Abstract
Fusobacterium nucleatum is a common oral anaerobe involved in periodontitis that is known to translocate and cause intrauterine infections. In the oral environment, F. nucleatum adheres to a large diversity of species, facilitating their colonization and creating biological bridges that stabilize the multispecies dental biofilm. Many of these interactions (called coadherences or coaggregations) are galactose sensitive. Galactose-sensitive interactions are also involved in the binding of F. nucleatum to host cells. Hemagglutination of some F. nucleatum strains is also galactose sensitive, suggesting that a single galactose-sensitive adhesin might mediate the interaction of fusobacteria with many partners and targets. In order to identify the fusobacterial galactose-sensitive adhesin, a system for transposon mutagenesis in fusobacteria was created. The mutant library was screened for hemagglutination deficiency, and three clones were isolated. All three clones were found to harbor the transposon in the gene coding for the Fap2 outer membrane autotransporter. The three fap2 mutants failed to show galactose-inhibitable coaggregation with Porphyromonas gingivalis and were defective in cell binding. A fap2 mutant also showed a 2-log reduction in murine placental colonization compared to that of the wild type. Our results suggest that Fap2 is a galactose-sensitive hemagglutinin and adhesin that is likely to play a role in the virulence of fusobacteria.
INTRODUCTION
Fusobacterium nucleatum is a non-spore-forming anaerobe (1) and is the Gram-negative species isolated most frequently from both healthy and diseased sites in the oral cavity (2, 3). Though highly associated with periodontitis, this bacterium is considered not to be a major periodontal pathogen but rather an agent hypothesized to impact the events leading to this disease (4). F. nucleatum is also a common isolate from extraoral infections (2, 5, 6), and recent evidence suggests that it is involved in colorectal carcinoma (7–10). Overabundance of F. nucleatum was observed in colorectal carcinomas and adenomas, activating Wnt signaling and oncogenes and generating a proinflammatory microenvironment conducive for colorectal neoplasia progression.
One of F. nucleatum's important virulence characteristics is its ability to adhere to early and late dental plaque colonizers and to bind a variety of mammalian cells.
Coadherence (specific binding to a surface-attached bacterium) is a major attachment mechanism of dental colonizers (11–13). Coadherence not only prevents the washout of oral colonizers by the saliva and gingival crevicular fluid but also creates spatial proximity that facilitates microbial communication and metabolic synergism (14–18). In vitro, F. nucleatum can mediate coaggregation (coadherence among planktonic bacteria) between many species of dental colonizers (19, 20) and therefore has been proposed to function as a bridging organism that stabilizes the developing dental plaque (21–23). Apart from its ability to bind multiple bacterial species, F. nucleatum is capable of adhering to (24) and invading (25–29) various mammalian cell types, inducing the secretion of proinflammatory cytokines that contribute to the initiation and progression of periodontal diseases (29–31). F. nucleatum was also shown to shuttle other noninvasive bacteria into epithelial cells (32).
F. nucleatum exhibits different types of adhesions and harbors several adhesins, including an arginine-inhibitable adhesin (20, 33), a mannose-sensitive lectin involved in fusobacterial coaggregation with Candida species (34, 35), and Fad, which is required for cell attachment and invasion (36–38) and was recently shown to bind E-cadherin on colorectal cancer cells and promote carcinogenesis (39). FadA was also shown to be involved in F. nucleatum 12230 colonization of the mouse placenta (36, 39, 40).
The ability to adhere to and invade host cells is believed to play a key role in F. nucleatum's oral and systemic virulence, as well as in its ability to colonize the placenta (28, 36, 38, 40). Periodontal disease is a risk factor for preterm labor (41–45), and F. nucleatum has been associated with preterm birth (46–50), stillbirth (51), and early-onset neonatal sepsis (52).
F. nucleatum's attachment to mammalian cells can be reversed by adding d-galactose, which also inhibits F. nucleatum's ability to coaggregate with the major periodontopathogen Porphyromonas gingivalis and with at least nine other oral bacterial species (19, 53, 54). Spontaneous F. nucleatum mutants defective in galactose-sensitive coaggregation with P. gingivalis were also defective in attachment to a variety of mammalian cells (24, 29), suggesting that a single galactose-inhibitable adhesin plays a key role in fusobacterial virulence associated with periodontal plaque and host cells. Though this adhesin appears to be involved in many interactions with bacterial and host cells, it has not yet been characterized.
Genetic manipulation in fusobacteria has been hampered possibly by anaerobic growth requirements, multiple fusobacterial restriction modification systems (including several restriction enzymes with four base recognition sites) (55), and the high AT content in its genome (56). Cloning of AT-rich genomic DNA is notoriously difficult (57). While the reasons are not clear, one suggestion is that the cloned sequences act as transcriptional promoters in Escherichia coli.
Though two shuttle vectors have been developed for fusobacteria (58–60) and several strains have been sequenced and annotated (56, 61, 62), research on this bacterium has been hindered by the lack of genetic systems to induce random mutations.
In this study, a random insertion-inactivation mutagenesis system was created for fusobacteria. Using this system, the Fap2 autotransporter was identified as the F. nucleatum galactose-sensitive adhesin.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
F. nucleatum ATCC 23726 and P. gingivalis PK 1924 were grown in Wilkins-Chalgren broth (Oxoid, United Kingdom) or on Columbia agar plates (Oxoid, United Kingdom) supplemented with 5% defibrinated sheep blood (Novamed, Israel). Both strains were grown in an anaerobic chamber (Bactron I-II; Shel Lab, USA) in an atmosphere of 90% N2, 5% CO2, and 5% H2 at 37°C.
Escherichia coli XL-1 was grown in LB broth (Difco, USA) or on LB agar plates (Difco, USA) under aerobic conditions at 37°C.
Streptococcus sanguinis NC 02863 was grown in brain heart infusion (Difco, USA) at 37°C in an atmosphere of 5% CO2.
All antimicrobials used in this study were purchased from Sigma-Aldrich Israel. Antibiotic concentrations (final) were as follows: ampicillin, 100 μg ml−1; chloramphenicol, 30 μg ml−1; and thiamphenicol, 5 μg ml−1. For fusobacteria, broth was supplemented with half these concentrations.
Electroporation of F. nucleatum.
Fusobacteria were electroporated as described previously (60). Briefly, fusobacterial cells were grown to log phase, washed three times in cold electroporation buffer (10% glycerol, 1 mM MgCl2), and concentrated to an optical density at 600 nm (OD600) of 6. One hundred microliters of competent cells was electroporated with 2 μl of DNA (containing 0.1 to 0.5 μg of DNA in double-distilled water) in a 0.1-cm-electrode-gap Gene Pulser cuvette (Bio-Rad), using the MicroPulser electroporator (Bio-Rad, USA) at settings of 2.5 kV, 200 Ω, and 25 μF. The cells were then quickly resuspended with 0.9 ml of prereduced Wilkins-Chalgren broth supplemented with 1 mM of MgCl2 and incubated for 5 h at 37°C in an anaerobic chamber. Bacteria were then spread on appropriate plates and incubated for 5 days.
