Abstract
Coxiella burnetii causes human Q fever, a zoonotic disease that presents with acute flu-like symptoms and can result in chronic life-threatening endocarditis. In human alveolar macrophages, C. burnetii uses a Dot/Icm type IV secretion system (T4SS) to generate a phagolysosome-like parasitophorous vacuole (PV) in which to replicate. The T4SS translocates effector proteins, or substrates, into the host cytosol, where they mediate critical cellular events, including interaction with autophagosomes, PV formation, and prevention of apoptosis. Over 100 C. burnetii Dot/Icm substrates have been identified, but the function of most remains undefined. Here, we identified a novel Dot/Icm substrate-encoding open reading frame (CbuD1884) present in all C. burnetii isolates except the Nine Mile reference isolate, where the gene is disrupted by a frameshift mutation, resulting in a pseudogene. The CbuD1884 protein contains two transmembrane helices (TMHs) and a coiled-coil domain predicted to mediate protein-protein interactions. The C-terminal region of the protein contains a predicted Dot/Icm translocation signal and was secreted by the T4SS, while the N-terminal portion of the protein was not secreted. When ectopically expressed in eukaryotic cells, the TMH-containing N-terminal region of the CbuD1884 protein trafficked to the endoplasmic reticulum (ER), with the C terminus dispersed nonspecifically in the host cytoplasm. This new Dot/Icm substrate is now termed ElpA (ER-localizing protein A). Full-length ElpA triggered substantial disruption of ER structure and host cell secretory transport. These results suggest that ElpA is a pathotype-specific T4SS effector that influences ER function during C. burnetii infection.
INTRODUCTION
Coxiella burnetii is a ubiquitous intracellular pathogen that infects macrophages and causes human Q fever. Humans are most often infected via contaminated aerosols while working with infected livestock. Q fever typically presents with acute flu-like symptoms, including high fever and pneumonia, but C. burnetii can also establish a chronic infection, causing life-threatening endocarditis (1). At the root of Q fever is the ability of C. burnetii to generate and replicate within a large, phagolysosome-like parasitophorous vacuole (PV) in macrophages (2). After engulfment in a tightly fitting early phagosome, organisms promote fusion with early and late phagosomes, endosomes, and ultimately lysosomes, wherein acidic conditions activate C. burnetii metabolism. The ability to thrive within this harsh environment makes the pathogen a unique example of an intracellular parasite that hijacks host cells to avoid immune detection. To establish the PV, C. burnetii uses a specialized Dot/Icm type IV secretion system (T4SS) to deliver bacterial effector proteins directly to the host cytosol, where they control critical infection events, many of which are uncharacterized but which include heterotypic fusion with autophagosomes and prevention of host cell death (3, 4).
Identification of C. burnetii effector-encoding genes has been achieved through comparative analysis of genomic content from isolates of disparate backgrounds. Additionally, bioinformatics approaches, using known secreted effector domains from other pathogens as a blueprint, have discovered many effectors with protein-protein interaction domains. A large family of ankyrin repeat domain-containing proteins (Ank proteins) contains the first identified C. burnetii effectors and shows substantial diversity among isolates that cause different forms of Q fever (5, 6). For example, the Nine Mile reference isolate encodes only four intact Ank proteins, while the Dugway isolate contains 11 full-length Ank genes. These studies and others demonstrate the presence of isolate-specific effector repertoires in addition to effectors conserved among all isolates. Further highlighting pathotype diversity, we recently discovered 12 T4SS effectors that vary among C. burnetii isolates depending on the plasmid maintained (7). A study by Beare et al. showed that C. burnetii isolates have undergone extensive transposon-mediated rearrangements resulting in disruption of genes encoding proteins with tetratricopeptide repeats, ankyrin repeats, and coiled-coil domains (8), all of which mediate protein-protein interactions (9), a key feature of many secreted effector proteins. Additionally, the Nine Mile isolate now contains the fewest intact effector-encoding genes, while the Dugway isolate contains the most, suggesting that effector repertoires may influence disease presentation.
