Abstract
Entomopathogenic nematodes (EPNs) in the genera Heterorhabditis and Steinernema are lethal parasites of insects that are of interest as models for understanding parasite-host interactions and as biocontrol agents for insect pests. EPNs harbor a bacterial endosymbiont in their gut that assists in insect killing. EPNs are capable of infecting and killing a wide range of insects, yet how the nematodes and their bacterial endosymbionts interact with the insect immune system is poorly understood. Here, we develop a versatile model system for understanding the insect immune response to parasitic nematode infection that consists of seven species of EPNs as model parasites and five species of Drosophila fruit flies as model hosts. We show that the EPN Steinernema carpocapsae, which is widely used for insect control, is capable of infecting and killing D. melanogaster larvae. S. carpocapsae is associated with the bacterium Xenorhabdus nematophila, and we show that X. nematophila induces expression of a subset of antimicrobial peptide genes and suppresses the melanization response to the nematode. We further show that EPNs vary in their virulence toward D. melanogaster and that Drosophila species vary in their susceptibilities to EPN infection. Differences in virulence among different EPN-host combinations result from differences in both rates of infection and rates of postinfection survival. Our results establish a powerful model system for understanding mechanisms of host-parasite interactions and the insect immune response to parasitic nematode infection.
INTRODUCTION
Entomopathogenic nematodes (EPNs) of the genera Steinernema and Heterorhabditis are insect-parasitic nematodes that are phylogenetically distant but share a similar life cycle as a result of convergent evolution (1). EPNs offer numerous advantages as model parasitic nematodes, including small size, short generation time, and amenability to in vitro culturing (2). EPN infective larvae are associated with bacterial endosymbionts: Steinernema species are associated with bacteria in the genus Xenorhabdus, and Heterorhabditis species are associated with bacteria in the genus Photorhabdus (1). At least some EPNs are capable of infecting the fruit fly Drosophila melanogaster, providing a genetically tractable system for understanding the immune response to parasitic nematodes and their bacterial endosymbionts (3–6). However, the insect immune response to EPN infection is poorly understood.
During a particular developmental stage called the infective juvenile (IJ), EPNs infect insects (Fig. 1A). IJs are developmentally arrested, third-stage larvae analogous to the dauer stage of free-living nematodes (7). IJs actively seek out insect hosts using chemosensory cues (8–10) and infect either by entering through natural body openings or by penetrating the insect cuticle (11). IJs harbor their bacterial endosymbiont in their gut and deposit it into the insect upon infection, where it assists the nematode in killing the insect, digesting insect tissues, and inhibiting the growth of other microorganisms (12–14). Following infection, the nematodes reproduce in the insect cadaver and feed on the bacterium-infested tissue until resources are depleted, at which point new IJs form and emerge from the cadaver to search for new hosts (Fig. 1B) (15).
FIG 1.
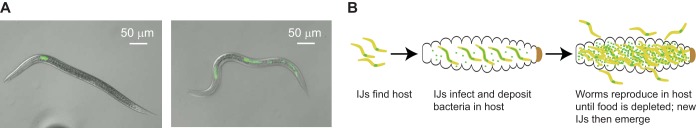
Life cycle of S. carpocapsae. (A) Photomicrographs of S. carpocapsae infective juveniles (IJs) with GFP-expressing X. nematophila. The left frame shows an IJ outside the host, and the right frame shows an IJ that was extracted from a host and that is defecating X. nematophila. (B) The life cycle of S. carpocapsae. IJs in the soil find a host, enter through a natural body opening, and defecate their symbiotic bacteria into the host. The bacteria play an important role in overcoming the host immune system (1). The nematodes develop and reproduce in the insect cadaver until resources are depleted. New IJs then form and exit the cadaver. Green dots represent the bacterial endosymbiont.
In response to EPN infection, insects mount an innate immune response that involves antimicrobial peptide (AMP) expression as well as activation of the melanization and encapsulation reactions (11). At the same time, the nematodes attempt to evade or suppress the insect immune response through a process that remains poorly understood. Both the nematode and its bacterial endosymbiont appear to inhibit some aspects of AMP production, melanization, encapsulation, and phagocytosis (11, 16–18). Studies using D. melanogaster larvae as a model host for the EPN Heterorhabditis bacteriophora and its endosymbiont Photorhabdus luminescens have shown that infection induces expression of a large number of immune genes, including AMPs, and that AMP expression is primarily a response to the bacterial endosymbiont rather than to the nematode (4, 19). Infection also stimulates clotting, and clotting mutants show decreased survival in response to H. bacteriophora-P. luminescens infection (20, 21). However, studies of the immune response of D. melanogaster to EPN infection have so far been limited to H. bacteriophora-P. luminescens, and the extent to which the immune response differs for different EPNs is unclear.
Here, we demonstrate that the distantly related EPN Steinernema carpocapsae and its bacterial endosymbiont Xenorhabdus nematophila are capable of infecting D. melanogaster larvae and are more virulent toward D. melanogaster than H. bacteriophora-P. luminescens. Infection with S. carpocapsae symbiont IJs (i.e., IJs harboring X. nematophila in their gut) induced expression of a subset of AMP genes. S. carpocapsae infection also activated the melanization pathway, and a higher rate of melanization occurred following infection with axenic IJs than with symbiont IJs, suggesting that X. nematophila suppresses the melanization response. Finally, exposure of D. melanogaster larvae to seven different EPN species revealed that EPNs vary in their virulence levels toward D. melanogaster, and exposure of five different Drosophila species to S. carpocapsae symbiont IJs revealed that Drosophila species vary in their susceptibilities to EPN infection. Our results establish the EPN-Drosophila system as a powerful model for investigating the insect immune response to nematode infection.
MATERIALS AND METHODS
Nematode strains.
The following EPN strains were used: S. carpocapsae ALL (8, 9), H. bacteriophora Baine (22), Steinernema glaseri NC (23), Steinernema scapterisci FL (24), Steinernema riobrave TX (25), Heterorhabditis indica HOM1 (22), and Steinernema feltiae SN (26).
Drosophila stocks.
Wild-type D. melanogaster larvae were from the Canton-S strain. Studies of AMP expression were conducted with strains of D. melanogaster containing either an attacinA::GFP, cecropinA1::GFP, metchnikowin::GFP, drosocin::GFP, drosomycin::GFP, diptericin::GFP, or defensin::GFP transgene (where GFP is green fluorescent protein) (27). Wild-type Drosophila virilis, Drosophila simulans, Drosophila yakuba, and Drosophila pseudoobscura were stocks 15010-1051.00, 14021-0251.006, 14021-0261.00, and 14011-0121.104 from the Drosophila Species Stock Center, respectively. We note that all D. melanogaster strains, as well as the wild-type Drosophila simulans strain, were confirmed to be infected with Wolbachia by PCR using previously described primers (28). However, Wolbachia status is unlikely to affect susceptibility to EPNs or their bacterial endosymbionts: although Wolbachia infection may protect against some viral infections (29, 30), it does not appear to protect against other types of infections and has little or no effect on AMP expression (29, 31–33).
Bacterial strains.