Creation of EZ::TnCat.
A catP gene of clostridial origin, conferring resistance to chloramphenicol on E. coli and thiamphenicol on fusobacteria, was amplified by PCR (Expand high-fidelity PCR system; Roche, USA) from the pHS30 plasmid (60) using the F-CatP (GGGGAATTCTAAAACCTTGGTTGTGTTGC) and R-CatP (GGGGAATTCAACGAGTGAAAAAGTGTCCC) primers. PCR conditions were as follows: denaturation at 94°C for 2 min, followed by 10 cycles of denaturing at 94°C for 15 s, annealing at 65°C for 45 s, and elongation at 72°C for 3 min, followed by an additional 20 cycles of denaturation at 94°C for 15 s, annealing at 60°C for 30 s, elongation at 72°C for 3 min, and a final 7-min extension at 72°C. The resulting fragment (∼0.6 kbp) was cloned into EcoRI-restricted pMOD-3<R6Kγori/MCS> (Epicentre) to generate pMODCat carrying the EZ::TnCat transposon, which inserts into any target DNA (63, 64). Transposons were amplified by PCR according to the manufacturer's instructions using the F-pMOD PCR (ATTCAGGCTGCGCAACTGT) and R-pMOD PCR (GTCAGTGAGCGAGGAAGCGGAAG) primers. Transposase (Epicentre) was added to the transposons, and they were incubated in the absence of magnesium to create stable transposomes in accordance with the manufacturer's instructions.
The transposomes (2 μl) were electroporated into competent fusobacterial cells (ATCC 23726 or ATCC 10953), and clones with transposon inserts were selected on Columbia blood agar plates supplemented with 5 μg ml−1 of thiamphenicol.
Clones were collected and the partial library (1,200 clones, each in Wilkins-Chalgren broth supplemented with 10% [final concentration] glycerol) was stored in 96-well plates at −80°C until used.
Determination of the transposon insertion site in the fusobacterial genome.
Genomic DNA was purified (GenElute; Sigma-Aldrich, Germany) from selected clones, restricted with the endonucleases ScaI, PvuII, and AhdI (New England BioLabs), which do not cleave within the transposon, and self-ligated using T4 ligase (TaKaRa, Japan), and the transposon-flanking sequences were amplified by inverse PCR (Herculase II; Agilent) using the FpMODSq (GCCAACGACTACGCACTAGCCAAC) and RpMODSq (GAGCCAATATGCGAGAACACCCGAGAA) primer pair. Whenever necessary, nested primers FNes3 (CAAGAGCTTCAGGGTTGAG) and RNes3 (ACCCGAGAAAATTCATCGATG) were used.
PCR conditions were as follows: denaturation at 95°C for 2 min, followed by 30 cycles of denaturing at 95°C for 20 s, annealing at 50°C for 30 s, and elongation at 72°C for 1.5 min and then a final 3-min extension at 72°C.
PCR products were sequenced (The Center for Genomic Technologies), and the insertion site was determined using BLAST software.
Hemagglutination assays.
Fusobacterial clones were grown overnight, washed twice in phosphate-buffered saline (PBS), and brought to an OD600 of 1 (∼109 CFU ml−1). Sheep erythrocytes (RBCs) were washed twice in PBS and brought to a concentration of 2% (vol/vol). Fifty microliters of fusobacterial cells was mixed with 50 μl of sheep erythrocytes (2% in PBS) in round-bottom 96-well plates (Nunc, Denmark) and incubated at room temperature for 2 h. Hemagglutination was determined visually and clones were selected based on their inability to hemagglutinate RBCs.
For inhibition assays, washed bacteria were preincubated with 6 mM d-galactose (Sigma-Aldrich, Germany) or 50 mM l-arginine (Sigma-Aldrich, Germany) (final concentrations) for 30 min prior to incubation with RBCs.
Membrane protein extraction.
Membrane proteins were prepared as described previously (65), with minor changes. Bacterial cells (0.5 liter) were grown overnight and washed in sodium phosphate buffer (pH 7.2) supplemented with 3 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich, Germany).
A French press (8,000 lb/in2) was used three times to disrupt the cells, and unbroken cells were removed by sedimentation at 10,000 × g at 4°C for 10 min. The supernatant was collected and subjected to high-speed centrifugation (150,000 × g at 4°C for 1 h), and the resulting pellet containing the cell walls was washed twice, resuspended in sodium phosphate buffer, and kept at −80°C until used. A sample of each pelleted membrane was boiled in SDS-PAGE sample buffer for 10 min and subjected to SDS-PAGE (4%). Coomassie blue was used to visualize protein bands.
Coaggregation assays. (i) Visual coaggregation.
Bacteria were grown overnight, washed twice in coaggregation buffer (0.1 mM CaCl2, 0.1 mM MgCl2, 0.15 M NaCl, and 0.02% NaN3 dissolved in 1 mM Tris and adjusted to pH 8.0) (66), and brought to an OD600 of 1. Both partners (200 μl of each) were mixed in a glass test tube and incubated at room temperature for 30 min. Visual coaggregation of each mutant was evaluated and compared to wild-type (WT) coaggregation. When testing the inhibitory effect of galactose, fusobacteria were preincubated with d-galactose 60 mM (final concentration) for 15 min.
(ii) Quantitative coaggregation.
As described previously (20), briefly, F. nucleatum and P. gingivalis or S. sanguinis were brought to an OD600 of 1 in coaggregation buffer and mixed (100 μl each) in round-bottom 96-well plates with or without 60 mM d-galactose (final concentration). The plates were incubated at room temperature for 30 min, and coaggregating particles were then sedimented by centrifugation at low speed (100 × g for 1 min). The supernatant containing noncoaggregating cells was transferred to a flat-bottom 96-well plate, and optical density at 595 nm was measured (Genios; Tecan Systems Austria). For each mutant, percent galactose-dependent coaggregation was calculated by dividing the difference between the mutant's supernatant optical density in the presence of 60 mM d-galactose and in the absence of galactose by the difference between the optical density of the wild-type strain in the presence of 60 mM d-galactose and in the absence of galactose, as follows: (mutant without galactose − mutant with galactose)/(wild type without galactose − wild type with galactose) × 100.
Galactose-independent coaggregation was calculated by dividing the mutant's supernatant optical density by that of the wild type.
The differences between the groups were compared using the Student t test. A P value of <0.05 was considered significant.
Cell attachment assay.
Bacteria were grown overnight, washed in sterile PBS, and brought to an OD600 of 1 in coaggregation buffer (66). The bacteria were then stained with carboxyfluorescein succinimidyl ester (CFSE) CellTrace (Molecular Probes, OR) according to the manufacturer's instructions and brought to a concentration of 3 × 106 cells ml−1 in coaggregation buffer. Human embryonic kidney (HEK) 293T cells (a kind gift from Avi-Hai Hovav) were grown in Dulbecco's modified Eagle medium (DMEM) (Biological Industries, Israel) supplemented with 10% fetal calf serum (FCS; Biological Industries), 2 mM l-glutamine (Biological Industries), 100 U ml−1 of penicillin, and 100 μg ml−1 of streptomycin (Biological Industries) (all final concentrations) until they reached confluence. The cells were scraped, washed once in DMEM, and brought to a concentration of 3 × 105 ml−1 in DMEM. The cells were then incubated with the stained bacteria at a multiplicity of infection (MOI) of 10 with gentle shaking for 30 min, followed by three washes in coaggregation buffer and resuspension in PBS supplemented with 2% FCS (final concentration).