Over 100 C. burnetii T4SS effectors have been identified to date, but the activity of most effectors remains elusive. A limited number of effectors have known activity, with AnkG, CaeA, and CaeB preventing host cell death, while CvpA intercepts vesicular trafficking pathways needed for PV development (10–12). C. burnetii lacking CvpA is unable to efficiently form a PV or replicate within host cells, demonstrating absolute reliance on Dot/Icm T4SS effectors for host cell manipulation and infection. Additionally, we recently found that the T4SS is responsible for C. burnetii interactions with autophagosomes (13), an event needed for delivery of nutrients to replicating organisms. Supporting this finding, recently identified Cig2 promotes fusion of autophagosomes with the PV during infection (14). Identification and characterization of additional effectors involved in autophagosome recruitment and PV formation will allow enhanced modeling of the C. burnetii infectious process at the cellular level.
In a recent screen of potential T4SS effectors containing coiled-coil domains (CCDs), we discovered a transmembrane helix- and CCD-containing protein that is translocated into host cells by the Dot/Icm T4SS. Interestingly, the gene encoding this effector is disrupted in the Nine Mile reference isolate yet intact in all other sequenced genomes. We characterized the regions of this effector, now termed endoplasmic reticulum (ER)-localizing protein A (ElpA), and found that distinct portions of the protein are responsible for Dot/Icm-mediated translocation and subcellular trafficking in eukaryotic cells. These results suggest that ElpA is a pathotype-specific effector that impacts ER function during C. burnetii infection.
MATERIALS AND METHODS
Bacteria and eukaryotic cell culture.
Coxiella burnetii Dugway (7E65-68) and Nine Mile I (RSA493) isolates were grown in acidified citrate cysteine medium (ACCM) for 7 days at 37°C, 5% CO2, and 2.5% O2 prior to infection of mammalian cells (15). Cultures were washed three times in sucrose phosphate buffer prior to use. All C. burnetii work was performed in the University of Arkansas for Medical Sciences (UAMS) biosafety level 3 laboratory approved by the Centers for Disease Control and Prevention. Legionella pneumophila was cultured on charcoal yeast extract agar plates, and transformant plates contained chloramphenicol (10 μg/ml). For plating LELA3118, kanamycin (25 μg/ml) was included in media. L. pneumophila transformations were conducted as previously described (5) and Escherichia coli TOP10 cells (Invitrogen) were used for all recombinant DNA procedures. All bacterial strains are described in Table 1.
TABLE 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| C. burnetii | ||
| Dugway (7E65-68) | Phase I, Utah, rodent, 1958 | 36 |
| Nine Mile (RSA493) | Phase I, Montana, tick, 1936 | 37 |
| L. pneumophila | ||
| JR32 | Salt-sensitive isolate of AM511 | 38 |
| LELA3118 | JR32 dotA::Tn903dIIlacZ3118 DotA− Kmr | 38 |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pJB2581 | cyaA fusion vector, Cmr | 39 |
| pDHVAnkN | ankN in pJB2581 | 5 |
| pDHVElpA1–343 | elpA N-terminal fragment in pJB2581 | This study |
| pDHVElpA341–418 | elpA C-terminal fragment in pJB2581 | This study |
| pCR2.1-TOPO | TA TOPO vector, Ampr | This study |
| pENTR-D/TOPO | Gateway entry vector, Kmr | This study |
| pcDNA6.2/N-EmGFP | N-terminal EmGFP fusion vector, Ampr | This study |
| GFP-ElpA | EmGFP::elpA | This study |
| GFP-ElpA1–343 | EmGFP::elpA N-terminal fragment | This study |
| GFP-ElpA-C341–418 | EmGFP::elpA C-terminal fragment | This study |
HeLa (human epithelial carcinoma) cells (CCL-2; American Type Culture Collection [ATCC]) and THP-1 human monocytic (TIB-202; ATCC) cells were cultured in RPMI 1640, and HEK293T (human kidney) cells (ACS-4500; ATCC) were cultured in Dulbecco's modified Eagle medium (DMEM)/F12 with 10% fetal bovine serum (Invitrogen) at 37°C and 5% CO2. THP-1 cells were differentiated into macrophage-like cells by incubation with phorbol 12-myristate 13-acetate (200 nM; PMA; EMD Biosciences) overnight as previously described (16, 17) prior to use in CyaA translocation assays.
Plasmid construction.
