Abstract
Clostridium difficile infection (CDI), one of the most common hospital-acquired infections, is increasing in incidence and severity with the emergence and diffusion of hypervirulent strains. CDI is precipitated by antibiotic treatment that destroys the equilibrium of the gut microbiota. Human α-defensin 5 (HD5), the most abundant enteric antimicrobial peptide, is a key regulator of gut microbiota homeostasis, yet it is still unknown if C. difficile, which successfully evades killing by other host microbicidal peptides, is susceptible to HD5. We evaluated, by means of viability assay, fluorescence-activated cell sorter (FACS) analysis, and electron microscopy, the antimicrobial activities of α-defensins 1 and 5 against a panel of C. difficile strains encompassing the most prevalent epidemic and hypervirulent PCR ribotypes in Europe (012, 014/020, 106, 018, 027, and 078). Here we show that (i) concentrations of HD5 within the intestinal physiological range produced massive C. difficile cell killing; (ii) HD5 bactericidal activity was mediated by membrane depolarization and bacterial fragmentation with a pattern of damage peculiar to C. difficile bacilli, compared to commensals like Escherichia coli and Enterococcus faecalis; and (iii) unexpectedly, hypervirulent ribotypes were among the most susceptible to both defensins. These results support the notion that HD5, naturally present at very high concentrations in the mucosa of the small intestine, could indeed control the very early steps of CDI by killing C. difficile bacilli at their germination site. As a consequence, HD5 can be regarded as a good candidate for the containment of hypervirulent C. difficile strains, and it could be exploited in the therapy of CDI and relapsing C. difficile-associated disease.
INTRODUCTION
Clostridium difficile, a Gram-positive, spore-forming anaerobic bacterium, is considered the major known cause of health care-associated infectious diarrhea in Western countries (1). The disease spectrum caused by C. difficile infection (CDI) ranges from mild diarrhea to severe pseudomembranous enterocolitis, sepsis, and death (2). Hospitalization and old age are major risk factors, and recent reports reveal an alarming association of pediatric CDI with increased mortality in hospitalized children (3).
In the past decade, increasing rates of CDI have been reported in North America and Europe, with a larger proportion of severe and recurrent cases of C. difficile-associated disease (CDAD) than previously reported (4). The most frequently reported toxigenic isolates belong to PCR ribotype 001 (RT001), RT012, RT014/020, RT017, RT106, RT027, RT078, and RT018 (5). In particular, PCR ribotypes 027 and 078 and the more recently described ribotype 018 (6, 7) are referred to as hypervirulent. Each of them carries one or more virulence factors, such as production of binary toxin, mutation in regulatory toxin genes tcdC and tcdD, or fluoroquinolone (FQ) resistance, and all are strongly associated with increased severity of CDI and higher attributable mortality (4, 5, 8).
C. difficile is transmitted via endospores that resist the acidity of the stomach and germinate in the small intestine; the resulting vegetative cells colonize the colon and can reside there asymptomatically for a long time (9). Disruption of the normal gut microflora by broad-spectrum antibiotics (10) allows C. difficile to proliferate and cause disease through the production of cytotoxic toxins A and B (11). Host protection against bacterial pathogens in the intestinal environment is largely mediated by a number of gene-encoded antimicrobial proteins and peptides (AMPs) (12). In mammals, defensins are the major group of AMPs (13); myeloid α-defensins 1 to 3 (HNP1 to HNP3) are expressed predominantly by neutrophils and kill pathogens at the sites of inflammation, while enteric α-defensins 5 and 6 (HD5 and HD6) are released by Paneth cells in the small intestine and patrol the intestinal mucosa (14). HD5 is the most abundant enteric AMP: it has been estimated that up to 450 μg/cm2 is stored in the ileal mucosa, with concentrations of 14 to 70 μM (15). Both HNP1 and HD5 have documented microbicidal activity against bacteria, fungi, and viruses (13, 16), as well as enhancing activity on certain adaptive immune responses (17). Such a broad spectrum of activity is based on their distinctive 6-cysteine motif, which results in a characteristic β-sheet structure and a net positive charge which allow α-defensins to target the negatively charged outermost leaflet of most pathogens (13, 18).
Intestinal microbiota homeostasis is maintained by the dynamic interplay between AMPs, mainly HD5, and commensal bacteria (19). HD5 controls the enteric microbiota composition by selective killing of bacterial pathogens while preserving commensals (20, 21); in turn, resident bacteria stimulate HD5 production via Toll-like receptor (TLR)-MyD88 signaling (22). In mice, oral antibiotic treatment results in a dramatic drop of HD5 gene transcription, which is correlated with loss of commensal microbiota diversity (23). Reduction or absence of HD5 release in the intestinal mucosa has been associated with Crohn's disease (24), susceptibility to enteric pathogens (25), changes in composition of the microbiota, and disruption of intestinal immune homeostasis (19). On the other hand, excess HD5 accumulation in the intestinal mucus of cystic fibrosis patients has been associated with resistance to CDAD (26). In addition, HD5 at concentrations commonly found in the small intestine efficiently neutralizes C. difficile toxin B, one of the most potent virulence factors of C. difficile (27).
In the course of CDI, the interaction of C. difficile toxins with colonic cells triggers a significant inflammatory response and neutrophil accumulation at the site of epithelial damage, with massive release of HNP1 (12, 28). Several recent studies have demonstrated that neutrophils are critical for defense against C. difficile infection (29, 30).
Despite considerable evidence suggesting an influence of α-defensins on C. difficile intestinal colonization, it is still unknown if C. difficile vegetative cells, which are responsible for toxin production, are susceptible to bactericidal activity of α-defensins (28). As a matter of fact, bacteria have evolved numerous mechanisms to resist AMPs (31), and C. difficile has exploited different strategies, including alteration of surface charge and secretion of proteases, to evade bacterial and host-derived cationic AMPs (32, 33). This scenario prompted us to investigate the susceptibility of the vegetative morphotype of C. difficile to α-defensins 1 and 5. We also addressed the issue of different efficacies for a panel of clinical isolates characterized by different virulence and epidemic features.
Here we report for the first time that human α-defensins exert potent dose-dependent damage to the vegetative isoform of C. difficile, resulting in plasma membrane depolarization and bacterial fragmentation. All strains tested were highly susceptible to the microbicidal activity of both α-defensins, with epidemic hypervirulent isolates being among the most sensitive to the microbicidal activity of α-defensins.
MATERIALS AND METHODS
Bacterial strains and antimicrobial peptides.