For galactose inhibition assays, bacteria were incubated with 60 mM d-galactose (final concentration) for 30 min prior to incubation with the cells, and coaggregation buffer supplemented with 60 mM d-galactose was used to wash the cell-bacterium complex. Cell attachment was determined by flow cytometry (Accuri C6 flow cytometer; BD, USA), and data were analyzed using FlowJo 7.6.5 software (Tree Star, Ashland, OR).
In vivo placental colonization.
Seven- to eight-week-old outbred CF1 mice were caged together at a female-to-male ratio of 2:1, and mating was determined by the presence of a white vaginal plug. The day when the plug was detected was termed the first day of gestation. The pregnant mice were randomly distributed into study groups of six mice per group. The pregnant mice were inoculated with wild-type ATCC 23726 or with the fap2 mutant K50 on day 15 to 17 of gestation. For the inoculation, an aliquot of 100 μl of the bacterial suspension (4.0 × 107 ∼ 5.2 × 107 CFU) or sterile PBS (sham) was injected into the tail vein. After 24 h, the placentas were harvested from each pregnant mouse and homogenized under sterile conditions. Serial log dilutions were performed and plated. The plates were incubated anaerobically at 37°C for 96 h, followed by enumeration. The difference between different groups was analyzed using one-way analysis of variance (ANOVA) and the Kruskal-Wallis test. The difference between two groups was compared using the Student t test. A P value of <0.05 was considered significant. Data shown are representative of two repeated experiments.
Mass spectrometry.
Coomassie-stained protein bands were excised from the denaturing gel, and mass spectrometry was carried out with Orbitrap (Thermo Finnigen). Data analysis was done using the BioWorks 3.3 package, and database searches were performed against the NCBInr database using the Mascot package (Matrix Science, England).
Statistical analyses.
Unless otherwise mentioned, all data are means and standard deviations from three independent experiments performed in triplicate. The Student t test was used for statistical analyses; a P value of <0.05 was considered significant.
RESULTS
Creation of a randomly inserted transposon mutation library in F. nucleatum ATCC 23726.
For creation of a system for random insertional mutagenesis in fusobacteria, the Clostridium perfringens chloramphenicol acetyltransferase gene (catP) conferring resistance to thiamphenicol (a chloramphenicol analog) on fusobacteria (59) was cloned into pMOD-3<R6Kγori/MCS> (Epicentre) to generate pMODCat carrying the EZ::TnCat transposon. Transposase was added to the EZ::TnCat transposon to form transposome complexes, which were electroporated into competent fusobacterial cells. Thiamphenicol-resistant colonies were obtained at an average efficiency of 8 × 103 CFU/μg of DNA using F. nucleatum ATCC 23726 and at an order of magnitude lower using ATCC 10953.
Four independent clones were selected, and the transposon's location was determined by amplifying the flanking region of the transposon by PCR and BLASTing it against the genome of F. nucleatum ATCC 25586, the strain most similar to ATCC 23726.
Sequence analysis indicated that the transposons were inserted at unique positions in each of the four fusobacterial genomes (data not shown).
Identification of the F. nucleatum galactose-sensitive hemagglutinin.
Hemagglutination is an attachment mechanism that is considered a virulence trait of many pathogens. Hemagglutination in some F. nucleatum strains, including Fusobacterium nucleatum subsp. polymorphum ATCC 10953, was found to be inhibited by arginine (67), while in others (Fusobacterium nucleatum subsp. nucleatum ATCC 25586), it is inhibited by galactose (68–70). As can be seen in Fig. 1, hemagglutination of F. nucleatum subsp. nucleatum ATCC 23726 is also galactose sensitive. In order to identify the fusobacterial galactose-sensitive adhesin, 1,200 clones with transposon inserts were screened for hemagglutination deficiency.
FIG 1.
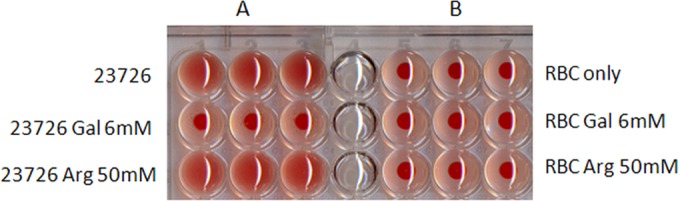
F. nucleatum ATCC 23726 hemagglutination is inhibited by d-galactose but not by l-arginine. (A) Bacteria incubated with 2% sheep red blood cells (RBC) in the absence and presence of 6 mM d-galactose (Gal) or 50 mM l-arginine (Arg). (B) Erythrocytes incubated without bacteria in the absence and presence of inhibitors. Precipitation of the red blood cells (seen as a red dot) represents a lack of hemagglutination.
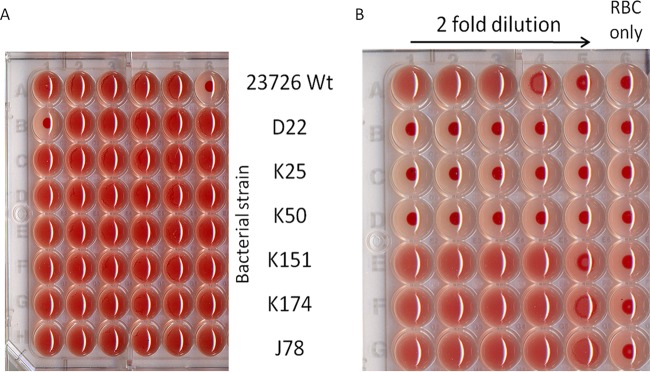
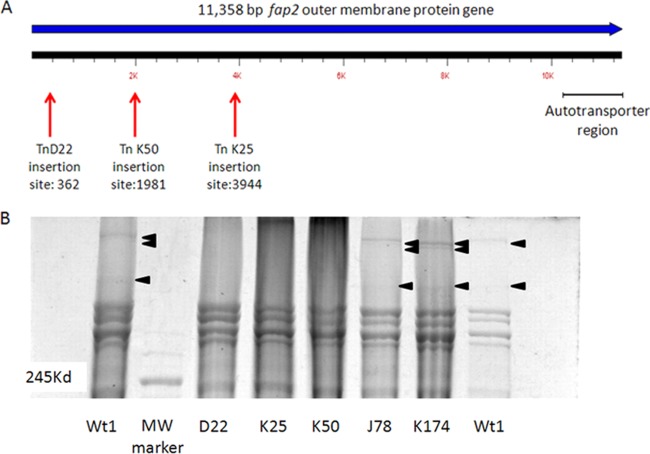
Three independent clones were found to be deficient in their ability to hemagglutinate sheep RBCs (Fig. 2). Analysis of the transposon location in those mutants determined that all three clones harbored the transposon in the same 11.3-kb gene, corresponding to FN1449, an autotransporter protein in F. nucleatum ATCC 25586 previously termed Fap2 for fusobacterial apoptosis protein (65) in ATCC 23726 (Fig. 3A).