For cyaA fusions, genes were amplified from C. burnetii Nine Mile genomic DNA by PCR using Accuprime Taq polymerase (Invitrogen) and gene-specific primers (Integrated DNA Technologies) containing 5′ BamHI and 3′ SalI sites (Table 2). Products were cloned into pCR2.1-TOPO (Invitrogen), plasmids were digested with BamHI/SalI (New England BioLabs), and gene-containing fragments were ligated to BamHI/SalI-digested pJB2581 using Ligate-IT (U.S. Biologicals). For green fluorescent protein (GFP) fusions, C. burnetii genes were amplified by PCR with gene-specific primers (Integrated DNA Technologies). Primers included a 5′ CACC motif for directional cloning and a Kozac sequence (ATGGGC) for eukaryotic expression. Genes and gene fragments were cloned into pENTR-D/TOPO (Invitrogen) and then subcloned into pcDNA6.2/N-GFP using LR Clonase II (Invitrogen). Plasmid constructs were confirmed by sequencing and are listed in Table 1.
TABLE 2.
Primers used in this study
| Purpose and primer | Sequencea |
|---|---|
| pJB2581 cloning | |
| ElpA-N-F | CATGCGGATCCGTGAGTCGAATAATTTATGATGAG |
| ElpA-N-R | CGCATGCGTCGACTTAATCAATAACGGGTGCTCC |
| ElpA-C-F | CATGCGGATCCTTGATTAATCTTCAGGAGGG |
| ElpA-C-R | CGCATGCGTCGACTTAAAAGTTAAATCTGGGGGC |
| pENTR cloning | |
| GFP-ElpA-N-F | CACCATGGGCGTGAGTCGAATAATTTATGATGAG |
| GFP-ElpA-N-R | TTAATCAATAACGGGTGCTCC |
| GFP-ElpA-C-F | CACCATGGGCTTGATTAATCTTCAGGAGGGTAATAATAG |
| GFP-ElpA-C-R | TTAAAAGTTAAATCTGGGGGCTTCTTC |
| RT-PCR | |
| ElpA-F | GAAACGAGAGAGAAGAAGAGGAAG |
| ElpA-R | GGTGTCTATTATTACCCTCCTGAAG |
| ElpA-probe | AAGATCCGGAGCACCCGTTATTGA |
Underlining indicates restriction sites.
CyaA translocation assays.
L. pneumophila transformants were incubated with IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM; ICN Biomedicals) for 2 h and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis using a CyaA-specific antibody (clone 3D1; Santa Cruz Biotechnology). Reacting proteins were detected using a horseradish peroxidase-conjugated anti-mouse IgG antibody (Cell Signaling Technology) and enhanced chemiluminescence (ECL Pico reagent; Pierce). CyaA assays were performed as previously described (5). Positive secretion of fusion proteins was scored as ≥2.5-fold more cytosolic cyclic AMP (cAMP) than in cells infected with L. pneumophila expressing CyaA alone (18, 19). CyaA fused to C. burnetii AnkN was used as a positive control (5), and Dot/Icm-mediated secretion was confirmed using the DotA− L. pneumophila mutant LELA3118.
Effector sequence analysis.
ElpA was subjected to in silico analysis using the programs i-TASSER, SMART, MacVector, and COILS. SMART (20) was used to identify eukaryotic-type domains, and i-TASSER (21, 22) was used to generate a predicted ElpA tertiary structure. i-TASSER-generated structures were further analyzed and rendered using iMOL (23) and ERdj5 was rendered using Protein Data Bank coordinates 3APO. MacVector (version 12.7.5) was used to analyze the hydrophobicity and transmembrane profiles of ElpA, and the COILS server (http://embnet.vital-it.ch/software/COILS_form.html) was used to identify coiled-coil domains in the protein.
Expression analysis of elpA during infection.
THP-1 cells were infected with Dugway, samples were harvested in TRIzol (Invitrogen), and RNA was isolated with a Qiagen RNeasy kit. cDNA was synthesized using random hexamers and the SuperScript III first-strand synthesis system (Invitrogen). Quantitative reverse transcription-PCR (qRT-PCR) was performed using 1 μg of total RNA, a StepOnePlus real-time system (Applied Biosystems), TaqMan primers and probe (Table 2) (Integrated DNA Technologies), and TaqMan Fast Advanced master mix reagents according to the manufacturer's protocol (Applied Biosystems).