The following bacterial strains were used: wild-type X. nematophila HGB800 (34), GFP-expressing X. nematophila HGB340 (13), colonization-defective X. nematophila HGB777 (35), Escherichia coli OP50-GFP, P. luminescens TT01-GFP (36), Photorhabdus temperata NC1-GFP (36), and colonization-defective P. temperata NC1-GFP TRN16 (36). Xenorhabdus was grown in LB broth containing 0.1% sodium pyruvate, and Photorhabdus was grown in PP3 broth as previously described (8).
Nematode culturing.
To generate symbiont IJs, nematodes were cultured in either the waxworm Galleria mellonella (for all species except S. scapterisci) or the house cricket Acheta domestica (for S. scapterisci) as previously described (8, 9). Briefly, five last-instar waxworms or one medium-sized cricket (American Cricket Ranch, Lakeside, CA) was placed in a 5-cm petri dish with a 55-mm Whatman 1 filter paper in the bottom of the dish. Approximately 500 to 1,000 IJs suspended in water were distributed on the filter paper and on the insects. Petri dishes were stored either at 25°C in the case of H. bacteriophora and S. riobrave or at room temperature (22 to 23°C) in the case of all other species. H. bacteriophora and S. riobrave infections were performed at 25°C because these species are found primarily in warm climates (37) and infect insects more efficiently at 25°C than at room temperature. Infections for the other species were performed at room temperature because these species infect more efficiently at room temperature than at 25°C. After ∼10 days the insect cadavers were placed on White traps (38) or, in the case of S. glaseri, on modified White traps containing plaster of Paris (9). Symbiont IJs were collected from traps within 10 days, stored at 15°C, and tested within 1 month of collection.
To generate axenic S. carpocapsae IJs, symbiont S. carpocapsae IJs were surface sterilized by incubation in 1% commercial bleach for 5 min. IJs were then rinsed three times in distilled H2O (dH2O), incubated in antibiotic solution (10 μg/ml gentamicin, 100 μg/ml streptomycin, 100 μg/ml carbenicillin, and 20 μg/ml kanamycin in dH2O) for 48 h, and plated onto 1× lipid agar-cholesterol plates (39) (final concentration of cholesterol, 5 mg/liter) containing 0.1% sodium pyruvate and seeded with X. nematophila HGB777 bacteria (35). Axenic nematodes were maintained on lipid agar-cholesterol plates seeded with HGB777, and IJs were collected from plates as previously described (8). IJs were incubated in 1% commercial bleach for 5 min and then rinsed three times in dH2O to surface sterilize them prior to testing. To verify that the IJs were axenic, 5 μl of IJ pellet was plated onto a lipid agar-cholesterol plate and incubated at 25°C. The absence of bacteria on the plate was confirmed after 2 to 3 days.
To generate symbiont S. carpocapsae IJs containing GFP-expressing X. nematophila, axenic IJs were plated onto lipid agar-cholesterol plates seeded with X. nematophila HGB340 bacteria. Nematodes were maintained on lipid agar-cholesterol plates seeded with HGB340, and IJs were collected from plates as previously described (8).
To generate symbiont H. bacteriophora IJs containing GFP-expressing P. luminescens, symbiont H. bacteriophora IJs that had emerged from waxworms were plated onto 1× lipid agar-cholesterol plates (39) containing 0.1% sodium pyruvate seeded with P. temperata NC1 TRN16 bacteria (36). IJs were collected from plates as previously described (8) and plated onto 1× lipid agar-cholesterol plates seeded with P. luminescens TT01-GFP bacteria (36). Nematodes were maintained on 1× lipid agar-cholesterol plates seeded with TT01-GFP, and IJs were collected from plates as previously described (8). Axenic H. bacteriophora IJs were generated as described above for axenic S. carpocapsae IJs, except that they were plated onto and maintained on TRN16.
Infection of Drosophila larvae with EPNs.
IJs used to assay survival were grown in waxworms; IJs used to assay infection were grown on GFP-expressing bacteria. IJs were rinsed three times in dH2O, and 10 μl of water containing 500 IJs was pipetted onto the center of a 5-cm petri dish containing nematode growth medium (NGM). For each trial, 20 third-instar Drosophila larvae were rinsed twice in 1× phosphate-buffered saline (PBS) and placed onto the NGM plate containing IJs. Drosophila larvae infected with H. bacteriophora and S. riobrave were kept in a 25°C incubator; larvae infected with all other strains were kept at room temperature. Different temperatures were used for H. bacteriophora and S. riobrave because these species are adapted for infection and growth at warmer temperatures than the other species, as described above. Metal rings were placed onto the plate lids as weights to prevent fly larvae from escaping. Infection and survival were scored at 24 and 48 h postexposure to IJs. Melanization was scored at 48 h postexposure to IJs to ensure that a majority of the population had been infected.
To score infection, fly larvae or pupae (in cases where the fly larvae pupated during the course of the experiment) were assayed under an epifluorescence dissecting microscope. IJs grown on GFP-expressing symbiotic bacteria were used to facilitate detection of worms inside the fly host. Fly larvae or pupae were considered infected if worms were visible inside the body. Although worms could be seen inside the host even without the presence of GFP-expressing symbiotic bacteria, worms could be identified more efficiently when they had GFP-expressing symbiotic bacteria. To score survival, fly larvae or pupae were assayed under a dissecting microscope at ×50 magnification. Animals were determined to be alive if they had a visible heartbeat or responded to gentle prodding.
To score melanization, fly larvae or pupae were first scored under a dissecting microscope at ×50 magnification for black spots on the cuticle and then dissected to determine whether they were infected. For each trial, the percentage of infected fly larvae or pupae with visible melanization was quantified. A value of 100% would indicate that all of the infected D. melanogaster larvae showed visible melanization; a value of 0% would indicate that none of the infected D. melanogaster larvae showed visible melanization.
Infection of Drosophila larvae with bacteria.
GFP-expressing X. nematophila, E. coli, or P. luminescens cells were used for infection assays; wild-type bacteria were used for all other assays. To generate each assay plate, 100 μl of a bacterial suspension (for X. nematophila and E. coli) or 200 μl of bacterial suspension (for P. luminescens) from a 1- or 2-day culture was spread onto a 5-cm plate containing LB supplemented with 100 μg/ml carbenicillin and 0.1% sodium pyruvate (X. nematophila), LB alone (E. coli), or 1× lipid agar with cholesterol plus 0.1% sodium pyruvate (P. luminescens). Plates were incubated at 25°C for 1 to 2 days (X. nematophila and P. luminescens) or at 37°C overnight (E. coli) to create a bacterial lawn. For each trial, 20 second-instar or early-third-instar D. melanogaster larvae were rinsed in 1× PBS and placed onto a plate containing a bacterial lawn. A second plate containing a bacterial lawn was then secured upside down on top of the first plate to prevent the fly larvae from avoiding the bacteria by crawling onto the plate lid. Survival was scored at 24, 48, and 72 h as described above. To assay infection, fly larvae were washed twice in 1×PBS, placed onto unseeded NGM plates, and scored for GFP expression under an epifluorescence dissecting microscope after 24 and 48 h. All flies with visible GFP expression inside the body were scored as GFP positive. The percentages of GFP-positive fly larvae were then calculated. Fly larvae that were GFP negative at 24 h were placed onto new plates seeded with bacteria and scored again at 48 h. No GFP expression was observed in control experiments where fly larvae were not exposed to bacteria.
AMP expression assay.
