cell Biol 14:589–593. doi: 10.1016/j.tcb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 48.McGee K, Zettl M, Way M, Fällman M. 2001. A role for N-WASP in invasin-promoted internalisation. FEBS Lett 509:59–65. doi: 10.1016/S0014-5793(01)03139-8. [DOI] [PubMed] [Google Scholar]
- 49.Hamon M, Bierne H, Cossart P. 2006. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 50.Hall A. 1998. G proteins and small GTPases: distant relatives keep in touch. Science 280:2074–2075. doi: 10.1126/science.280.5372.2074. [DOI] [PubMed] [Google Scholar]
- 51.Sit ST, Manser E. 2011. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci 124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 52.Ridley AJ. 2006. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Atre AN, Surve SV, Shouche YS, Joseph J, Patole MS, Deopurkar RL. 2009. Association of small Rho GTPases and actin ring formation in epithelial cells during the invasion by Candida albicans. FEMS Immunol Med Microbiol 55:74–84. doi: 10.1111/j.1574-695X.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- 54.Alrutz MA, Isberg RR. 1998. Involvement of focal adhesion kinase in invasin-mediated uptake. Proc Natl Acad Sci U S A 95:13658–13663. doi: 10.1073/pnas.95.23.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruce-Staskal PJ, Weidow CL, Gibson JJ, Bouton AH. 2002. Cas, Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J Cell Sci 115:2689–2700. [DOI] [PubMed] [Google Scholar]
- 56.Martinez JJ, Hultgren SJ. 2002. Requirement of Rho-family GTPases in the invasion of type 1-piliated uropathogenic Escherichia coli. Cell Microbiol 4:19–28. doi: 10.1046/j.1462-5822.2002.00166.x. [DOI] [PubMed] [Google Scholar]
- 57.Bougnères L, Girardin SE, Weed SA, Karginov AV, Olivo-Marin JC, Parsons JT, Sansonetti PJ, Van Nhieu GT. 2004. Cortactin and Crk cooperate to trigger actin polymerization during Shigella invasion of epithelial cells. J Cell Biol 166:225–235. doi: 10.1083/jcb.200402073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra SK, Hanson DA, Schlaepfer DD. 2005. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol 6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 59.Seong J, Lu S, Wang Y. 2011. Live cell imaging of Src/FAK signaling by FRET. Cell Mol Bioeng 2:138–147. doi: 10.1007/s12195-011-0161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotula L. 2012. Abi1, a critical molecule coordinating actin cytoskeleton reorganization with PI-3 kinase and growth signaling. FEBS Lett 586:2790–2794. doi: 10.1016/j.febslet.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