As summarized in Table 1, three reference strains of C. difficile were used in this study: CD630 (PCR ribotype 012, ATCC BAA-1382), obtained from LGC Standards (Teddington, United Kingdom), and R20291 (PCR ribotype 027, NCTC 13366) and N1 (PCR ribotype 001, NCTC 11204), obtained from the National Collection of Type Cultures (NCTC, Health Protection Agency, United Kingdom). Clinical isolates CD1470 and UP106 were kind gifts from M. Rupnik (University of Maribor, Slovenia) and from V. Pasquale (Parthenope University of Naples), respectively. All the other clinical isolates were collected at the Ospedale San Raffaele, Italy, and selected for the presence of toxins A and B by VIDAS assay (bioMérieux). C. difficile strains were characterized by standard PCR ribotyping (7) and detection of enterotoxin genes tcdA and tcdB, binary toxin (cdtA and cdtB) genes, and variations in the tcdC gene (7). The presence of the determinant for resistance to macrolide/lincosamide/streptogramin B (MLSB), ermB, was determined according to the method of Sutcliffe et al. (34), while the C245T mutation in gyrA, which confers high-level resistance to fluoroquinolones, was detected according to the method of Carman et al. (35). Escherichia coli ATCC 25922 and Enterococcus faecalis ATCC 29212 were both from the American Type Culture Collection (ATCC; Rockville, MD). All the experiments were conducted under anaerobic conditions (Concept 400 anaerobic chamber; Ruskinn Technologies, Leeds, United Kingdom) for C. difficile under both aerobic and anaerobic conditions for E. coli and E. faecalis for comparison purposes.
TABLE 1.
Molecular characterization of C. difficile strainsa
| Strain | PCR ribotype | Main virulence factors |
Sourcee | ||||
|---|---|---|---|---|---|---|---|
| tcdA/tcdB | Binary toxinb | Variations in tcdC | FQ resistancec | MLSB resistanced | |||
| C. difficile | |||||||
| N1 (NCTC 11204) | 001 | +/+ | − | − | − | − | NCTC |
| CD630 (ATCC BAA-1382) | 012 | +/+ | − | − | − | + | ATCC |
| CD496 | 014/020 | +/+ | − | − | − | − | OSR |
| CD501 | 014/020 | +/+ | − | − | − | − | OSR |
| UP106 | 106 | +/+ | − | − | − | − | Parthenope University of Naples, Italy |
| CD1470 | 017 | −/+ | + | − | − | − | University of Maribor, Slovenia |
| CD369 | 018 | +/+ | − | − | + | − | OSR |
| CD483 | 018 | +/+ | − | − | + | − | OSR |
| CD498 | 018 | +/+ | − | − | + | − | OSR |
| R20291 (NCTC 13366) | 027 | +/+ | + | Δ18 bp/Δ117 | + | − | NCTC |
| CD349 | 027 | +/+ | + | Δ18 bp/Δ117 | + | − | OSR |
| CD513 | 027 | +/+ | + | Δ18 bp/Δ117 | + | − | OSR |
| CD683 | 027 | +/+ | + | Δ18 bp/Δ117 | + | − | OSR |
| CD524 | 078 | +/+ | + | Δ39 bp/C184T | + | − | OSR |
| CD740 | 078 | +/+ | + | Δ39 bp/C184T | + | − | OSR |
| CD528 | 078 | +/+ | + | Δ39 bp/C184T | − | + | OSR |
| CD731 | 078 | +/+ | + | Δ39 bp/C184T | − | − | OSR |
| Human commensals | |||||||
| E. coli ATCC 25922 | ATCC | ||||||
| E. faecalis ATCC 29212 | ATCC | ||||||
+, present; −, absent.
PCR detection (cdtA and cdtB).
C245T mutation in gyrA.
Presence of ermB.
OSR, Ospedale San Raffaele, Italy.
Recombinant human α-defensin HNP1 and HD5 were from Peptides International, Inc. (Louisville, KY). The synthetic peptide RL26495 was used as control peptide since it shares with α-defensins similarities of molecular weight, isoelectric point, and net charge. Both α-defensins and control peptide were reconstituted in 0.01% acetic acid as recommended by the manufacturer and stored as single-use aliquots at −80°C.
CFU antimicrobial assay.
According to previous reports showing that high ionic strength of bacterial broths is a major inactivation factor for the microbicidal activity of α-defensins (12, 36) and to match the low ionic strength and pH of the intestinal mucous layer, we used Schaedler broth diluted 1:6 (SB6 medium) in sterile distilled water. This medium does not compromise bacterial growth, as previously reported (37–39). Bacteria (1 × 106) from late logarithmic (C. difficile) and mid-logarithmic (E. coli and E. faecalis) growth phase suspensions were incubated in the presence or absence of increasing concentrations of HNP1, HD5, or RL26495 for 2 h at 37°C in a final volume of 100 μl of SB6 medium. Each incubation mixture (treated and untreated) was serially diluted, spread in triplicate on Columbia blood agar plates (Becton Dickinson), and incubated at 37°C for 72 h for C. difficile and 18 h for E. coli and E. faecalis, followed by determination of CFU count. Each assay was repeated at least three times independently. The percentage of surviving bacteria was calculated as 100 × (1 − r/c), where c is the number of CFU in untreated controls (SB6 medium alone) and r is the number of CFU in treated samples. The concentration of defensin that caused 50% bacterial growth inhibition (IC50) was calculated from a plot of percent inhibition versus the logarithm of defensin concentration.
Flow cytometric antimicrobial assay.
Flow cytometric antimicrobial assay for measuring membrane depolarization of bacteria was carried out on all strains listed in Table 1 as previously described, with minor modifications (38). Briefly, after 2 h of incubation with defensins, RL26495, or SB6 medium only, DiBAC4(3) [bis-(1,3-dibutylbarbituric acid) trimethine oxonol; Sigma-Aldrich, St. Louis, MO], a dye sensitive to membrane potential, was added at a concentration of 1 mg/ml and incubated for 10 min at room temperature. Cell damage leads to the uptake of the dye in the bacterial cells and an increase in fluorescence. Bacterial suspensions were then centrifuged for 10 min at 4,500 × g, and the bacterial pellets were resuspended in 300 μl of phosphate-buffered solution (PBS) filtered through a 0.22-μm filter. Samples were kept on ice until fluorescence-activated cell sorter (FACS) acquisition. For each sample, 40,000 events were analyzed on a Gallios 775016 flow cytometer (Beckman Coulter, Brea, CA). The percentage of depolarized fluorescent bacteria and the percentage of bacterial cell fragments were determined by evaluation of forward scatter and fluorescence 1 parameters. The antimicrobial activity of defensins was expressed as percentage of depolarized bacteria in defensin- or RL26495-treated samples compared to medium-only-treated controls. Data obtained were analyzed using FlowJo software (Treestar, Ashland, OR).
Transmission electron microscopy studies.
Approximately 2 × 108 CFU of C. difficile strain CD630 was incubated anaerobically for 2 h at 37°C in SB6 medium in the presence or absence of 7 μM HNP1, HD5, or control peptide RL26495. For transmission electron microscopy, bacteria were fixed at 4°C for 15 min in 125 mM cacodylate buffer containing 4% paraformaldehyde and 2.5% glutaraldehyde. After centrifugation, the pellet was postfixed for 1 h in 2.0% osmium tetroxide in 125 mM cacodylate buffer, washed, and embedded in Epon resin (Sigma). Ultrathin sections (30 nm) were cut using an Ultracut microtome (Reichert, Austria), mounted on copper grids, double contrasted with uranyl and lead citrate, and analyzed using a LEO912 electron microscope (Leo Electron Microscopy Ltd., Cambridge, United Kingdom).