AMP expression was assayed in D. melanogaster larvae following infection with symbiont IJs, axenic IJs, or bacteria. For these experiments, both symbiont IJs and axenic IJs were grown in vitro on plates containing lawns of X. nematophila to eliminate any potential differences in D. melanogaster AMP expression resulting from differences in nematode culturing conditions. We used seven different transgenic D. melanogaster strains as hosts, each of which expressed a reporter construct in which GFP expression was driven by the promoter of a different AMP gene (27). Details of the transgenic D. melanogaster larvae expressing the AMP reporter constructs are described above under “Drosophila stocks.” We note that no GFP-expressing X. nematophila was present in this experiment; symbiont IJs contained bacteria that did not express GFP, and the bacteria used for bacterial infections also did not express GFP. Thus, the GFP expression observed in these experiments was from the transgenic D. melanogaster hosts and was a reflection of AMP gene expression.
Infection of transgenic D. melanogaster larvae was performed as described above; each trial consisted of 20 fly larvae. For the uninfected controls, fly larvae were placed onto either NGM plates without IJs (controls for infection with symbiont or axenic IJs) or LB plates without bacteria (control for infection with X. nematophila). AMP expression was scored at 24 h postexposure to IJs. We note that in initial experiments, AMP expression was scored at 8, 24, and 48 h postexposure to IJs. AMP expression levels at 8 h postexposure were low because most fly larvae had not yet been infected, and no difference was observed between AMP expression levels at 24 and 48 h postexposure (data not shown). We therefore focused on the 24-h time point for further experiments.
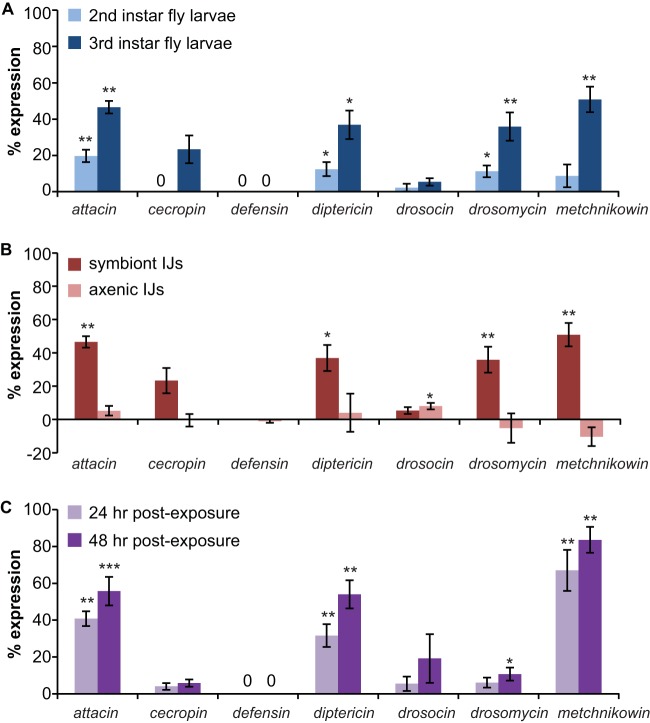
To score AMP expression, larvae were removed from the plates, rinsed twice in 1× PBS, placed onto unseeded NGM plates, and observed using the GFP filter of an epifluorescence dissecting microscope. For the attacin, cecropin, defensin, drosocin, and metchnikowin genes, fly larvae were scored as GFP positive if any GFP expression was observed. For the diptericin and drosomycin genes, fly larvae were scored as GFP positive if diffuse GFP expression was observed because small spots of GFP expression were often observed under normal culturing conditions. The percentage of fly larvae expressing GFP was then calculated. We note that we previously validated our visual GFP scoring method for these reporter lines by comparing expression data obtained by visual scoring versus quantification in ImageJ and determining that expression data obtained by both methods were consistent (4). To control for GFP expression not due to infection with IJs or bacteria, the percentage of GFP-positive fly larvae obtained from the uninfected control experiments was subtracted from the percentage of GFP-positive fly larvae obtained from the infection experiments. Thus, Fig. 3 reports the background-subtracted values for the percentage of fly larvae that express the indicated AMP reporter construct.
FIG 3.
Infection of D. melanogaster larvae with symbiont S. carpocapsae IJs or X. nematophila induces a humoral immune response. (A) AMP expression following infection with symbiont S. carpocapsae IJs. Infection of second- or third-instar D. melanogaster larvae with symbiont S. carpocapsae IJs induces expression of a subset of AMP genes. (B) AMP expression following infection with symbiont versus axenic S. carpocapsae IJs. Infection of third-instar D. melanogaster larvae with symbiont S. carpocapsae IJs results in AMP expression, while infection with axenic S. carpocapsae IJs results in little or no AMP expression. Data for symbiont IJ exposure are from panel A. For panels A and B, AMP expression was examined at 24 h postexposure to IJs using transgenic fly larvae containing reporter constructs in which an AMP gene promoter was used to drive expression of GFP (27). (C) Infection of second-instar D. melanogaster larvae with X. nematophila results in AMP expression. AMP expression was examined at 24 and 48 h postexposure to bacteria using the same transgenic fly larvae as used in the experiments described in panels A and B. *, P < 0.05; **, P < 0.01; ***, P < 0.001, unpaired t test or Mann-Whitney test (infected larvae versus uninfected control larvae of the same genotype; n = 5 to 12 trials for each condition). For all graphs, error bars represent standard errors of the means.
For the bacterial infection experiment, plates were observed at 24 and 48 h postexposure to X. nematophila. On plates where some or all of the fly larvae had burrowed into the agar by the 24-h time point, all larvae on that plate were transferred to a new X. nematophila plate to ensure that the fly larvae remained in contact with the bacteria for the duration of the experiment.
Examining the time course for infection and survival following EPN exposure.
For each trial, ∼20 fly larvae were exposed to symbiont IJs containing GFP-expressing X. nematophila or P. temperata. IJs were used within 1 week of collection. Infection was scored at 2, 4, 6, 8, 24, and 48 h postexposure to IJs and was visualized using the GFP filter on an epifluorescence microscope. At each time point, larvae were rinsed twice in 1×PBS and placed onto unseeded NGM plates prior to scoring. After scoring, infected larvae were placed onto unseeded NGM plates that did not contain IJs, while uninfected larvae were placed back onto the original plate containing IJs so that they could be scored for infection at later time points. To examine postinfection survival, all larvae infected by 8 h were scored for survival at 24 and 48 h. Trials in which fewer than three fly larvae became infected by 8 h were not included in the analysis. To assay long-term survival, animals were scored for infection and survival as described above, except that any animals still alive at 48 h were placed onto new unseeded NGM plates and monitored for survival to adulthood.
Statistical analysis.
Statistical analysis was performed using GraphPad Instat or Prism software. Standard statistical tests were used for all experiments, as described in the figure legends. All statistical comparisons are described in the relevant figure legends and supplemental tables. The value for sample size (n) used for statistical analysis refers to the number of trials performed for each treatment, condition, or genotype; each trial consisted of ∼20 fly larvae.