HeLa cell transfection and microscopy.
HeLa cells were either left uninfected or infected with Dugway (multiplicity of infection = 10) for 24 h and then transfected with plasmid constructs containing GFP fused to ElpA or ElpA fragments. Transfections were performed with Effectene transfection reagent (Qiagen) for 18 h. Cells were then processed for confocal fluorescence microscopy and incubated with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) to visualize the nucleus. An anticalnexin (Enzo Life Sciences) primary antibody was used to detect the ER, and anti-C. burnetii polyclonal antiserum was used to detect bacteria. Alexa Fluor-labeled secondary antibodies were used to detect primary antibodies (Invitrogen). Confocal fluorescence microscopy was performed using a Nikon Ti-U microscope and 100× oil immersion objective (Nikon). Images were analyzed and fluorescence intensity profiles were derived using NIS-Elements software (Nikon).
SEAP assay.
HEK293T cells plated in 24-well plates (3 × 105 cells/well) were cotransfected with 200 ng of CMV-SEAP (secreted alkaline phosphatase) and GFP (negative control) or a GFP-ElpA plasmid encoding full-length or truncated ElpA. Transfection efficiencies were optimized for each construct to obtain similar efficiencies using Lipofectamine LTX (Invitrogen) according to the manufacturer's protocol. Cells were cotransfected for 18 h, and medium was replaced at 18 h posttransfection (time zero). Cells were incubated for 7 h, and then supernatants and cell lysates were harvested. SEAP levels in each cell lysate and corresponding supernatant were determined using the Phospha-Light system according to the manufacturer's protocol (Applied Biosystems). Results are presented as ratios of extracellular to intracellular SEAP activity.
RESULTS
Identification of ElpA as a Dot/Icm substrate.
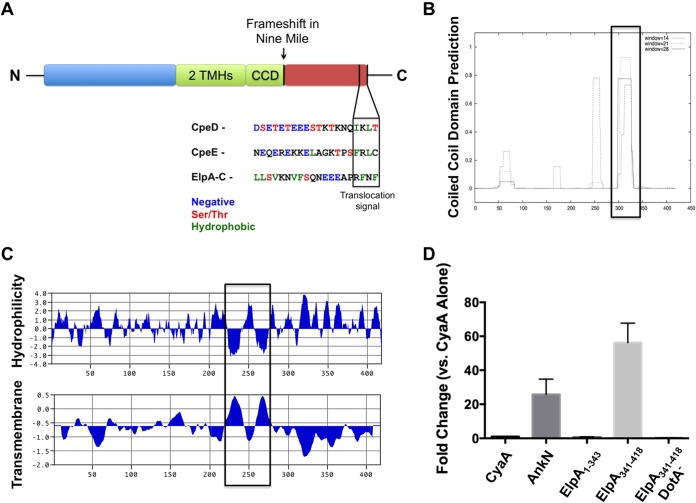
In a screen of genes encoding potential T4SS effectors with coiled-coil domains (CCDs) or transmembrane helices (TMHs) (data not shown), we assessed CbuD1884, a gene that is intact in the Dugway, G (CbuG1796), and K (CbuK0401) C. burnetii genomes but is disrupted in the Nine Mile reference isolate genome. Full-length CbuD1884 encodes a protein now termed ER-localizing protein A (ElpA). Comparative analyses showed that, instead of a full-length ElpA gene, CBU0211 and CBU0212 comprise a disrupted gene in Nine Mile, resulting in a pseudogene. The presence of an additional base at the 3′ end of CBU0211 results in a frameshift mutation that would prevent expression of both genes as a single product, making Nine Mile a naturally occurring mutant. As shown in Fig. 1D, using the established L. pneumophila model for identifying C. burnetii Dot/Icm substrates (6, 18, 19), the protein encoded by CBU0211 (ElpA1–343) was not secreted by wild-type L. pneumophila, while the protein encoded by CBU0212 (ElpA341–418) was secreted in a Dot/Icm-dependent manner as evidenced by lack of secretion by a DotA mutant. Amino acid analysis revealed two hydrophobic (Phe) residues in the C-terminal three amino acids of ElpA and an E-block motif (24) within the 10 C-terminal residues (Fig. 1A), consistent with previous C-terminal Dot/Icm translocation signal predictions (25–27).