Statistical analysis.
We used the Wilcoxon Mann-Whitney rank sum 2-tailed test to determine statistical significance for experiments on the susceptibilities of C. difficile strains belonging to different PCR ribotypes to α-defensins (see Fig. 4). Gaussian distribution was determined using the Kolmogorov-Smirnov test. Fisher's exact test was used for the comparison of categorical variables for experiment of dose-dependent bacterial damage (see Fig. 3B). Results are presented as means ± standard errors. P values of <0.05 were considered statistically significant. Data were analyzed using GraphPad Prism 5.0 software (San Diego, CA).
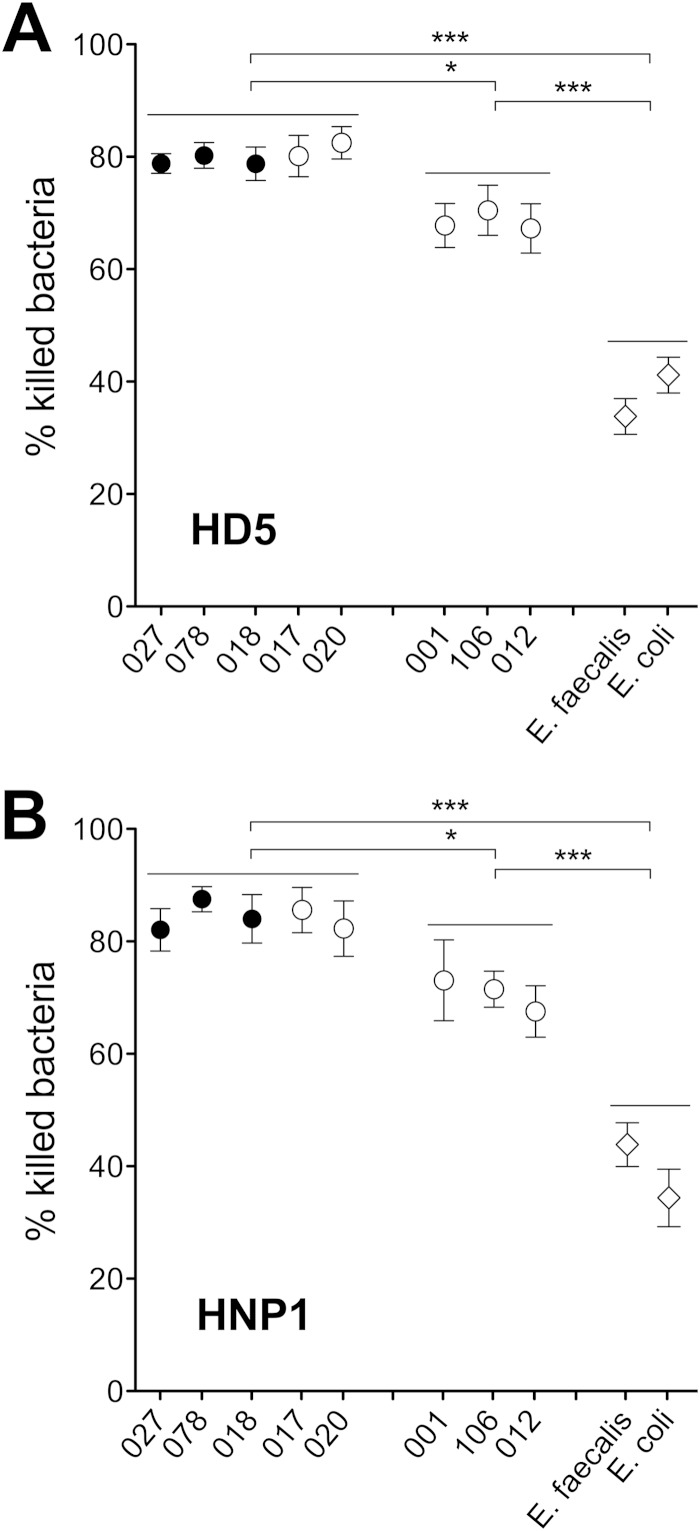
FIG 4.
Susceptibility to α-defensins of C. difficile strains belonging to different PCR ribotypes. Bacterial suspensions were tested against 7 μM HD5 (A) and HNP1 (B) or control peptide (data not shown) for 2 h at 37°C. Values are means ± SEMs for C. difficile hypervirulent and other epidemic PCR ribotypes, E. coli ATCC 25922, and E. faecalis ATCC 29212. P values were calculated by the Wilcoxon Mann-Whitney rank sum 2-tailed test.
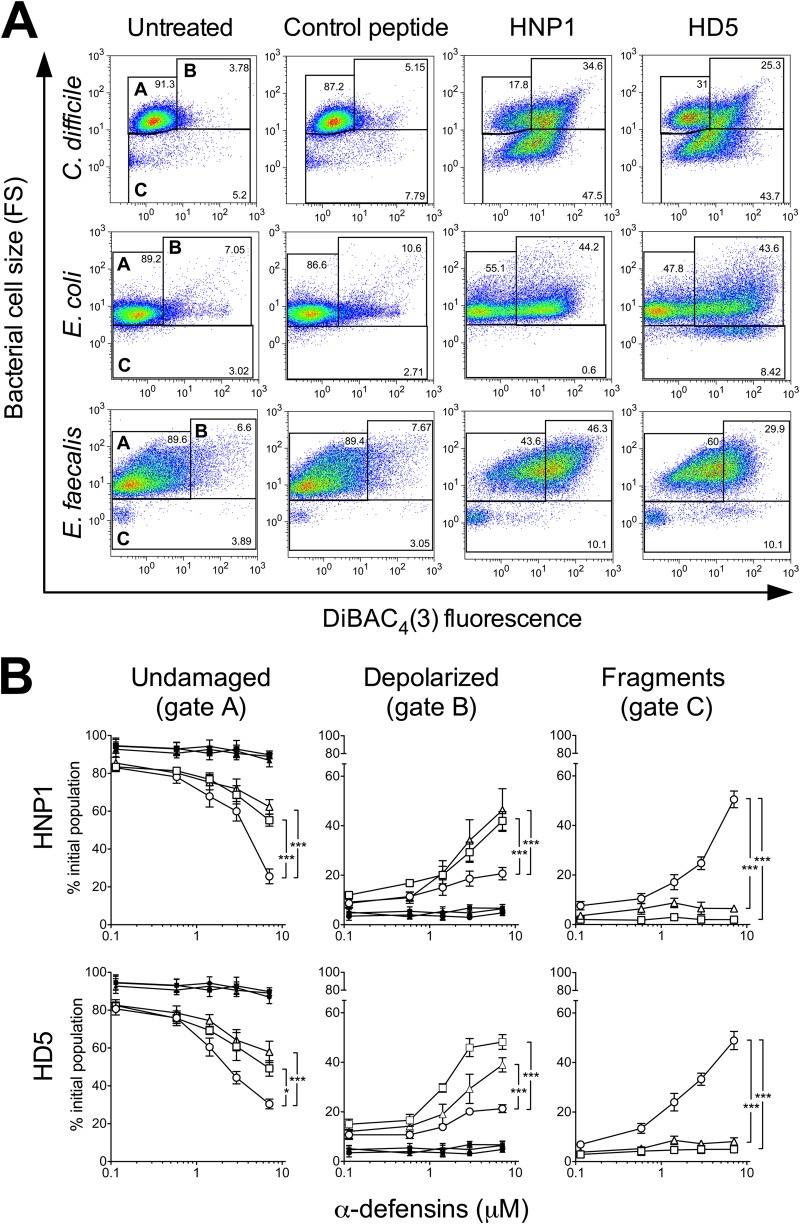
FIG 3.