RESULTS AND DISCUSSION
S. carpocapsae infects and kills D. melanogaster.
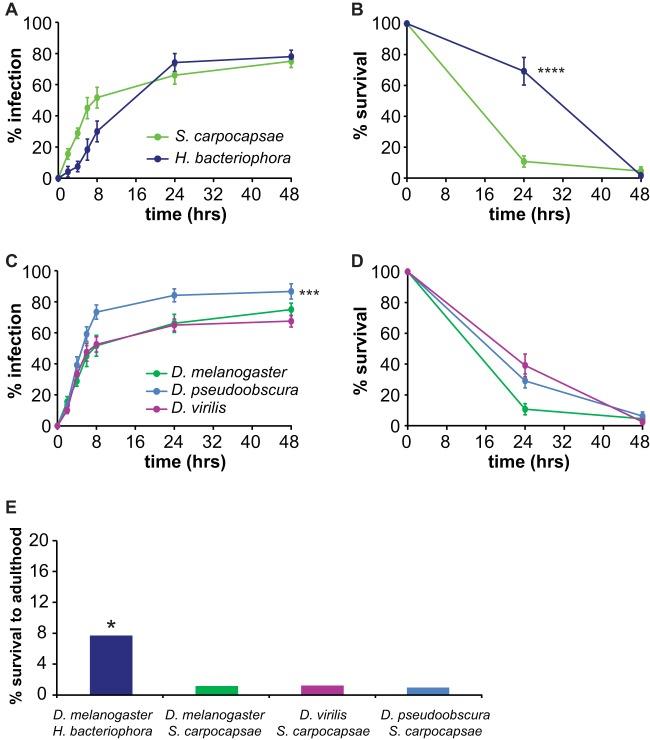
S. carpocapsae has a wide geographical distribution and a broad host range and is used as a biocontrol agent for numerous insect pests (40). To determine whether S. carpocapsae is pathogenic for D. melanogaster, we used an infection assay in which we exposed third-instar fly larvae to symbiont IJs. Third-instar larvae were used for this assay because EPNs typically infect late-stage insect larvae (41). We scored infection and survival at 24 and 48 h postexposure since EPNs generally kill hosts within 48 h (41). We found that approximately 60% of the fly larvae exposed to symbiont IJs died within 24 h (Fig. 2A). Thus, S. carpocapsae is capable of infecting and killing D. melanogaster larvae. To determine whether pathogenicity was conferred primarily by S. carpocapsae or X. nematophila, we exposed fly larvae to axenic IJs. We found that axenic IJs were also capable of infecting and killing D. melanogaster larvae, although with less efficiency than symbiont IJs (Fig. 2A). Thus, S. carpocapsae IJs are pathogenic for D. melanogaster even in the absence of X. nematophila, consistent with previous studies of S. carpocapsae infection in larger insects, such as Galleria mellonella (42).
FIG 2.
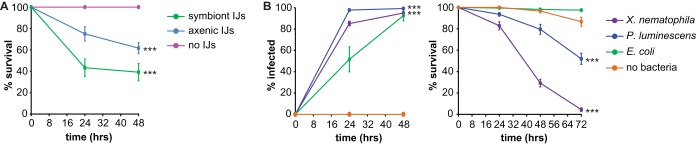
S. carpocapsae and its bacterial endosymbiont X. nematophila are pathogenic toward D. melanogaster. (A) Survival of D. melanogaster larvae exposed to symbiont S. carpocapsae IJs, axenic S. carpocapsae IJs, or no IJs. All three survival curves are significantly different (P < 0.0001, log rank test). ***, P < 0.001 relative to the no-IJ control (log rank test with Bonferroni correction; n = 6 trials for each condition). (B) Infection (left) and survival (right) of D. melanogaster larvae exposed to either X. nematophila, P. luminescens, E. coli, or no bacteria. All three species of bacteria successfully infected D. melanogaster (left graph) although infection rates were significantly different for each species (P < 0.01, log rank test). ***, P < 0.001 for X. nematophila and P. luminescens relative to E. coli (log rank test with Bonferroni correction). The survival curve for fly larvae exposed to E. coli was not significantly different from the survival curve for the no-bacteria control (right graph); all other survival curves were significantly different from each other (P < 0.001, log rank test with Bonferroni correction). ***, P < 0.001 relative to E. coli and the no-bacteria control (log rank test with Bonferroni correction). The no-bacteria control shown in the graph was performed on LB plates; a no-bacteria control was also performed on lipid-agar plates (for comparison to P. luminescens), and results were not significantly different from those of the control on LB plates (P = 0.2728, log rank test; n = 5 to 9 trials for each condition). For all graphs, the x axis refers to time postexposure, and error bars represent standard errors of the means. In some cases, error bars are too small to be visible.
We then examined the pathogenicity of X. nematophila in the absence of its nematode vector by exposing fly larvae to agar plates containing lawns of X. nematophila. For comparison, we also exposed fly larvae to lawns of P. luminescens and E. coli. We found that exposure to all three bacteria resulted in infection of fly larvae, as determined by counting the number of GFP-positive fly larvae postexposure to GFP-labeled bacteria (Fig. 2B, left graph). X. nematophila was pathogenic for D. melanogaster larvae: 95% of fly larvae exposed to X. nematophila were dead by 72 h (Fig. 2B, right graph). X. nematophila was significantly more virulent than P. luminescens, which killed only approximately 50% of fly larvae by 72 h (Fig. 2B). In contrast, E. coli was not pathogenic for D. melanogaster larvae. These results are consistent with a previous study which found that X. nematophila is more virulent than P. luminescens when injected into D. melanogaster adults (43). Taken together, these results suggest that both P. luminescens and X. nematophila are pathogenic for D. melanogaster larvae but differ in their virulence levels.
Infection of fly larvae with X. nematophila most likely occurred by ingestion since GFP-expressing bacteria appeared to localize initially to the digestive tract (see Fig. S1 in the supplemental material). This is consistent with our previous observations of exposure of fly larvae to Photorhabdus bacteria (4). Susceptibility to X. nematophila is not likely to be the result of exposure to external toxins, since nonfeeding third-instar larvae did not become infected with X. nematophila in our assay. However, we cannot exclude the possibility that X. nematophila secretes toxins that have external effects on second- and early-third-instar larvae but not older third-instar larvae.
Infection induces expression of antimicrobial peptide (AMP) genes.
A major component of the insect innate immune response is AMP production by the fat body, a structure similar to the mammalian liver and adipose tissue (44). Studies of a number of insects, including the cecropia moth Hyalophora cecropia, the beet armyworm Spodoptera exigua, and the tobacco hornworm Manduca sexta have shown that EPN infection can induce expression of AMP genes and that both the nematode and the bacteria can suppress AMP activity (45–49). We previously showed that infection of D. melanogaster larvae with H. bacteriophora symbiont IJs resulted in expression of four AMP genes, attacin, diptericin, drosomycin, and metchnikowin, and that this expression was a specific response to P. luminescens (4). Similar results were subsequently observed for infection of M. sexta with H. bacteriophora symbiont IJs, thus validating D. melanogaster as a model for other insect hosts (45). The AMP genes diptericin and drosomycin have also been shown to be upregulated following direct injection of either P. luminescens or X. nematophila into D. melanogaster adults (43). However, the AMP response of D. melanogaster to infection with S. carpocapsae symbiont IJs had not yet been examined, and the extent to which AMP expression is induced by EPNs versus their bacterial endosymbionts remains unclear (4, 6, 45).