FIG 1.
CbuD1884 encodes a Dot/Icm substrate with protein-protein interaction domains. (A) Schematic of the CbuD1884 protein (ElpA) containing two transmembrane helices (TMHs), a coiled-coil domain (CCD), and a putative C-terminal translocation signal. ElpA is full length in all C. burnetii isolates except the Nine Mile reference isolate, where a frameshift mutation (arrow) results in a pseudogene. The Dot/Icm substrates CpeD and CpeE are shown for C-terminal comparisons, and the putative Dot/Icm translocation signal is shown in the boxed region. The ElpA amino acid sequence was analyzed with COILS (B) and MacVector (C). ElpA contains two hydrophobic regions between residues 224 and 280, and these regions correspond to the presence of two TMHs. COILS analysis shows a CCD at residues 298 to 331. Boxes denote regions of interest. These results suggest the functional domains of ElpA reside in the N-terminal region (amino acids 1 to 331) of the protein. (D) In an established Dot/Icm translocation assay, L. pneumophila expressing the indicated proteins fused to CyaA was allowed to infect THP-1 cells for 30 min, and then cAMP levels were determined as a reporter for delivery to the cytosol. CyaA was used as a nonsecreted negative control and AnkN was a positive secreted control (5). Experiments were performed in triplicate, and error bars represent the standard deviations from the means. Elevated levels of cAMP indicative of secretion were observed for ElpA341–418 when it was expressed by wild-type L. pneumophila but not the DotA mutant (DotA−). These results show that only the C-terminal region of ElpA, not the N-terminal portion (ElpA1–343), contains the Dot/Icm T4SS translocation signal.
In non-Nine Mile isolates, ElpA is a predicted 47.6-kDa protein of 418 amino acids with a pI of 4.6. ElpA contains two TMHs between amino acid residues 224 and 280 and a predicted CCD from residues 298 to 331 (Fig. 1B and C). Structural prediction analysis using i-TASSER (21, 22, 28) showed that ElpA is similar to the endoplasmic reticulum (ER) protein ERdj5 (data not shown), a TMH-containing, ER-resident disulfide reductase that functions in retrotranslocation of proteins destined for ubiquitin-mediated degradation (29). ElpA contains a predicted thioredoxin fold similar to ERdj5 in the N-terminal region of the effector. This prediction suggests that ElpA may manipulate secretory processes during C. burnetii infection by mimicking ER-related proteins.
elpA is expressed during C. burnetii growth in mammalian cells.
To determine if elpA is expressed during infection, Dugway isolate-infected THP-1 macrophage-like cells were subjected to RT-PCR analysis. As shown in Fig. 2, elpA transcripts were detected by 24 h postinfection (hpi), and expression increased substantially from 48 to 120 hpi. This timing corresponds to PV expansion, suggesting that elpA is highly expressed at times after PV maturation to a phagolysosome-like compartment. elpA expression followed a similar trend during Dugway isolate infection of primary human alveolar macrophages (data not shown), suggesting that the effector is produced during human disease. These results differ from dotA expression, which is needed for functional T4SS activity that promotes PV generation and occurs throughout infection (30), suggesting that ElpA activity is needed at later times of infection and is not required for initial PV maturation events.
FIG 2.
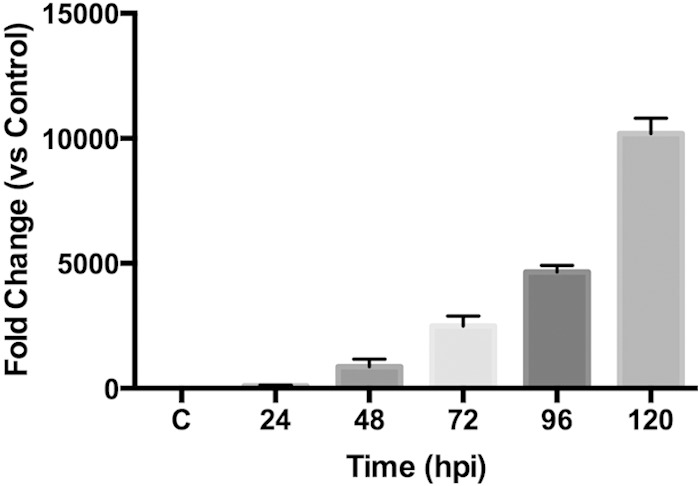
elpA is expressed during Dugway isolate growth in mammalian cells. THP-1 macrophage-like cells were infected for the indicated time postinfection (hpi) with the C. burnetii Dugway isolate, RNA was harvested and converted to cDNA, and samples were subjected to RT-PCR analysis. Each sample was compared to the 2-hpi sample (C), and results are representative of two independent experiments performed in triplicate. Error bars represent the standard deviations from the means. The results show that elpA is expressed throughout Dugway intracellular growth.