FIG 2.
Isolation of F. nucleatum mutants deficient in hemagglutination. (A) Microtiter plate screening for library clones defective in hemagglutination. (B) Hemagglutination assay with 2-fold dilutions of the hemagglutination-deficient mutants (D22, K25, and K50) compared to wild-type F. nucleatum ATCC 23726 and the randomly selected transposon insertion controls: K151, K174, and J78.
FIG 3.
All three hemagglutination-deficient mutants are defective in Fap2. (A) Locations of transposon insertions in the fap2 genes of three hemagglutination-deficient mutants. (B) SDS-PAGE demonstrating the absence of the Fap2 protein bands in membrane proteins extracted from all three hemagglutination-deficient mutants compared to the wild type and the randomly selected mutants J78 and K174. The clear band seen in the MW marker lane indicates 245 kDa.
In order to verify that the mutation affected the Fap2 protein, membrane proteins were extracted from wild-type F. nucleatum, from the three hemagglutination-deficient mutants (D22, K25, and K50), and from randomly selected mutants (J78 and K174) used as controls (20, 65) The same high-molecular-mass bands were missing in all three mutants. The corresponding bands from wild-type F. nucleatum were sent for mass spectrometry analysis, which identified peptides belonging to the FN1449 protein of F. nucleatum ATCC 25586 (Fig. 3B).
The Fap2 adhesin involved in hemagglutination is also involved in coaggregation.
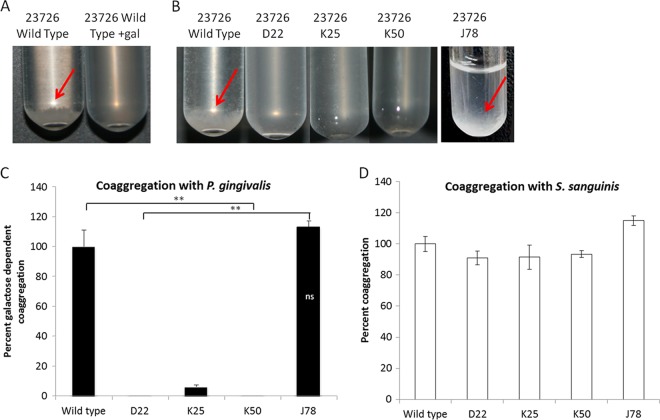
Similar to hemagglutination, coaggregation of F. nucleatum ATCC 23726 with P. gingivalis is galactose sensitive (19, 66). All three hemagglutination-deficient mutants (but not the randomly selected J78 mutant) failed to show galactose-sensitive coaggregation with P. gingivalis (Fig. 4A to C). However, the mutants retained their ability to coaggregate with Streptococcus sanguinis; this coaggregation is mediated by the fusobacterial arginine-inhibitable RadD adhesin (20) (Fig. 4D).
FIG 4.
All three hemagglutination-deficient mutants fail to coaggregate with P. gingivalis. F. nucleatum and P. gingivalis were brought to an OD600 of 1 in coaggregation buffer mixed and incubated in a glass tube at room temperature for 30 min. Coaggregation (pellet), which is indicated with a red arrow, is absent in the presence of 60 mM d-galactose (gal) (A) and with the hemagglutinin mutants D22, K25, and K50 compared to the wild type and the randomly selected mutant J78 (B). (C and D) Coaggregation was quantified as described in Materials and Methods. All three hemagglutination-deficient mutants (but not the randomly selected mutant J78) failed to coaggregate with P. gingivalis but not with S. sanguinis. **, P < 0.01 compared to the wild-type control. ns, not significant.
Fap2-deficient mutants are impaired in cell binding.
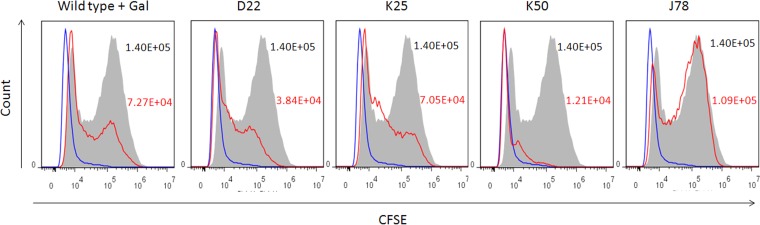
As galactose was previously demonstrated to inhibit the attachment of some F. nucleatum strains to mammalian cells (66, 70), we next tested whether the hemagglutination- and coaggregation-deficient clones are also deficient in attachment to nonerythrocyte mammalian cells. Wild-type F. nucleatum ATCC 23726, the three hemagglutination-deficient mutants, and a randomly selected control mutant, J78, were stained with carboxyfluorescein succinimidyl ester (CFSE) and incubated with HEK 293T cells. Cell attachment was determined by flow cytometry. Adherence to cells in all three fap2 mutants was 3- to 10-fold lower than that of the wild type or the control mutant. The Fap2 mutants retained some cell binding activity (similar to the galactose-treated cells), presumably due to the remaining FadA adhesin (Fig. 5). The presence of the FadA adhesin was verified in each of the tested mutants and in the wild-type strain (data not shown).
FIG 5.
All three hemagglutination-deficient mutants are impaired in binding to mammalian cells. Shown are the results of fluorescence-activated cell sorter (FACS) analysis of HEK 293T cells (blue outline) incubated with carboxyfluorescein succinimidyl ester (CFSE)-labeled wild-type ATCC 23726 in the presence of d-galactose (gal) or with labeled hemagglutinin-deficient mutants (red outline) D22, K25, and K50 and J78, a randomly selected mutant used as control. Wild-type ATCC 23726 control is represented by the gray-filled histogram. Mean fluorescence intensity values are indicated for each histogram. Values for the wild type are in black, and those for the galactose-treated bacteria and the mutants are in red. Data are representative of three independent experiments.
Fap2 is involved in placental colonization.
Fusobacteria have been implicated in preterm births, and FadA, a fusobacterial adhesin unique to oral bacteria, was shown to be involved (36). In order to determine the role of the Fap2 galactose-sensitive adhesin in adverse pregnancy outcomes, wild-type F. nucleatum and the K50 hemagglutination-deficient mutant were injected into the tail veins of 7- to 8-week-old outbred CF1 female mice at day 15 to 17 of gestation. After 24 h, the placentas were harvested from the pregnant mice and homogenized. Serial dilutions were plated, and fusobacterial colonies were enumerated after 96 h of incubation. As can be seen in Fig. 6, the mutation in fap2 reduced placental colonization by 2 orders of magnitude.
FIG 6.