To determine whether infection of D. melanogaster larvae with S. carpocapsae-X. nematophila induces AMP expression, we exposed both second-instar and third-instar fly larvae to symbiont IJs and monitored AMP expression at 24 h postexposure. To monitor AMP expression, we used seven different transgenic fly lines, each of which contained a reporter construct that expressed GFP under the control of a different AMP gene promoter. For each transgenic line, AMP expression was determined by scoring the fly larvae or pupae (in cases where the fly larvae pupated during the course of the experiment) for GFP expression at 24 h postexposure to IJs and quantifying the percentage of fly larvae or pupae expressing GFP. We found that exposure of third-instar fly larvae to symbiont IJs induced significant expression of four AMP genes: attacin, diptericin, drosomycin, and metchnikowin (Fig. 3A). Thus, the same subset of AMP genes is induced by exposure to S. carpocapsae symbiont IJs and H. bacteriophora symbiont IJs (4). The percentage of animals showing AMP expression was higher for third-instar larvae exposed to symbiont IJs than for second-instar larvae exposed to symbiont IJs (Fig. 3A), most likely because symbiont IJs were more effective at killing third-instar than second-instar larvae (see Fig. S2 in the supplemental material).
To determine whether AMP expression is a response to the nematode or the bacteria, we first exposed fly larvae to axenic IJs. Third-instar fly larvae were used for this experiment since a higher rate of AMP expression was observed with third-instar larvae than with second-instar larvae (Fig. 3A). We found that whereas fly larvae exposed to symbiont IJs showed AMP expression, fly larvae exposed to axenic IJs showed little or no AMP expression (Fig. 3B). Thus, the AMP response observed upon infection with symbiont IJs is not observed upon infection with axenic IJs.
We then exposed fly larvae to bacteria alone by placing fly larvae on a plate containing a lawn of X. nematophila. Second-instar or early-third-instar larvae were used for these experiments since older third-instar larvae did not become infected with bacteria in this assay. Exposure to X. nematophila induced expression of the same four AMP genes that were induced by infection with symbiont IJs (Fig. 3C). The routes of infection differ for fly larvae exposed to symbiont IJs and bacteria, and we cannot exclude the possibility that AMP expression might vary based on the route of infection. However, our results suggest that AMP expression is primarily a response to X. nematophila rather than S. carpocapsae.
We note that for comparison of AMP expression following infection of D. melanogaster larvae with either symbiont IJs or axenic IJs, we used nematodes grown in vitro on plates containing lawns of X. nematophila rather than nematodes grown in waxworms (see Materials and Methods). Growing nematodes in vitro was necessary to obtain axenic IJs, and thus both symbiont IJs and axenic IJs were grown in vitro for these experiments so that differences in AMP expression between axenic IJs and symbiont IJs could not be attributed to differences in nematode culturing conditions. However, we also directly tested whether AMP expression levels in fly larvae differed following infection with IJs cultured in vitro versus in vivo. We compared AMP expression in third-instar fly larvae infected with symbiont IJs grown on plates of X. nematophila and symbiont IJs grown in waxworms. No significant differences in AMP expression levels were observed following infection with IJs cultured in vitro versus in vivo (see Fig. S3 in the supplemental material), suggesting that the D. melanogaster immune response to S. carpocapsae symbiont IJs is similar, regardless of whether the IJs are cultured in vitro versus in vivo.
X. nematophila and P. luminescens suppress the melanization response of D. melanogaster.
Melanization is a cellular immune response of arthropods that results in melanin production at the wound site and that contributes to pathogen killing and wound healing (11, 50). Previous studies have shown that EPN infection of some insects results in rapid melanization and encapsulation of IJs although in permissive hosts IJs can escape the capsule and kill the insect (51–53). Both Xenorhabdus and Photorhabdus produce specific inhibitors of phospholipase A2, a key component of the melanization and encapsulation reactions, suggesting that the bacterial endosymbionts of EPNs promote nematode survival by suppressing these reactions (54, 55). To test whether infection with S. carpocapsae-X. nematophila or H. bacteriophora-P. luminescens activates the melanization response, we exposed fly larvae to either symbiont or axenic IJs and scored infected larvae for the presence of visible melanin spots (Fig. 4A). We found that both symbiont and axenic IJs induced melanization but that axenic IJs induced a higher rate of melanization than symbiont IJs (Fig. 4B). These results suggest that X. nematophila and P. luminescens facilitate the killing of D. melanogaster larvae by partially suppressing the melanization response.
FIG 4.
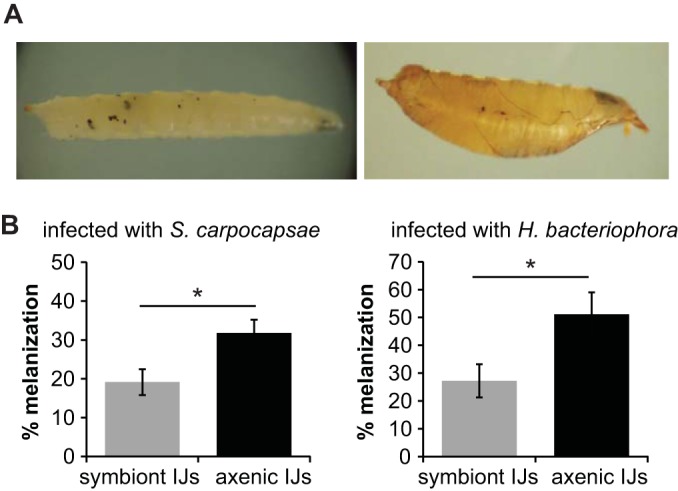
X. nematophila and P. luminescens partially suppress the melanization response of D. melanogaster. (A) Melanization from S. carpocapsae infection. The panel shows a representative D. melanogaster larva (left) and pupa (right) melanized by axenic S. carpocapsae IJs. (B) X. nematophila and P. luminescens inhibit melanization. Infection of D. melanogaster larvae with axenic S. carpocapsae (left graph) or H. bacteriophora (right graph) IJs resulted in a higher percentage of melanized fly larvae or pupae than infection with symbiont IJs. *, P < 0.05, unpaired t test. No melanization was observed in fly larvae not exposed to IJs. Melanization was scored at 48 h postexposure to IJs. Error bars represent standard errors of the means (n = 7 to 8 trials).
Virulence differs for different EPN-Drosophila combinations.
To examine the versatility of the fruit fly-EPN model system, we compared the ability of symbiont IJs from seven EPN species—S. carpocapsae, S. scapterisci, S. riobrave, S. glaseri, S. feltiae, H. bacteriophora, and H. indica—to infect and kill D. melanogaster larvae. These EPN species differ dramatically in their host ranges: S. carpocapsae and S. feltiae have broad host ranges that include insects from multiple orders, S. scapterisci has a narrow host range that is limited to orthopterans, and the other species have intermediate host ranges (24, 56–59). S. feltiae was also recently shown to be virulent toward D. melanogaster (3). We exposed D. melanogaster larvae to symbiont IJs of the different EPN species and scored survival at 24 and 48 h. We found that virulence differed greatly among species: S. scapterisci and S. riobrave were not virulent toward D. melanogaster larvae, S. carpocapsae and S. feltiae were highly virulent, and the other species displayed intermediate virulence (Fig. 5A). Thus, EPNs vary in their virulence toward D. melanogaster larvae.
FIG 5.