Killing of C. difficile by α-defensins results in a peculiar pattern of damage. (A) Dot blots of C. difficile CD630 (RT012), E. coli ATCC 25922, and E. faecalis ATCC 29212 populations after incubation with 7 μM HD5, HNP1, or control peptide RL26495. Membrane depolarization and cell size were recorded by FACS analysis. (B) Quantification of dose-dependent bacterial damage. Percentages of undamaged bacteria (gate A), depolarized fluorescent bacteria (gate B), and bacterial cell fragments (gate C) were obtained by treatment with increasing concentrations of α-defensins (open symbols) or control peptide RL26495 (solid symbols) and analyzed by FACS. Circles, CD630; squares, E. coli ATCC 25922; triangles, E. faecalis ATCC 29212. Each data point is representative of 4 to 6 independent experiments. Error bars show mean values ± SEMs. *, P < 0.05; ***, P < 0.001 (compared to C. difficile) according to Fisher's exact test.
RESULTS
Selection and characterization of C. difficile clinical isolates.
In this study, we compared the sensitivities to HD5 and HNP1 of eight of the most common PCR ribotypes in Europe (001, 012, 014/020, 106, 017, 018, 027, and 078), including three PCR ribotypes (018, 027, and 078) associated with more severe outcomes and greater attributable mortality and therefore considered hypervirulent (4, 5, 40). As illustrated in Table 1, all isolates were toxigenic, i.e., tested positive for the presence of toxins A and B. CD1470 belongs to PCR ribotype 017 and, as expected, tested singly positive for toxin B (41). PCR ribotype 027 included R20291, a reference strain entirely sequenced and characterized phenotypically (42, 43), and three clinical isolates (CD349, CD513, and CD683) from the stools of CDI patients. All these strains were positive for several virulence factors at the same time: FQ resistance (C245T mutation in gyrA), deletions in tcdC (18-bp deletion and point deletion in 117), and the presence of binary toxin genes (cdtA and cdtB) (Table 1). Similar to the PCR ribotype 027 strains, the four clinical isolates belonging to PCR ribotype 078 (CD524, CD740, CD528, and CD731) were positive for C. difficile binary toxin genes and for variation in tcdC (point mutation C184T and 39-bp deletion), but not all were FQ resistant. In addition, CD528, which is not FQ resistant, carried resistance to MLSB (mutation in ermB). PCR ribotype 018 is highly diffused in Italy and has been significantly associated with complicated outcomes (5); isolates CD369, CD483, and CD498 were all positive for FQ resistance, but none of them had the genes for the binary toxins or mutations in tcdC. PCR ribotypes 001, 014/020, and 106, which are common but not considered epidemic, were negative for all considered virulence factors and lacked the determinants associated with resistance to FQs and MLSB.
C. difficile bacilli are highly susceptible to α-defensins 1 and 5.
As the first step, we investigated the effects of HNP1 and HD5 on the viability of C. difficile reference strain CD630 PCR ribotype 012 in comparison with the effects on E. coli and E. faecalis, two prominent species of the intestinal flora. We used the reference strains E. coli ATCC 25922 and E. faecalis ATCC 29212 (Fig. 1B), which were previously shown to be, respectively, susceptible and partially resistant to several defensins (37, 44, 45). Preliminary studies on α-defensin bactericidal activity indicated that the maximal effect occurred within 2 h (data not shown), consistent with previously reported data (37).
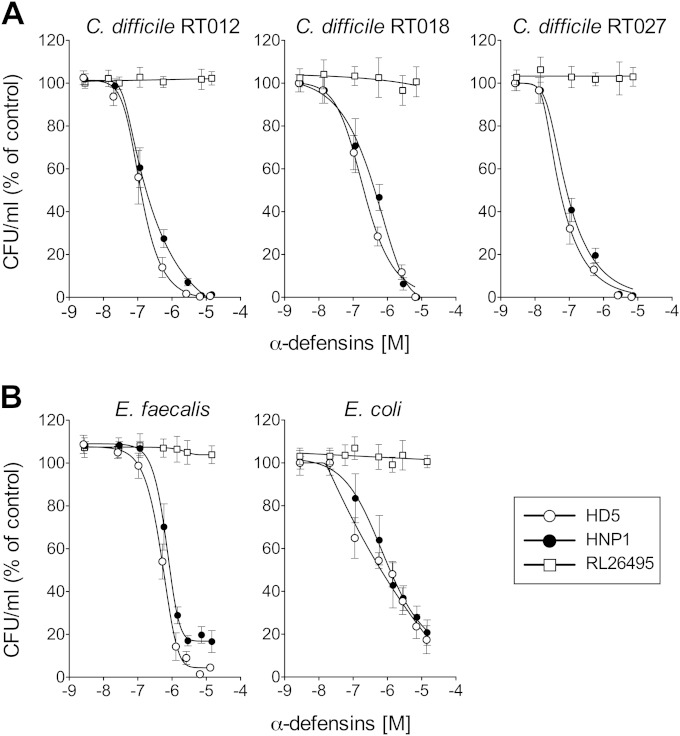
FIG 1.
HD5 and HNP1 inhibit C. difficile growth in vitro. (A) C. difficile reference strain CD630 (RT012) and clinical epidemic strains CD369 (RT018) and CD349 (RT027). (B) Comparison with α-defensin activity against E. coli ATCC 25922 and E. faecalis ATCC 29212. All strains were exposed for 2 h at 37°C to HD5, HNP1, or unrelated control peptide RL26495 at concentrations ranging from 0.1 to 14 μM. The number of CFU was determined in triplicate and expressed as percentage of the initial inoculum. The means ± SEMs for at least three independent experiments are shown.