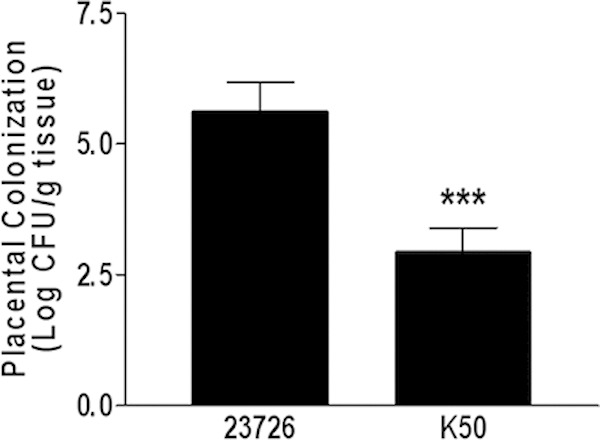
The Fap2 adhesin is involved in placental colonization. Wild-type F. nucleatum ATCC 23726 or the hemagglutination-deficient mutant K50 was injected into the tail veins of pregnant mice. After 24 h, the placentas were harvested and homogenized, and bacterial CFU were determined. ***, P < 0.001.
DISCUSSION
Fusobacterium nucleatum is a significant pathogen in human infections, involved in periodontitis, in a variety of systemic diseases (71–74), in colorectal carcinoma (7, 8, 10, 39, 75, 76), and in preterm births (47, 77). Its role in oral disease is of particular importance, because the oral cavity is the suspected port of entry and reservoir of this organism in systemic diseases (51). In spite of its implication in numerous diseases, little is known about how F. nucleatum might act to cause disease.
F. nucleatum is not known to harbor significant virulence factors. However, it is capable of inducing the secretion of proinflammatory cytokines, downregulating host immunity by inhibiting T cells (78), and inducing apoptosis in lymphocytes (79). It also expresses an IgG Fc binding protein (80) and a weak serine protease (81). It is suggested that the adherence capability is this bacterium's primary virulence trait, conferring on it the means to survive in the host.
Genetic manipulation in fusobacteria is difficult, due to their anaerobic growth conditions, the low GC content in their genome (56), and their multiple restriction systems (55). The two shuttle vectors developed for fusobacteria (58–60) and the sequencing and annotation of three strains representing subspecies F. nucleatum subsp. nucleatum, Fusobacterium nucleatum subsp. vincentii, and F. nucleatum subsp. polymorphum (56, 61, 62) have opened new avenues of research. However, though site-directed mutagenesis (40, 60, 65) and complementation assays (36) have been performed with this bacterium, research has been hindered by the lack of genetic systems to induce random insertion mutagenesis. Here we present a system for random insertion mutagenesis in F. nucleatum.
Transposomes electroporated into F. nucleatum ATCC 23726 or F. nucleatum ATCC 10953 (data not shown) enabled random integration of EZ::TnCat into the fusobacterial genome and the creation of the F. nucleatum ATCC 23726 library. Phenotypic screening of the mutant library identified three mutants that did not hemagglutinate sheep RBCs (Fig. 2).
The three isolated hemagglutination mutants were found to be defective in the same gene that codes for a fusobacterial outer membrane autotransporter (FN1449 in F. nucleatum ATCC 25586), previously termed fap2 (Fig. 3). The multiple high-molecular-mass bands visualized (in agreement with the predicted Fap2 molecular mass of 389.8 kDa) are hypothesized to be due to some sort of modification such as glycosylation or truncation.
F. nucleatum ATCC 23726 is predicted to have approximately 2,100 encoded proteins, of which Fap2 is the largest one. Though only 1,200 clones were screened, three fap2 mutants were identified. This might be explained by the large size of fap2 (11.3 kb). Comparative analysis of the sequenced strain, F. nucleatum ATCC 25586, and the preliminary genome annotation of F. nucleatum ATCC 49256 suggested that the major protein secretion systems, such as type II, type III, and type IV, were missing (61). However, several autotransporters belonging to the type V secretion system family have been identified (13). This large and diverse superfamily of polypeptides which is produced by pathogenic Gram-negative bacteria is often associated with virulence traits such as aggregation, adherence, toxicity, biofilm formation, and invasion (82, 83).
Interestingly, one of the characteristics of autotransporter proteins is a low abundance of cysteine residues in the passenger domain, presumably to prevent disulfide bond formation of the protein while it is in the periplasm and facilitate its translocation (84). The Fap2 hemagglutinin contains over 3,000 amino acids and has no cysteine residues.
Structural analysis revealed that the predicted structure of Fap2 contains domains homologous to the Hmw1 secretion domain in Haemophilus influenzae (30% identity) (85), which contains a carbohydrate-dependent hemagglutination domain found in various hemagglutinins and hemolysins such as the Bordetella pertussis hemagglutinin (86).
Hemagglutination is a characteristic feature of oral fusobacteria (67). It has been previously shown that fusobacterial hemagglutination is mediated by a protein moiety on the bacterial cell surface (87) and that in some strains, the reaction is galactose sensitive (68). A fusobacterial hemagglutinin was previously characterized as a high-molecular-mass protein, which corresponds with our findings. A high-molecular-mass arginine-sensitive hemagglutinin was purified from F. nucleatum subsp. polymorphum ATCC 10953 and has been shown to be involved in coaggregation of streptococci (88). Similarly, Fap2 is another high-molecular-mass protein which appears to be involved in F. nucleatum ATCC 23726 galactose-sensitive hemagglutination and coaggregation.
Coaggregation is defined as a specific binding of two genetically distinct microorganisms (89). In the oral cavity, this interbacterial attachment mechanism serves to anchor the bacteria to a surface and withstand the salivary flow that will otherwise wash them away into the digestive tract. Coaggregation also enables bacterial communication between the coaggregating partners and facilitates a mutual metabolic relationship (14, 15, 90).
It has been shown that coaggregation is a key factor in periodontitis and that dual infection with F. nucleatum and P. gingivalis aggravates periodontal disease, which is manifested in greater alveolar bone loss (91).
So far, an arginine-inhibitable adhesin, RadD, has been identified in F. nucleatum (20), mediating coaggregation with streptococci and implicated in apoptosis of human lymphocytes (65).
Coaggregation with P. gingivalis and cell adhesion were shown by others (92) and by us (66) to be reversed in some F. nucleatum subspecies by adding galactose, leading to the hypothesis that the galactose-sensitive coaggregation and cell adhesion may be mediated by the same adhesin.
Hemagglutination in F. nucleatum ATCC 23726 is also galactose inhibitable (Fig. 1), and the three hemagglutination-deficient mutants were found to be deficient in galactose-sensitive coaggregation with P. gingivalis but not in arginine-inhibitable coaggregation with S. sanguinis (Fig. 4). They are also defective in adherence to HEK 293T cells (Fig. 5), supporting the aforementioned hypothesis that a single adhesin mediates fusobacterial attachment to host cells and galactose-sensitive coaggregation (19). Many known pathogens express a variety of structures on their surface, which function as bacterial adhesins (93). These adhesins, such as type I fimbriae (94), YadA expressed by Yersinia enterocolitica (95), and the filamentous hemagglutinin expressed by Bordetella pertussis (96–98), have lectin-like properties and appear to act as multifactorial adhesins capable of mediating bacterium-host cell interactions, as well as promoting bacterial autoaggregation. Many microbial lectins were originally detected based on their hemagglutinating ability (99).