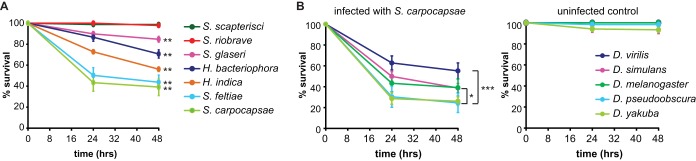
Species specificity of Drosophila-EPN interactions. (A) Infection of D. melanogaster with EPNs. EPNs vary in their virulence toward D. melanogaster larvae. Fly larvae were infected with symbiont IJs. Survival curves are significantly different (P < 0.0001, log rank test). Posttest results for each pairwise comparison are shown in Table S1 in the supplemental material. **, P < 0.01 relative to S. scapterisci (log rank test with Bonferroni correction; n = 6 to 8 trials for each condition). (B) Drosophila species vary in their susceptibility to S. carpocapsae symbiont IJ infection. Survival curves (left graph) are significantly different (P < 0.01, log rank test). Posttest results for each pairwise comparison are shown in Table S2 in the supplemental material. ***, P < 0.001 for D. virilis versus D. pseudoobscura and D. virilis versus D. yakuba; *, P < 0.05 for D. pseudoobscura versus D. simulans. The right graph shows survival curves of uninfected control larvae (n = 6 to 7 trials for each condition). For all graphs, the x axis refers to time postexposure, and error bars represent standard errors of the means.
We then examined the ability of S. carpocapsae, one of the most virulent EPNs for D. melanogaster, to infect and kill four phylogenetically and ecologically diverse Drosophila species: D. virilis, D. simulans, D. pseudoobscura, and D. yakuba. We found that S. carpocapsae symbiont IJs were capable of infecting and killing all Drosophila species tested (Fig. 5B). However, survival rates following exposure to S. carpocapsae symbiont IJs varied across species, with D. virilis showing the highest survival rate and D. pseudoobscura and D. yakuba showing the lowest survival rates (Fig. 5B). Thus, Drosophila species vary in their susceptibility to EPN infection.
Studies of larger insects, such as the Japanese beetle Popillia japonica, the house cricket Acheta domesticus, and the Colorado potato beetle Leptinotarsa decemlineata, have suggested that differences in virulence among EPNs can be attributed to differences in the abilities of EPNs to infect different hosts as well as differences in the host immune response to infection (51–53). To investigate the cause of differences in survival rates among Drosophila species exposed to different EPNs, we examined infection rates and postinfection survival rates of selected Drosophila-EPN combinations. Infection rates were examined by using IJs containing GFP-expressing endosymbionts to facilitate detection of IJs within the host. Fly larvae were considered infected if nematodes were visible inside the body. We found that different Drosophila-EPN combinations varied in both the rates at which the fly larvae became infected and the rates at which they succumbed to the infection (Fig. 6). For example, although S. carpocapsae symbiont IJs and H. bacteriophora symbiont IJs infected D. melanogaster at the same rate, S. carpocapsae symbiont IJs killed D. melanogaster more rapidly than H. bacteriophora symbiont IJs (Fig. 6A and B). In contrast, S. carpocapsae symbiont IJs infected D. pseudoobscura more rapidly than D. melanogaster or D. virilis, but all three fly species succumbed to infection at the same rate (Fig. 6C and D). Thus, the Drosophila-EPN model system can be used to study differences in both nematode infectivity and the host immune response to nematode infection.
FIG 6.
Virulence of EPNs for Drosophila species. (A) D. melanogaster infection. S. carpocapsae and H. bacteriophora symbiont IJs infect D. melanogaster larvae at the same rate (P > 0.05, log rank test; n = 6 to 9 trials for each condition). (B) Survival of D. melanogaster. S. carpocapsae symbiont IJs kill D. melanogaster larvae more rapidly than H. bacteriophora symbiont IJs. ****, P < 0.0001, log rank test. Survival was scored for fly larvae that became infected within 8 h of exposure to IJs (n = 4 to 9 trials for each condition). (C) Infection with S. carpocapsae. S. carpocapsae symbiont IJs infect D. pseudoobscura larvae more rapidly than D. melanogaster and D. virilis larvae. ***, P < 0.001 for D. pseudoobscura relative to D. melanogaster and D. virilis (log rank test with Bonferroni correction; n = 6 to 9 trials for each condition). (D) Survival from S. carpocapsae infection. S. carpocapsae symbiont IJs kill D. melanogaster, D. pseudoobscura, and D. virilis larvae at the same rate (P > 0.05, log rank test; n = 6 to 9 trials for each condition). Note that panels B and D show survival rates only of infected fly larvae rather than of all fly larvae exposed to EPNs. (E) Long-term survival of EPN-infected fly larvae. D. melanogaster larvae infected with H. bacteriophora symbiont IJs show a higher rate of long-term survival than the other Drosophila-EPN combinations. *, P < 0.05, chi-square test (n = 52 to 104 fly larvae for each condition). For graphs in panels A to D, error bars show standard errors of the means.
We also assayed the long-term survival of EPN-infected Drosophila larvae by exposing fly larvae to symbiont IJs, separating out all fly larvae that became infected by 8 h postexposure to IJs, and monitoring their survival until death or adulthood. We found that Drosophila larvae were capable of surviving EPN infection at low levels (Fig. 6E). Moreover, survival rates varied for different EPN species: the long-term survival rate was 1% for D. melanogaster, D. virilis, and D. pseudoobscura infected with S. carpocapsae but 8% for D. melanogaster infected with H. bacteriophora (Fig. 6E). Thus, Drosophila larvae are more successful at overcoming some EPN infections than others. Whether long-term survival occurs because nematodes exit the host shortly after infection or because the host immune system overcomes the infection remains to be determined.
Conclusions.
Our results demonstrate that both S. carpocapsae and its bacterial endosymbiont X. nematophila are pathogenic for D. melanogaster larvae. We also show that EPN species vary in their virulence toward D. melanogaster and that Drosophila species vary in their susceptibility to EPN infection. These differences in virulence reflect differences in both the rates at which fly larvae become infected with EPNs and the rates at which infected fly larvae succumb to the infection. All five of the Drosophila species tested have sequenced genomes (60), and six of the seven nematode species tested have sequenced or nearly sequenced genomes (61, 62). A comparison of Drosophila genomes revealed that many immune-related genes evolve more rapidly than other genes and identified numerous species-specific differences in immune-related genes, including copy number differences in AMP genes (63). Our results establish a versatile model system for investigating at a genome-wide level how genetic differences contribute to the diverse immune responses of insects to parasitic nematode infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Heidi Goodrich-Blair, David Shapiro-Ilan, Adler Dillman, Paul Sternberg, Utpal Banerjee, Cathy Clarke, and the Drosophila Species Stock Center for nematode, fly, and bacterial strains. We also thank Adler Dillman, Michelle Castelletto, Joon Ha Lee, Manon Guillermin, and Kristen Yankura for comments on the manuscript.
J.M.P. was a Howard Hughes Undergraduate Research Program Scholar and was supported by National Institute of General Medical Sciences grant R25GM055052. M.A.C. is an NSF Predoctoral Fellow and a Eugene V. Cota-Robles Fellow. E.A.H. is a MacArthur Fellow, a McKnight Scholar, a Rita Allen Foundation Scholar, and a Searle Scholar.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02740-14.