Although reported to be resistant to several cationic AMPs (46), C. difficile CD630 (RT012) was highly susceptible to both defensins, with IC50s in the nanomolar range, 224 nM for HNP1 and 131 nM for HD5. These values were in the same range as those obtained with E. coli (HNP1, IC50 of 473 nM, and HD5, IC50 of 168 nM), which is described to be highly susceptible to α-defensins (37, 45). E. faecalis showed a lower sensitivity to α-defensins, with IC50s of 685 nM and 548 nM, respectively, consistent with its lower susceptibility to enteric antimicrobials (38) (Fig. 1). No effect was seen with the control peptide, indicating the specificity of the activities of the defensins (Fig. 1). HD5 was slightly more effective than HNP1 against all bacteria tested, but the difference did not reach statistical significance.
Next, we extended the study to two epidemic strains of C. difficile: R20291 (RT027) and CD369 (RT018). As shown in Fig. 1A, both strains showed a much higher susceptibility to killing by α-defensins: HNP1 had an IC50 of 402 nM and HD5 had an IC50 of 218 nM for CD369, and HNP1 had an IC50 of 83 nM and HD5 had an IC50 of 56 nM for R20291.
α-Defensins induce severe and widespread damage to the C. difficile cell wall and plasma membrane.
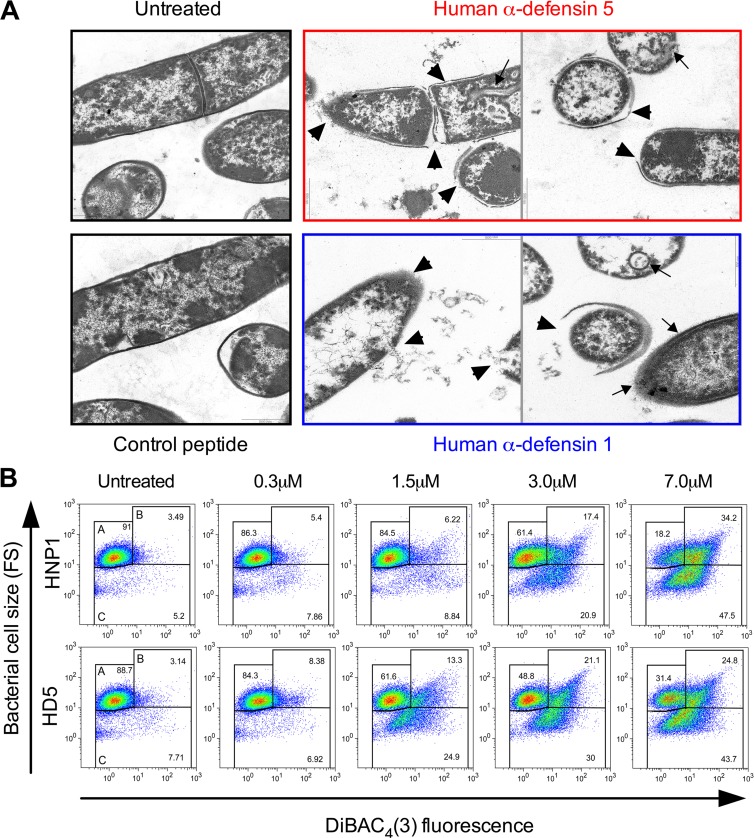
To gain insight into the mechanism of inhibition of C. difficile by α-defensins, we analyzed by transmission electron microscopy C. difficile CD630 (RT012) bacilli incubated in the presence or absence of 7 μM HNP1, HD5, or control peptide RL26495. α-Defensins used at concentrations that can be normally found in the small intestine (15) caused severe and widespread morphological changes in treated samples. As illustrated in Fig. 2A, both defensins caused strong damage of the bacterial cell wall, which appeared to be detached or missing in the majority of bacterial cells analyzed. Several breaches were visible in the plasma membrane, with leakage of cytoplasmic content (arrowheads). Spherical double-layered mesosome-like structures and fibers extending from the cell surface were also frequently detected (arrows). Mesosomes are produced by the lateral expansion of the membrane area occurring upon binding and insertion of the AMPs (47). In contrast, control peptide RL26495 did not cause any morphological damage of bacterial cells (Fig. 2A).
FIG 2.
Bactericidal effects of HD5 and HNP1 on C. difficile. (A) Transmission electron micrographs of α-defensin-treated C. difficile. Suspensions of C. difficile CD630 (RT012) bacilli were incubated in the absence or in the presence of 7 μM HD5, HNP1, or control peptide RL26495. Arrowheads indicate cell wall detachment or severe leakage of cytoplasmic contents; arrows indicate mesosome-like structures and fibers extending from the cell surface. (B) Dose-dependent bactericidal effects of HD5 and HNPl against C. difficile R20291 (RT027). Bacteria were incubated in the presence or absence of increasing concentrations of α-defensins, and the degrees of membrane depolarization [DiBAC4(3) binding (FLl)] and bacterial fragmentation (FS) were quantified by FACS analysis. Gate A, undamaged bacteria; gate B, depolarized bacteria; gate C, bacterial cell fragments. All plots are representative of at least four independent experiments.
α-Defensins cause dose-dependent plasma membrane depolarization and bacterial cell fragmentation.
The bactericidal activity of AMPs is mainly based on the attachment of cationic peptides to the negatively charged bacterial cell surface. This results in electrostatic charge-based depolarization and disruption of the plasma membrane (12). Thus, to further characterize and quantify the bactericidal activity of α-defensins against C. difficile, we analyzed by flow cytometry at the single-cell level the effects of increasing concentrations of HNP1 and HD5 on four C. difficile strains of different ribotypes (CD630 [RT012], CD369 [RT018], R20291 [RT027], and CD524 [RT078]) by evaluating the simultaneous uptake of the membrane potential-sensitive dye DiBAC4(3) (48) and the change in bacterial cell size. As shown in Fig. 2B, in a representative experiment with reference strain R20291, this assay enabled us to discriminate intact nonfluorescent bacteria (gate A; low fluorescence and medium particle size), depolarized fluorescent bacteria and aggregates of damaged bacteria (gate B; high fluorescence and medium to large particle size), and bacterial fragments (gate C; variable fluorescence and very small particle size). Figure 2B illustrates that the percentage of depolarized bacteria (green fluorescence channel, FL1, gate B) and the amount of bacterial fragments (forward scatter [FS], gate C) increased proportionally to the defensin concentration. Smaller particles (gate C) also carried bound DiBAC4(3), thus indicating their bacterial origin. The minimal active concentration of α-defensins was 0.3 μM, and the maximal effect was observed at 7 μM. HD5 demonstrated higher bactericidal activity than HNP1 in the range of concentrations between 0.3 μM and 3 μM, while maximal concentrations of both defensins resulted in similar bactericidal activities. This was consistent with CFU data (Fig. 1A). Treatment with control peptide RL26495 resulted in minimal physiological depolarization and fragmentation levels (Fig. 3A).