Fap2 of F. nucleatum ATCC 23726 was previously shown to be involved in induction of cell death (65). However, in light of our results, it is plausible that its involvement in apoptosis is due to its ability to enable fusobacterial adherence to host cells. In theory, an open reading frame (FN1448) downstream of the fap2 gene could potentially be involved in the hemagglutination deficiency phenotype. However, Kaplan et al. (65) showed that an insertion mutation in fap2 did not cause a polar effect.
Fusobacterium is prevalent in intrauterine infections, and its role in preterm birth has been documented previously (47, 50, 51, 100). Inactivation and complementation of FadA, a 12-kDa outer membrane protein, demonstrated its essential role in host cell attachment and invasion by F. nucleatum subsp. polymorphum 12230, as well as in the promotion of colorectal carcinogenesis by binding the E-cadherin receptor and inducing clathrin-mediated endocytosis and β-catenin signaling. The role of FadA in placental colonization was also demonstrated in F. nucleatum 12230 (36); however, its inactivation did not completely abolish placental colonization, suggesting the involvement of an additional adhesin(s).
Here (Fig. 6), we show that a Fap2 mutant (K50) of F. nucleatum subsp. nucleatum ATCC 23726 was also impaired in its ability to colonize the placenta, demonstrating for the first time that this fusobacterial subspecies can also colonize the placenta and that Fap2 is involved in placenta colonization.
This study would have greatly benefitted from a complementation mutant, which would have verified fap2's role in the aforementioned interactions. However, multiple attempts to clone this gene were unsuccessful due to spontaneous deletions. These deletions seemed to be insert size dependent, because cloning of the 5′ half of fap2 and of the 3′ half of fap2 (∼6 kbp each) was successful. However, attempts to clone one half with the other (in both high- and low-copy-number plasmids) resulted in deletions of random sizes (data not shown), in agreement with previous reports of difficulties with cloning AT-rich genomic DNA (57).
In this study, we used a transposon-based mutagenesis system in fusobacteria and created a library which enables phenotypic screening and the identification of virulence traits.
To the best of our knowledge, this is the first report of a transposon insertion mutagenesis system in fusobacteria. We have also identified Fap2 as a galactose-sensitive adhesin involved in hemagglutination, coaggregation, and adherence to mammalian cells in F. nucleatum subsp. nucleatum ATCC 23726. A better understanding of F. nucleatum's interactions with periodontal bacteria and with host cells will perhaps enable us to improve control of periodontal disease and to reduce F. nucleatum-related systemic conditions, adverse pregnancy outcome in particular.
ACKNOWLEDGMENTS
We thank Avi-Hai Hovav for valuable comments and discussion.
This research was supported by the United States-Israel Binational Science Foundation (grant number 2005084) and by the Israel Science Foundation (grant number 208/10).
REFERENCES
- 1.Bolstad AI, Jensen HB, Bakken V. 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev 9:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore WE, Moore LV. 1994. The bacteria of periodontal diseases. Periodontol 2000 5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 3.Teles FR, Teles RP, Siegelin Y, Paster B, Haffajee AD, Socransky SS. 2011. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Mol Oral Microbiol 26:127–139. doi: 10.1111/j.2041-1014.2010.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 5.Swidsinski A, Dorffel Y, Loening-Baucke V, Theissig F, Ruckert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kuhn S, Schilling J, Dorffel WV. 2011. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 6.Han YW, Wang X. 2013. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res 92:485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. 2012. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 10.Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. 2011. Towards the human colorectal cancer microbiome. PLoS One 6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. 1993. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J 7:406–413. [DOI] [PubMed] [Google Scholar]
- 12.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627–1636. [DOI] [PubMed] [Google Scholar]
- 13.Desvaux M, Khan A, Beatson SA, Scott-Tucker A, Henderson IR. 2005. Protein secretion systems in Fusobacterium nucleatum: genomic identification of type 4 piliation and complete type V pathways brings new insight into mechanisms of pathogenesis. Biochim Biophys Acta 1713:92–112. doi: 10.1016/j.bbamem.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Egland PG, Palmer RJ Jr, Kolenbrander PE. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci U S A 101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ Jr. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol 13:11–15. doi: 10.1016/j.tim.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Hansen SK, Rainey PB, Haagensen JA, Molin S. 2007. Evolution of species interactions in a biofilm community. Nature 445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 17.Kolenbrander PE, Jakubovics NS, Chalmers NI, Bachrach G. 2007. Coaggregation and distance-critical communication, p 89–100. In Brogden KA, Minion FC, Cornick N, Stanton TB, Zhang K, Nolan LK, Wannemuehler MJ (ed), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC. [Google Scholar]
- 18.Liu PF, Shi W, Zhu W, Smith JW, Hsieh SL, Gallo RL, Huang CM. 2010. Vaccination targeting surface FomA of Fusobacterium nucleatum against bacterial co-aggregation: implication for treatment of periodontal infection and halitosis. Vaccine 28:3496–3505. doi: 10.1016/j.vaccine.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander PE, London J. 1993. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol 175:3247–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan CW, Lux R, Haake SK, Shi W. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr. 2002. Communication among oral bacteria. Microbiol Mol Biol Rev 66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachrach G, Coppenhagen-Glazer S, Simon I. 2011. Fusobacteria: bridging between health and disease a metatranscriptomic approach, p 93–102. In Kolenbrander PE. (ed), Genomic inquiries into oral bacterial communities. ASM Press, Washington, DC. [Google Scholar]
- 23.Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 24.Weiss EI, Shaniztki B, Dotan M, Ganeshkumar N, Kolenbrander PE, Metzger Z. 2000. Attachment of Fusobacterium nucleatum PK1594 to mammalian cells and its coaggregation with periodontopathogenic bacteria are mediated by the same galactose-binding adhesin. Oral Microbiol Immunol 15:371–377. doi: 10.1034/j.1399-302x.2000.150606.x. [DOI] [PubMed] [Google Scholar]
- 25.Gursoy UK, Pollanen M, Kononen E, Uitto VJ. 2010. Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J Periodontol 81:1084–1091. doi: 10.1902/jop.2010.090664. [DOI] [PubMed] [Google Scholar]
- 26.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 27.Han YW. 2011. Fusobacterium nucleatum interaction with host cells, p 221–232. In Kolenbrander PE. (ed), Oral microbial communities: genomic inquiry and interspecies communication. ASM Press, Washington, DC. [Google Scholar]