REFERENCES
- 1.Dillman AR, Sternberg PW. 2012. Entomopathogenic nematodes. Curr Biol 22:R430–R431. doi: 10.1016/j.cub.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciche T. 20 February 2007. The biology and genome of Heterorhabditis bacteriophora. In The C. elegans Research Community (ed), WormBook. doi: 10.1895/wormbook.1.135.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobes P, Wang Z, Markus R, Theopold U, Hyrsl P. 2012. An improved method for nematode infection assays in Drosophila larvae. Fly 6:75–79. doi: 10.4161/fly.19553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallem EA, Rengarajan M, Ciche TA, Sternberg PW. 2007. Nematodes, bacteria, and flies: a tripartite model for nematode parasitism. Curr Biol 17:898–904. doi: 10.1016/j.cub.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Castillo JC, Shokal U, Eleftherianos I. 2012. A novel method for infecting Drosophila adult flies with insect pathogenic nematodes. Virulence 3:339–347. doi: 10.4161/viru.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castillo JC, Shokal U, Eleftherianos I. 2013. Immune gene transcription in Drosophila adult flies infected by entomopathogenic nematodes and their mutualistic bacteria. J Insect Physiol 59:179–185. doi: 10.1016/j.jinsphys.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Hotez P, Hawdon J, Schad GA. 1993. Hookworm larval infectivity, arrest and amphiparatenesis: the Caenorhabditis elegans Daf-c paradigm. Parasitol Today 9:23–26. doi: 10.1016/0169-4758(93)90159-D. [DOI] [PubMed] [Google Scholar]
- 8.Hallem EA, Dillman AR, Hong AV, Zhang Y, Yano JM, DeMarco SF, Sternberg PW. 2011. A sensory code for host seeking in parasitic nematodes. Curr Biol 21:377–383. doi: 10.1016/j.cub.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillman AR, Guillermin ML, Lee JH, Kim B, Sternberg PW, Hallem EA. 2012. Olfaction shapes host-parasite interactions in parasitic nematodes. Proc Natl Acad Sci U S A 109:E2324–E2333. doi: 10.1073/pnas.1211436109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmann S, Ali JG, Helder J, van der Putten WH. 2012. Ecology and evolution of soil nematode chemotaxis. J Chem Ecol 38:615–628. doi: 10.1007/s10886-012-0118-6. [DOI] [PubMed] [Google Scholar]
- 11.Castillo JC, Reynolds SE, Eleftherianos I. 2011. Insect immune responses to nematode parasites. Trends Parasitol 27:537–547. doi: 10.1016/j.pt.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Ciche TA, Ensign JC. 2003. For the insect pathogen Photorhabdus luminescens, which end of a nematode is out? Appl Environ Microbiol 69:1890–1897. doi: 10.1128/AEM.69.4.1890-1897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martens EC, Heungens K, Goodrich-Blair H. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol 185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhurst RJ. 1982. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol 128:3061–3065. [DOI] [PubMed] [Google Scholar]
- 15.Adams BJ, Nguyen KB. 2002. Taxonomy and systematics, p 1–33. In Gaugler R. (ed), Entomopathogenic nematology. CABI Publishing, New York, NY. [Google Scholar]
- 16.Eleftherianos I, ffrench-Constant RH, Clarke DJ, Dowling AJ, Reynolds SE. 2010. Dissecting the immune response to the entomopathogen Photorhabdus. Trends Microbiol 18:552–560. doi: 10.1016/j.tim.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Richards GR, Goodrich-Blair H. 2009. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol 11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brivio MF, Mastore M, Nappi AJ. 2010. A pathogenic parasite interferes with phagocytosis of insect immunocompetent cells. Dev Comp Immunol 34:991–998. doi: 10.1016/j.dci.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Arefin B, Kucerova L, Dobes P, Markus R, Strnad H, Wang Z, Hyrsl P, Zurovec M, Theopold U. 2014. Genome-wide transcriptional analysis of Drosophila larvae infected by entomopathogenic nematodes shows involvement of complement, recognition and extracellular matrix proteins. J Innate Immun 6:192–204. doi: 10.1159/000353734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyrsl P, Dobes P, Wang Z, Hauling T, Wilhelmsson C, Theopold U. 2011. Clotting factors and eicosanoids protect against nematode infections. J Innate Immun 3:65–70. doi: 10.1159/000320634. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, Loseva O, Morgelin M, Ikle J, Cripps RM, Herwald H, Theopold U. 2010. Pathogen entrapment by transglutaminase—a conserved early innate immune mechanism. PLoS Pathog 6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro-Ilan DI, Mizell RF. 2012. Laboratory virulence of entomopathogenic nematodes to two ornamental plant pests, Corythucha ciliata (Hemiptera: Tingidae) and Stethobaris nemesis (Coleoptera: Curculionidae). Fla Entomol 95:922–927. doi: 10.1653/024.095.0415. [DOI] [Google Scholar]
- 23.Li XY, Cowles EA, Cowles RS, Gaugler R, Cox-Foster DL. 2009. Characterization of immunosuppressive surface coat proteins from Steinernema glaseri that selectively kill blood cells in susceptible hosts. Mol Biochem Parasitol 165:162–169. doi: 10.1016/j.molbiopara.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen KB, Smart GC. 1991. Pathogenicity of Steinernema scapterisci to selected invertebrate hosts. J Nematol 23:7–11. [PMC free article] [PubMed] [Google Scholar]
- 25.Canhilal R, Reid W, Kutuk H, El-Bouhssini M. 2007. Susceptibility of sunn pest, Eurygaster integriceps Puton (Hemiptera: Scutelleridae), to various entomopathogenic nematodes (Rhabditida: Steinernematidae and Heterorhabditidae). J Agric Urban Entomol 24:19–26. doi: 10.3954/1523-5475-24.1.19. [DOI] [Google Scholar]
- 26.Campbell JF, Gaugler R. 1997. Inter-specific variation in entomopathogenic nematode foraging strategy: dichotomy or variation along a continuum? Fundam Appl Nematol 20:393–398. [Google Scholar]
- 27.Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737–748. doi: 10.1016/S1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 28.O'Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A 89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton PT, Perlman SJ. 2013. Host defense via symbiosis in Drosophila. PLoS Pathog 9:e1003808. doi: 10.1371/journal.ppat.1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez J, Longdon B, Bauer S, Chan Y-S, Miller WJ, Bourtzis K, Teixeira L, Jiggins FM. 2014. Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog 10:e1004369. doi: 10.1371/journal.ppat.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourtzis K, Pettigrew MM, O'Neill SL. 2000. Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Mol Biol 9:635–639. doi: 10.1046/j.1365-2583.2000.00224.x. [DOI] [PubMed] [Google Scholar]