We then compared by FACS analysis the morphological changes produced by HNP1, HD5, and control peptide RL26495 in strain R20291 (RT027) in comparison with E. coli ATCC 25922 and E. faecalis ATCC 29212. As shown in representative plots (Fig. 3A) and quantified in dose-dependent assays (Fig. 3B), both defensins caused dose-dependent killing, i.e., decreased numbers of viable cells (Fig. 3B, gate A) in all the species considered. However, while α-defensin treatment of E. coli and E. faecalis mainly induced a shift of the bacterial cell population toward higher green fluorescence, i.e., depolarized bacteria (Fig. 3A, gate B), the C. difficile cell population was mostly fragmented, as indicated by a dramatic decrease in particle size (Fig. 3A, gate C). These differences were statistically significant, as shown in quantitative dose-dependent assays (Fig. 3B).
Broad-spectrum inhibitory activity of α-defensins 1 and 5 on C. difficile strains belonging to different PCR ribotypes.
In order to understand the potential roles of HD5 and HNP1 in the host protection against C. difficile epidemic strains with different virulence features, we quantified by flow cytometry the susceptibilities to α-defensin-mediated killing of a panel of C. difficile strains representative of the most diffuse epidemic PCR ribotypes (Fig. 4 and Table 1). We found that both HD5 and HNP1, used at 7 μM, exhibited broad-spectrum antimicrobial activity against all the C. difficile strains tested, with percentages of killed bacteria ranging from 67.5% ± 4.6% to 87.5% ± 2.2% for HNP1 and from 67.3% ± 4.3% to 82.4% ± 2.8% for HD5. Both of them resulted in significantly more damage to C. difficile than for E. faecalis (HNP1, P < 0.05, and HD5, P < 0.01) but also than for the susceptible control bacterium, E. coli (HNP1 and HD5 P < 0.001), as shown in Fig. 4. As a group, hypervirulent PCR ribotypes 027, 078, and 018 and epidemic ribotypes 017 and 020 were the most susceptible to HNP1 (82.0% ± 3.7% to 87.5% ± 2.2%) and HD5 (73.8% ± 1.7% to 80.4% ± 2.3%). Interestingly, among all the C. difficile PCR ribotypes studied, strains N1 (RT001), CD630 (RT012), and UP106 (RT106) (Fig. 4) showed the lowest susceptibilities, although the susceptibilities were greater than those of E. coli and E. faecalis. The commensal E. faecalis, in agreement with CFU data (Fig. 1B) and with previously published studies (49), showed very low susceptibility to α-defensins (HNP1, 43.7% ± 3.9%, and HD5, 33.8% ± 3.2%), thus confirming itself as a partially resistant species. All tested strains were unaffected by treatment with control peptide RL26495 tested at 7 μM (data not shown).
DISCUSSION
The mechanisms underlying the ability of C. difficile to colonize the intestine and to evade host innate immune responses are still poorly understood. Here we report for the first time that neutrophil HNP1 and enteric HD5 exert potent inhibitory activities against C. difficile epidemic strains belonging to PCR ribotypes characterized by different virulence features.
The observation that C. difficile can reside asymptomatically in the intestines of immunocompetent individuals, whereas severe CDAD occurs mainly in immunocompromised or elderly subjects (2), strongly suggests that host immune responses are important determinants of disease pathogenesis (11). HD5 not only restricts C. difficile colonization by maintaining microbiota homeostasis and inactivating C. difficile toxin B (27) but also, as described here, directly kills C. difficile bacilli. This was not an obvious finding: cationic AMPs share a mechanism of action, and C. difficile has evolved numerous strategies to evade their attack (28). C. difficile has been described to be resistant to bacterially derived AMPs, like bacitracin, nisin, gallidermin, vancomycin, and polymyxin B, but also to host-derived AMPs, like lysozyme (46, 50). Furthermore, resistance to mammalian SMAP-29 and LL-37 was reported for epidemic-associated PCR ribotype 027 isolates (33). We found that C. difficile is susceptible to both HD5 and HNP1, with IC50s in the nanomolar range; thus, HD5, which in the small intestine can reach concentrations of 70 μM (15), is likely to largely block the replication of clostridia at the site of germination (9), suggesting a protective role against C. difficile colonization. Indeed, proof of principle for HD5-associated antimicrobial activity in vivo has been obtained with transgenic mice by showing a direct cause-effect relation between the presence or absence of HD5 expression and survival of infection with the enteric pathogen Salmonella enterica serovar Typhimurium (51). Most importantly, all strains belonging to PCR ribotype 027 were shown to be twice as susceptible to both HNP1 and HD5 as reference strain CD630 and almost 10 times more susceptible to HD5 than was E. faecalis (Fig. 1). These results clearly suggest that α-defensins can circumvent the mechanisms of evasion adopted by C. difficile to resist cathelicidin LL-37 (33). A plausible explanation relies on cationic peptide structure: LL-37 is characterized by an extended α-helical structure and can be cleaved and inactivated by bacterial proteases (31), while mature HD5 and HNP1, due to their tightly folded structure, are inherently resistant to proteolysis (52, 53).
Our findings also address the notion of a peculiar mechanism of interaction between α-defensins and C. difficile bacilli. Indeed, transmission electron microscopy analysis showed morphological alterations of the bacterial cell wall and cytoplasmic membrane consistent with the cationic, amphiphilic nature of α-defensins, which are electrostatically attracted by the negatively charged bacterial surface layers and get embedded into the hydrophobic regions of the lipid membranes (54, 55). However, we did not observe blisters, protruding bubbles, and overall moderate damage to the bacterial wall as reported for other Gram-positive bacteria (47, 56, 57); rather, we found massive damage of the bacterial cell wall and plasma membrane, with leakage of cytoplasmic content and widespread cell fragmentation. Accordingly, data from FACS analysis showed a pattern of damage peculiar to C. difficile: bacterial fragmentation was completely absent in E. coli, consistent with its Gram-negative nature (38, 57), but was also absent in E. faecalis, a Gram-positive organism also characterized by a thick peptidoglycan wall. HD5 was more potent than HNP1 at a lower range of concentrations, i.e., 0.3 and 3 μM, whereas maximal concentrations of these two defensins resulted in similar bactericidal activities, with slightly more strength for HNP1. This agrees with the propensity of HD5 to form aggregates at high concentrations, thus losing available sites to interact with the cell membrane (58). Furthermore, such a different range of activities is compatible with the different physiological roles of HD5 and HNP1. Indeed, HD5, which is secreted at high concentrations in the intestinal crypts, gets diluted in the mucous layer and still maintains its bactericidal activity (52, 59). On the other hand, HNP1, whose role is to intervene once the inflammatory process is initiated and a massive bacterial invasion has to be tackled, is more active at the highest levels of the range (7 μM). Accordingly, physiological concentrations of HNP1 are very high, both in neutrophils (above 10 mg/ml) and in neutrophils nets, where it kills, respectively, engulfed and trapped bacteria (12), thus indicating that levels of HNP1 microbicidal for C. difficile bacilli can be easily reached in the extracellular milieu in the vicinity of activated neutrophils.