- 28.Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun 72:2272–2279. doi: 10.1128/IAI.72.4.2272-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, Genco RJ. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun 68:3140–3146. doi: 10.1128/IAI.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji S, Shin JE, Kim YS, Oh JE, Min BM, Choi Y. 2009. Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by Fusobacterium nucleatum in gingival epithelial cells. Infect Immun 77:1044–1052. doi: 10.1128/IAI.00449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grenier D, Grignon L. 2006. Response of human macrophage-like cells to stimulation by Fusobacterium nucleatum ssp. nucleatum lipopolysaccharide. Oral Microbiol Immunol 21:190–196. doi: 10.1111/j.1399-302X.2006.00278.x. [DOI] [PubMed] [Google Scholar]
- 32.Edwards AM, Grossman TJ, Rudney JD. 2006. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun 74:654–662. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards AM, Grossman TJ, Rudney JD. 2007. Association of a high-molecular weight arginine-binding protein of Fusobacterium nucleatum ATCC 10953 with adhesion to secretory immunoglobulin A and coaggregation with Streptococcus cristatus. Oral Microbiol Immunol 22:217–224. doi: 10.1111/j.1399-302X.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 34.Grimaudo NJ, Nesbitt WE. 1997. Coaggregation of Candida albicans with oral Fusobacterium species. Oral Microbiol Immunol 12:168–173. doi: 10.1111/j.1399-302X.1997.tb00374.x. [DOI] [PubMed] [Google Scholar]
- 35.Jabra-Rizk MA, Falkler WA Jr, Merz WG, Kelley JI, Baqui AA, Meiller TF. 1999. Coaggregation of Candida dubliniensis with Fusobacterium nucleatum. J Clin Microbiol 37:1464–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikegami A, Chung P, Han YW. 2009. Complementation of the fadA mutation in Fusobacterium nucleatum demonstrates that the surface-exposed adhesin promotes cellular invasion and placental colonization. Infect Immun 77:3075–3079. doi: 10.1128/IAI.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. 2007. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem 282:25000–25009. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 38.Fardini Y, Wang X, Temoin S, Nithianantham S, Lee D, Shoham M, Han YW. 15 November 2011. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA Adhesin. Cell Host Microbe 14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. 2005. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol 187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Offenbacher S, Katz V, Fertik G, Collins J, Boyd D, Maynor G, McKaig R, Beck J. 1996. Periodontal infection as a possible risk factor for preterm low birth weight. J Periodontol 67:1103–1113. doi: 10.1902/jop.1996.67.10s.1103. [DOI] [PubMed] [Google Scholar]
- 42.Guimarães AN, Silva-Mato A, Miranda Cota LO, Siqueira FM, Costa FO. 2010. Maternal periodontal disease and preterm or extreme preterm birth: an ordinal logistic regression analysis. J Periodontol 81:350–358. doi: 10.1902/jop.2009.090527. [DOI] [PubMed] [Google Scholar]
- 43.Ebersole JL, Novak MJ, Michalowicz BS, Hodges JS, Steffen MJ, Ferguson JE, Diangelis A, Buchanan W, Mitchell DA, Papapanou PN. 2009. Systemic immune responses in pregnancy and periodontitis: relationship to pregnancy outcomes in the Obstetrics and Periodontal Therapy (OPT) study. J Periodontol 80:953–960. doi: 10.1902/jop.2009.080464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han YW. 2011. Oral health and adverse pregnancy outcomes—what's next? J Dent Res 90:289–293. doi: 10.1177/0022034510381905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fardini Y, Chung P, Dumm R, Joshi N, Han YW. 2010. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun 78:1789–1796. doi: 10.1128/IAI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill GB. 1993. Investigating the source of amniotic fluid isolates of fusobacteria. Clin Infect Dis 16(Suppl 4):S423–S424. [DOI] [PubMed] [Google Scholar]
- 47.Hill GB. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann Periodontol 3:222–232. doi: 10.1902/annals.1998.3.1.222. [DOI] [PubMed] [Google Scholar]
- 48.Chaim W, Mazor M. 1992. Intraamniotic infection with fusobacteria. Arch Gynecol Obstet 251:1–7. doi: 10.1007/BF02718272. [DOI] [PubMed] [Google Scholar]
- 49.Cahill RJ, Tan S, Dougan G, O'Gaora P, Pickard D, Kennea N, Sullivan MH, Feldman RG, Edwards AD. 2005. Universal DNA primers amplify bacterial DNA from human fetal membranes and link Fusobacterium nucleatum with prolonged preterm membrane rupture. Mol Hum Reprod 11:761–766. doi: 10.1093/molehr/gah234. [DOI] [PubMed] [Google Scholar]
- 50.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. 2009. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol 47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM, Redline RW. 2010. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet Gynecol 115:442–445. doi: 10.1097/AOG.0b013e3181cb9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Buhimschi CS, Temoin S, Bhandari V, Han YW, Buhimschi IA. 2013. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One 8:e56131. doi: 10.1371/journal.pone.0056131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosen G, Nisimov I, Helcer M, Sela MN. 2003. Actinobacillus actinomycetemcomitans serotype b lipopolysaccharide mediates coaggregation with Fusobacterium nucleatum. Infect Immun 71:3652–3656. doi: 10.1128/IAI.71.6.3652-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosen G, Sela MN. 2006. Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol Lett 256:304–310. doi: 10.1111/j.1574-6968.2006.00131.x. [DOI] [PubMed] [Google Scholar]
- 55.Lui AC, McBride BC, Vovis GF, Smith M. 1979. Site specific endonuclease from Fusobacterium nucleatum. Nucleic Acids Res 6:1–15. doi: 10.1093/nar/6.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D'Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol 184:2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dillard JP, Yother J. 1991. Analysis of Streptococcus pneumoniae sequences cloned into Escherichia coli: effect of promoter strength and transcription terminators. J Bacteriol 173:5105–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haake SK, Yoder SC, Attarian G, Podkaminer K. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J Bacteriol 182:1176–1180. doi: 10.1128/JB.182.4.1176-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bachrach G, Haake SK, Glick A, Hazan R, Naor R, Andersen RN, Kolenbrander PE. 2004. Characterization of the novel Fusobacterium nucleatum plasmid pKH9 and evidence of an addiction system. Appl Environ Microbiol 70:6957–6962. doi: 10.1128/AEM.70.12.6957-6962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kinder Haake S, Yoder S, Gerardo SH. 2006. Efficient gene transfer and targeted mutagenesis in Fusobacterium nucleatum. Plasmid 55:27–38. doi: 10.1016/j.plasmid.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapatral V, Ivanova N, Anderson I, Reznik G, Bhattacharyya A, Gardner WL, Mikhailova N, Lapidus A, Larsen N, D'Souza M, Walunas T, Haselkorn R, Overbeek R, Kyrpides N. 2003. Genome analysis of F. nucleatum sub spp vincentii and its comparison with the genome of F. nucleatum ATCC 25586. Genome Res 13:1180–1189. doi: 10.1101/gr.566003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, Yerrapragada S, Fox GE, Haake SK, Weinstock GM, Highlander SK. 2007. Genome sequence of Fusobacterium nucleatum subspecies polymorphum—a genetically tractable fusobacterium. PLoS One 2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goryshin IY, Reznikoff WS. 1998. Tn5 in vitro transposition. J Biol Chem 273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 64.Hoffman LM, Jendrisak JJ, Meis RJ, Goryshin IY, Reznikof SW. 2000. Transposome insertional mutagenesis and direct sequencing of microbial genomes. Genetica 108:19–24. doi: 10.1023/A:1004083307819. [DOI] [PubMed] [Google Scholar]