- 32.Chrostek E, Marialva MSP, Yamada R, O'Neill SL, Teixeira L. 2014. High anti-viral protection without immune upregulation after interspecies Wolbachia transfer. PLoS One 9:e99025. doi: 10.1371/journal.pone.0099025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong ZS, Hedges LM, Brownlie JC, Johnson KN. 2011. Wolbachia-mediated antibacterial protection and immune gene regulation in Drosophila. PLoS One 6:e25430. doi: 10.1371/journal.pone.0025430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards GR, Vivas EI, Andersen AW, Rivera-Santos D, Gilmore S, Suen G, Goodrich-Blair H. 2009. Isolation and characterization of Xenorhabdus nematophila transposon insertion mutants defective in lipase activity against Tween. J Bacteriol 191:5325–5331. doi: 10.1128/JB.00173-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowles CE, Goodrich-Blair H. 2004. Characterization of a lipoprotein, NilC, required by Xenorhabdus nematophila for mutualism with its nematode host. Mol Microbiol 54:464–477. doi: 10.1111/j.1365-2958.2004.04271.x. [DOI] [PubMed] [Google Scholar]
- 36.Ciche TA, Kim KS, Kaufmann-Daszczuk B, Nguyen KC, Hall DH. 2008. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl Environ Microbiol 74:2275–2287. doi: 10.1128/AEM.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hominick WM. 2002. Biogeography, p 115–143. In Gaugler R. (ed), Entomopathogenic nematology. CABI Publishing, New York, NY. [Google Scholar]
- 38.White GF. 1927. A method for obtaining infective nematode larvae from cultures. Science 66:302–303. doi: 10.1126/science.66.1709.302. [DOI] [PubMed] [Google Scholar]
- 39.Vivas EI, Goodrich-Blair H. 2001. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J Bacteriol 183:4687–4693. doi: 10.1128/JB.183.16.4687-4693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro-Ilan DI, Gouge DH, Koppenhofer AM. 2002. Factors affecting commercial success: case studies in cotton, turf, and citrus, p 333–355. In Gaugler R. (ed), Entomopathogenic nematology. CABI Publishing, New York, NY. [Google Scholar]
- 41.Kaya HK, Gaugler R. 1993. Entomopathogenic nematodes. Annu Rev Entomol 38:181–206. doi: 10.1146/annurev.en.38.010193.001145. [DOI] [Google Scholar]
- 42.Dowds BC, Peters A. 2002. Virulence mechanisms, p 79–98. In Gaugler R. (ed), Entomopathogenic nematology. CABI Publishing, New York, NY. [Google Scholar]
- 43.Aymeric JL, Givaudan A, Duvic B. 2010. Imd pathway is involved in the interaction of Drosophila melanogaster with the entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus luminescens. Mol Immunol 47:2342–2348. doi: 10.1016/j.molimm.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Liu H, Liu S, Wang S, Jiang RJ, Li S. 2009. Hormonal and nutritional regulation of insect fat body development and function. Arch Insect Biochem Physiol 71:16–30. doi: 10.1002/arch.20290. [DOI] [PubMed] [Google Scholar]
- 45.Eleftherianos I, Joyce S, Ffrench-Constant RH, Clarke DJ, Reynolds SE. 2010. Probing the tri-trophic interaction between insects, nematodes and Photorhabdus. Parasitol 137:1695–1706. doi: 10.1017/S0031182010000508. [DOI] [PubMed] [Google Scholar]
- 46.Gotz P, Boman A, Boman HG. 1981. Interactions between insect immunity and an insect-pathogenic nematode with symbiotic bacteria. Proc R Soc Lond B 212:333–350. doi: 10.1098/rspb.1981.0043. [DOI] [Google Scholar]
- 47.Ji D, Kim Y. 2004. An entomopathogenic bacterium, Xenorhabdus nematophila, inhibits the expression of an antibacterial peptide, cecropin, of the beet armyworm, Spodoptera exigua. J Insect Physiol 50:489–496. doi: 10.1016/j.jinsphys.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Park Y, Herbert EE, Cowles CE, Cowles KN, Menard ML, Orchard SS, Goodrich-Blair H. 2007. Clonal variation in Xenorhabdus nematophila virulence and suppression of Manduca sexta immunity. Cell Microbiol 9:645–656. doi: 10.1111/j.1462-5822.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- 49.Hwang J, Park Y, Kim Y, Hwang J, Lee D. 2013. An entomopathogenic bacterium, Xenorhabdus nematophila, suppresses expression of antimicrobial peptides controlled by Toll and Imd pathways by blocking eicosanoid biosynthesis. Arch Insect Biochem Physiol 83:151–169. doi: 10.1002/arch.21103. [DOI] [PubMed] [Google Scholar]
- 50.Cerenius L, Lee BL, Soderhall K. 2008. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol 29:263–271. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Ebrahimi L, Niknam G, Dunphy GB. 2011. Hemocyte responses of the Colorado potato beetle, Leptinotarsa decemlineata, and the greater wax moth, Galleria mellonella, to the entomopathogenic nematodes, Steinernema feltiae and Heterorhabditis bacteriophora. J Insect Sci 11:75. doi: 10.1673/031.011.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X-Y, Cowles RS, Cowles EA, Gaugler R, Cox-Foster DL. 2007. Relationship between the successful infection by entomopathogenic nematodes and the host immune response. Int J Parasitol 37:365–374. doi: 10.1016/j.ijpara.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Gaugler R, Cui L. 1994. Variations in immune response of Popillia japonica and Acheta domesticus to Heterorhabditis bacteriophora and Steinernema species. J Nematol 26:11–18. [PMC free article] [PubMed] [Google Scholar]
- 54.Song CJ, Seo S, Shrestha S, Kim Y. 2011. Bacterial metabolites of an entomopathogenic bacterium, Xenorhabdus nematophila, inhibit a catalytic activity of phenoloxidase of the diamondback moth, Plutella xylostella. J Microbiol Biotechnol 21:317–322. [PubMed] [Google Scholar]
- 55.Seo S, Lee S, Hong Y, Kim Y. 2012. Phospholipase A2 inhibitors synthesized by two entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus temperata subsp. temperata. Appl Environ Microbiol 78:3816–3823. doi: 10.1128/AEM.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poinar GO., Jr 1979. Nematodes for biological control of insects. CRC Press, Boca Raton, FL. [Google Scholar]
- 57.Hodson AK, Friedman ML, Wu LN, Lewis EE. 2011. European earwig (Forficula auricularia) as a novel host for the entomopathogenic nematode Steinernema carpocapsae. J Invertebr Pathol 107:60–64. doi: 10.1016/j.jip.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen KB, Smart GC. 1990. Steinernema scapterisci n. sp (Rhabditida: Steinernematidae). J Nematol 22:187–199. [PMC free article] [PubMed] [Google Scholar]
- 59.Frank JH. 2009. Steinernema scapterisci as a biological control agent of Scapteriscus mole crickets, p115–132. In Hajek AE, Glare TR, O'Callaghan M (ed), Use of microbes for control and eradication of invasive arthropods. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 60.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, Roark M, Wiley KL Jr, Kulathinal RJ, Zhang P, Myrick KV, Antone JV, Celniker SE, Gelbart WM, Kellis M. 2007. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dillman AR, Mortazavi A, Sternberg PW. 2012. Incorporating genomics into the toolkit of nematology. J Nematol 44:191–205. [PMC free article] [PubMed] [Google Scholar]
- 62.Bai X, Adams BJ, Ciche TA, Clifton S, Gaugler R, Kim KS, Spieth J, Sternberg PW, Wilson RK, Grewal PS. 2013. A lover and a fighter: the genome sequence of an entomopathogenic nematode Heterorhabditis bacteriophora. PLoS One 8:e69618. doi: 10.1371/journal.pone.0069618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat Genet 39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.