Variable susceptibilities to α-defensins have been reported among different strains of the same species (38, 49); thus, we exploited FACS analysis to quantify the effect of defensins on C. difficile strains belonging to different PCR ribotypes. Very recent studies have investigated the relationships among strain types, biomarkers, other risk factors, and mortality and demonstrated unequivocally that RT027, RT078, and RT018 strains are associated with a worse prognosis and/or greater mortality (4, 5). HD5 and HNP1 exhibited broad-spectrum antimicrobial activity against all C. difficile strains tested, independently from their epidemic and virulence features. As a matter of fact, the strains belonging to hypervirulent ribotypes 027, 078, and 018 were among the most susceptible to α-defensins, while the least sensitive were represented by ribotypes 001, 012, and 106, which are characterized by lower virulence (5). This moderate resistance suggests that the interaction between α-defensins and C. difficile could follow distinctive mechanisms related to differences in the molecular wall composition, i.e., the presence of a bacterial capsule or of an S-layer (60, 61). We observed that bacterial susceptibility to α-defensins quantified by CFU assays was consistently higher than quantification of damaged cells by FACS analysis, nonetheless maintaining a good correlation. This was consistent with previously published observations (38, 48) and further validated in our experimental system using E. faecalis, E. coli, and C. difficile strains CD630 (RT012), CD369 (RT018), and CD349 (RT027).
Hence, we can speculate that in immunocompetent individuals, C. difficile spores germinate in the small intestine, where bacilli encounter HD5 at concentrations more than 50 times higher than their IC50s (15). Then the surviving bacilli travel to the colon carried by the mucous flow, rich with HD5 and other AMPs (62), which constrains their replicative capacity and protects the host from C. difficile colonization. This hypothesis is supported by experiments showing that HD5 persists in an intact and functional form throughout the all intestinal tract, including the colon (52), and is active throughout a broad pH range (pH 5.5 to 8.0) (36). Upon oral antibiotic treatment, the expression of HD5 and other AMPs is downregulated (23) and pathogenic C. difficile can thrive and produce large quantities of cytotoxic toxin A and B, leading to CDI and CDAD (9). In immunodeficient patients or in the absence of a proper bacterial repopulation, HD5 deficiency persists and leads to recurrent CDAD (11). Concurrently, during early stages of CDI, neutrophil infiltration and release of HNP1 at the site of infection (30) play a beneficial role for the clearance C. difficile bacilli, whereas in the case of advanced stages of CDAD, massive neutrophil infiltration enhances the inflammatory response and leads to host damage (28). Notably, neutrophils, unlike macrophages and lymphocytes, are also resistant to C. difficile toxin A-mediated apoptosis (11).
From this point of view, the fact that C. difficile, especially highly virulent epidemic strains like ribotypes 018, 078, and 027, is highly susceptible to both HD5 and HNP1 could be exploited to prevent and/or treat CDI. From a therapeutic perspective, HD5 used in combination with fecal microbiota transplant therapies (63) would contribute, with its antitoxin, bactericidal, and immunostimulatory actions, to the treatment of detrimental forms of recurrent CDAD (64).
ACKNOWLEDGMENTS
We are grateful to Alessio Palini (Flow Cytometry Core Facility) and Maria C. Panzeri (ALEMBIC, Advanced Microscopy Facility), both at San Raffaele Scientific Institute of Milan, for technical help and guidance.
This study was supported by European Grant (ATP) 2009, Amicrotex project ID 13587782.
There are no conflicts of interests to disclose.
REFERENCES
- 1.Dubberke ER, Olsen MA. 2012. Burden of Clostridium difficile on the healthcare system. Clin Infect Dis 55(Suppl 2):S88–S92. doi: 10.1093/cid/cis335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ananthakrishnan AN. 2011. Clostridium difficile infection: epidemiology, risk factors and management. Nat Rev Gastroenterol Hepatol 8:17–26. doi: 10.1038/nrgastro.2010.190. [DOI] [PubMed] [Google Scholar]
- 3.Sammons JS, Localio R, Xiao R, Coffin SE, Zaoutis T. 2013. Clostridium difficile infection is associated with increased risk of death and prolonged hospitalization in children. Clin Infect Dis 57:1–8. doi: 10.1093/cid/cit155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker AS, Eyre DW, Wyllie DH, Dingle KE, Griffiths D, Shine B, Oakley S, O'Connor L, Finney J, Vaughan A, Crook DW, Wilcox MH, Peto TE. 2013. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis 56:1589–1600. doi: 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ. 2011. Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 6.Spigaglia P, Barbanti F, Dionisi AM, Mastrantonio P. 2010. Clostridium difficile isolates resistant to fluoroquinolones in Italy: emergence of PCR ribotype 018. J Clin Microbiol 48:2892–2896. doi: 10.1128/JCM.02482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldan R, Cavallerio P, Tuscano A, Parlato C, Fossati L, Moro M, Serra R, Cirillo DM. 2010. First report of hypervirulent strains polymerase chain reaction ribotypes 027 and 078 causing severe Clostridium difficile infection in Italy. Clin Infect Dis 50:126–127. doi: 10.1086/649011. [DOI] [PubMed] [Google Scholar]
- 8.Tenover FC, Akerlund T, Gerding DN, Goering RV, Bostrom T, Jonsson AM, Wong E, Wortman AT, Persing DH. 2011. Comparison of strain typing results for Clostridium difficile isolates from North America. J Clin Microbiol 49:1831–1837. doi: 10.1128/JCM.02446-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kachrimanidou M, Malisiovas N. 2011. Clostridium difficile infection: a comprehensive review. Crit Rev Microbiol 37:178–187. doi: 10.3109/1040841X.2011.556598. [DOI] [PubMed] [Google Scholar]
- 10.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 11.Madan R, Petri WA Jr. 2012. Immune responses to Clostridium difficile infection. Trends Mol Med 18:658–666. doi: 10.1016/j.molmed.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiemstra PS, Zaat SA. 2013. Antimicrobial peptides and innate immunity. Springer, Basel, Switzerland. [Google Scholar]
- 13.Lehrer RI, Lu W. 2012. α-Defensins in human innate immunity. Immunol Rev 245:84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 14.Salzman NH. 2011. Microbiota-immune system interaction: an uneasy alliance. Curr Opin Microbiol 14:99–105. doi: 10.1016/j.mib.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. 2002. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol 3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 16.Furci L, Sironi F, Tolazzi M, Vassena L, Lusso P. 2007. Alpha-defensins block the early steps of HIV-1 infection: interference with the binding of gp120 to CD4. Blood 109:2928–2935. doi: 10.1182/blood-2006-05-024489. [DOI] [PubMed] [Google Scholar]
- 17.Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. 2004. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol 22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- 18.Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. 2006. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci 15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eriguchi Y, Takashima S, Oka H, Shimoji S, Nakamura K, Uryu H, Shimoda S, Iwasaki H, Shimono N, Ayabe T, Akashi K, Teshima T. 2012. Graft-versus-host disease disrupts intestinal microbial ecology by inhibiting Paneth cell production of alpha-defensins. Blood 120:223–231. doi: 10.1182/blood-2011-12-401166. [DOI] [PubMed] [Google Scholar]