- 65.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. 2010. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun 78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachrach G, Ianculovici C, Naor R, Weiss EI. 2005. Fluorescence based measurements of Fusobacterium nucleatum coaggregation and of fusobacterial attachment to mammalian cells. FEMS Microbiol Lett 248:235–240. doi: 10.1016/j.femsle.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 67.Ozaki M, Miyake Y, Shirakawa M, Takemoto T, Okamoto H, Suginaka H. 1990. Binding specificity of Fusobacterium nucleatum to human erythrocytes, polymorphonuclear leukocytes, fibroblasts, and HeLa cells. J Periodontal Res 25:129–134. doi: 10.1111/j.1600-0765.1990.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 68.Mongiello JR, Falkler WA Jr. 1979. Sugar inhibition of oral Fusobacterium nucleatum haemagglutination and cell binding. Arch Oral Biol 24:539–545. doi: 10.1016/0003-9969(79)90133-X. [DOI] [PubMed] [Google Scholar]
- 69.Falkler WA Jr, Burger BW. 1981. Microbial surface interactions: reduction of the haemagglutination activity of the oral bacterium Fusobacterium nucleatum by absorption with Streptococcus and Bacteroides. Arch Oral Biol 26:1015–1025. doi: 10.1016/0003-9969(81)90112-6. [DOI] [PubMed] [Google Scholar]
- 70.Shaniztki B, Hurwitz D, Smorodinsky N, Ganeshkumar N, Weiss EI. 1997. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect Immun 65:5231–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koornstra JJ, Veenendaal D, Bruyn GA, de Graaf H. 1998. Septic arthritis due to Fusobacterium nucleatum. Br J Rheumatol 37:1249. doi: 10.1093/rheumatology/37.11.1249. [DOI] [PubMed] [Google Scholar]
- 72.Botha SJ, Senekal R, Steyn PL, Coetzee WJ. 1993. Anaerobic bacteria in orofacial abscesses. J Dent Assoc S Afr 48:445–449. [PubMed] [Google Scholar]
- 73.Bauer C, Schoonbroodt D, Wagner C, Horsmans Y. 2000. Liver abscesses due to Fusobacterium species. Liver 20:267–268. doi: 10.1034/j.1600-0676.2000.020003267.x. [DOI] [PubMed] [Google Scholar]
- 74.Roberts GL. 2000. Fusobacterial infections: an underestimated threat. Br J Biomed Sci 57:156–162. [PubMed] [Google Scholar]
- 75.McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO. 2013. Fusobacterium is associated with colorectal adenomas. PLoS One 8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han YW. 2011. Can oral bacteria cause pregnancy complications? Womens Health (Lond Engl) 7:401–404. doi: 10.2217/whe.11.37. [DOI] [PubMed] [Google Scholar]
- 78.Demuth DR, Savary R, Golub E, Shenker BJ. 1996. Identification and analysis of fipA, a Fusobacterium nucleatum immunosuppressive factor gene. Infect Immun 64:1335–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, Haake SK. 2005. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J Dent Res 84:700–704. doi: 10.1177/154405910508400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo M, Han YW, Sharma A, De Nardin E. 2000. Identification and characterization of human immunoglobulin G Fc receptors of Fusobacterium nucleatum. Oral Microbiol Immunol 15:119–123. doi: 10.1034/j.1399-302x.2000.150208.x. [DOI] [PubMed] [Google Scholar]
- 81.Bachrach G, Rosen G, Bellalou M, Naor R, Sela MN. 2004. Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol Immunol 19:155–159. doi: 10.1111/j.0902-0055.2004.00132.x. [DOI] [PubMed] [Google Scholar]
- 82.Henderson IR, Nataro JP. 2001. Virulence functions of autotransporter proteins. Infect Immun 69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wells TJ, Tree JJ, Ulett GC, Schembri MA. 2007. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol Lett 274:163–172. doi: 10.1111/j.1574-6968.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 84.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, Zhang N, Bryant SH. 2007. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res 35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, Jacob-Dubuisson F, Locht C, Steven AC. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol 42:279–292. doi: 10.1046/j.1365-2958.2001.02598.x. [DOI] [PubMed] [Google Scholar]
- 87.Falkler WA Jr, Hawley CE. 1977. Hemagglutinating activity of Fusobacterium nucleatum. Infect Immun 15:230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takemoto T, Ozaki M, Shirakawa M, Hino T, Okamoto H. 1993. Purification of arginine-sensitive hemagglutinin from Fusobacterium nucleatum and its role in coaggregation. J Periodontal Res 28:21–26. doi: 10.1111/j.1600-0765.1993.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 89.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol 54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 90.Diaz PI, Zilm PS, Rogers AH. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 148:467–472. [DOI] [PubMed] [Google Scholar]
- 91.Polak D, Wilensky A, Shapira L, Halabi A, Goldstein D, Weiss EI, Houri-Haddad Y. 2009. Mouse model of experimental periodontitis induced by Porphyromonas gingivalis/Fusobacterium nucleatum infection: bone loss and host response. J Clin Periodontol 36:406–410. doi: 10.1111/j.1600-051X.2009.01393.x. [DOI] [PubMed] [Google Scholar]
- 92.Kolenbrander PE, Andersen RN. 1989. Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect Immun 57:3204–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beachey EH. 1981. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J Infect Dis 143:325–345. doi: 10.1093/infdis/143.3.325. [DOI] [PubMed] [Google Scholar]
- 94.Lindberg F, Lund B, Johansson L, Normark S. 1987. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature 328:84–87. doi: 10.1038/328084a0. [DOI] [PubMed] [Google Scholar]
- 95.Balligand G, Laroche Y, Cornelis G. 1985. Genetic analysis of virulence plasmid from a serogroup 9 Yersinia enterocolitica strain: role of outer membrane protein P1 in resistance to human serum and autoagglutination. Infect Immun 48:782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brennan MJ, Shahin RD. 1996. Pertussis antigens that abrogate bacterial adherence and elicit immunity. Am J Respir Crit Care Med 154:S145–S149. doi: 10.1164/ajrccm/154.4_Pt_2.S145. [DOI] [PubMed] [Google Scholar]
- 97.Locht C, Bertin P, Menozzi FD, Renauld G. 1993. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol Microbiol 9:653–660. doi: 10.1111/j.1365-2958.1993.tb01725.x. [DOI] [PubMed] [Google Scholar]
- 98.Makhov AM, Hannah JH, Brennan MJ, Trus BL, Kocsis E, Conway JF, Wingfield PT, Simon MN, Steven AC. 1994. Filamentous hemagglutinin of Bordetella pertussis. A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in beta strands and turns. J Mol Biol 241:110–124. [DOI] [PubMed] [Google Scholar]
- 99.Sharon N, Lis H. 2004. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology 14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 100.Gonzales-Marin C, Spratt DA, Allaker RP. 2013. Maternal oral origin of Fusobacterium nucleatum in adverse pregnancy outcomes as determined using the 16S-23S rRNA gene intergenic transcribed spacer region. J Med Microbiol 62:133–144. doi: 10.1099/jmm.0.049452-0. [DOI] [PubMed] [Google Scholar]