- 21.Bevins CL, Salzman NH. 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 22.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menendez A, Willing BP, Montero M, Wlodarska M, So CC, Bhinder G, Vallance BA, Finlay BB. 2013. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell alpha-defensins. J Innate Immun 5:39–49. doi: 10.1159/000341630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H Jr, Fellermann K, Ganz T, Stange EF, Bevins CL. 2005. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A 102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 26.Welkon CJ, Long SS, Thompson CM Jr, Gilligan PH. 1985. Clostridium difficile in patients with cystic fibrosis. Am J Dis Child 139:805–808. [DOI] [PubMed] [Google Scholar]
- 27.Giesemann T, Guttenberg G, Aktories K. 2008. Human alpha-defensins inhibit Clostridium difficile toxin B. Gastroenterology 134:2049–2058. doi: 10.1053/j.gastro.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. 2012. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 3:121–134. doi: 10.4161/gmic.19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasegawa M, Yamazaki T, Kamada N, Tawaratsumida K, Kim YG, Nunez G, Inohara N. 2011. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol 186:4872–4880. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 30.Jarchum I, Liu M, Shi C, Equinda M, Pamer EG. 2012. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect Immun 80:2989–2996. doi: 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koprivnjak T, Peschel A. 2011. Bacterial resistance mechanisms against host defense peptides. Cell Mol Life Sci 68:2243–2254. doi: 10.1007/s00018-011-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride SM, Sonenshein AL. 2011. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McQuade R, Roxas B, Viswanathan VK, Vedantam G. 2012. Clostridium difficile clinical isolates exhibit variable susceptibility and proteome alterations upon exposure to mammalian cationic antimicrobial peptides. Anaerobe 18:614–620. doi: 10.1016/j.anaerobe.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 40:2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carman RJ, Genheimer CW, Rafii F, Park M, Hiltonsmith MF, Lyerly DM. 2009. Diversity of moxifloxacin resistance during a nosocomial outbreak of a predominantly ribotype ARU 027 Clostridium difficile diarrhea. Anaerobe 15:244–248. doi: 10.1016/j.anaerobe.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Porter EM, van Dam E, Valore EV, Ganz T. 1997. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun 65:2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest 84:553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuding S, Fellermann K, Wehkamp J, Mueller HA, Stange EF. 2006. A flow cytometric assay to monitor antimicrobial activity of defensins and cationic tissue extracts. J Microbiol Methods 65:335–345. doi: 10.1016/j.mimet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Wehkamp J, Schmid M, Stange EF. 2007. Defensins and other antimicrobial peptides in inflammatory bowel disease. Curr Opin Gastroenterol 23:370–378. doi: 10.1097/MOG.0b013e328136c580. [DOI] [PubMed] [Google Scholar]
- 40.Miller M, Gravel D, Mulvey M, Taylor G, Boyd D, Simor A, Gardam M, McGeer A, Hutchinson J, Moore D, Kelly S. 2010. Health care-associated Clostridium difficile infection in Canada: patient age and infecting strain type are highly predictive of severe outcome and mortality. Clin Infect Dis 50:194–201. doi: 10.1086/649213. [DOI] [PubMed] [Google Scholar]
- 41.Drudy D, Harnedy N, Fanning S, Hannan M, Kyne L. 2007. Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect Control Hosp Epidemiol 28:932–940. doi: 10.1086/519181. [DOI] [PubMed] [Google Scholar]
- 42.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuding S, Fellermann K, Wehkamp J, Stange EF. 2007. Reduced mucosal antimicrobial activity in Crohn's disease of the colon. Gut 56:1240–1247. doi: 10.1136/gut.2006.118646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ericksen B, Wu Z, Lu W, Lehrer RI. 2005. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother 49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride SM, Sonenshein AL. 2011. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedrich CL, Moyles D, Beveridge TJ, Hancock RE. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob Agents Chemother 44:2086–2092. doi: 10.1128/AAC.44.8.2086-2092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deere D, Porter J, Edwards C, Pickup R. 1995. Evaluation of the suitability of bis-(1,3-dibutylbarbituric acid) trimethine oxonol, (diBA-C4(3) −), for the flow cytometric assessment of bacterial viability. FEMS Microbiol Lett 130:165–169. doi: 10.1016/0378-1097(95)00199-F. [DOI] [PubMed] [Google Scholar]
- 49.Nuding S, Zabel LT, Enders C, Porter E, Fellermann K, Wehkamp J, Mueller HA, Stange EF. 2009. Antibacterial activity of human defensins on anaerobic intestinal bacterial species: a major role of HBD-3. Microbes Infect 11:384–393. doi: 10.1016/j.micinf.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Ho TD, Ellermeier CD. 2011. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect Immun 79:3229–3238. doi: 10.1128/IAI.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salzman NH, Chou MM, de Jong H, Liu L, Porter EM, Paterson Y. 2003. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect Immun 71:1109–1115. doi: 10.1128/IAI.71.3.1109-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mastroianni JR, Ouellette AJ. 2009. Alpha-defensins in enteric innate immunity: functional Paneth cell alpha-defensins in mouse colonic lumen. J Biol Chem 284:27848–27856. doi: 10.1074/jbc.M109.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peschel A, Sahl HG. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol 4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 54.Lohner K, Prossnigg F. 2009. Biological activity and structural aspects of PGLa interaction with membrane mimetic systems. Biochim Biophys Acta 1788:1656–1666. doi: 10.1016/j.bbamem.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Huang HW. 2009. Free energies of molecular bound states in lipid bilayers: lethal concentrations of antimicrobial peptides. Biophys J 96:3263–3272. doi: 10.1016/j.bpj.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimoda M, Ohki K, Shimamoto Y, Kohashi O. 1995. Morphology of defensin-treated Staphylococcus aureus. Infect Immun 63:2886–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother 54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajabi M, Ericksen B, Wu X, de Leeuw E, Zhao L, Pazgier M, Lu W. 2012. Functional determinants of human enteric alpha-defensin HD5: crucial role for hydrophobicity at dimer interface. J Biol Chem 287:21615–21627. doi: 10.1074/jbc.M112.367995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. 2000. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol 1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 60.Davies HA, Borriello SP. 1990. Detection of capsule in strains of Clostridium difficile of varying virulence and toxigenicity. Microb Pathog 9:141–146. doi: 10.1016/0882-4010(90)90088-8. [DOI] [PubMed] [Google Scholar]
- 61.Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, Dell A, Dougan G, Fairweather N. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol Microbiol 40:1187–1199. doi: 10.1046/j.1365-2958.2001.02461.x. [DOI] [PubMed] [Google Scholar]
- 62.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, Andersson M. 2008. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 63.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 64.Yeung AT, Gellatly SL, Hancock RE. 2011. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci 68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]