ElpA requires a transmembrane helical region to traffic to the ER.
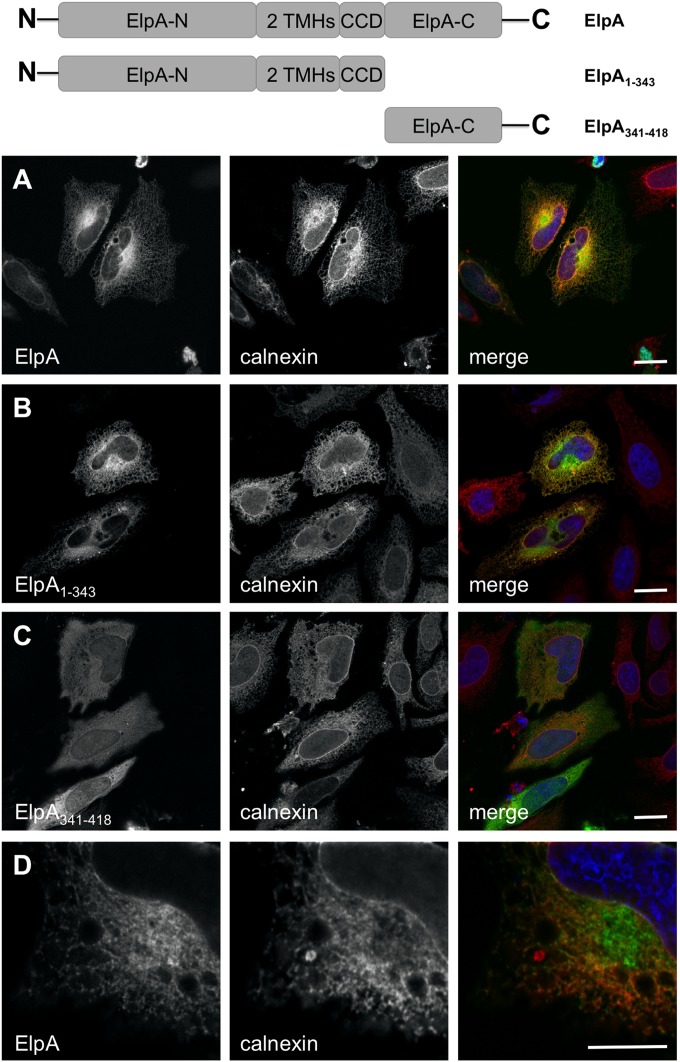
Structural predictions (data not shown) suggested that ElpA contains a thioredoxin fold found in proteins that localize to the ER. Additionally, Brucella T4SS-secreted effectors containing TMHs interact with the host ER and disrupt secretory transport (9). Therefore, we assessed subcellular trafficking of ElpA fused to GFP in HeLa cells. As seen in Fig. 3, full-length GFP-ElpA colocalizes with the ER protein calnexin. Once distinct domains (TMHs and a CCD) were identified within the ElpA sequence (Fig. 1), truncation mutants were constructed and subcellular localization was assessed to determine the contribution of each region to ER association. While the C-terminal region of ElpA (lacking TMHs and CCDs) necessary for Dot/Icm-mediated translocation localized nonspecifically in the host cell cytoplasm, any fusion containing the TMHs colocalized with calnexin at the ER (Fig. 3A to C and data not shown), indicating that these regions are critical for proper ElpA subcellular localization. Additionally, fusions containing the CCD, but no TMH, localized nonspecifically in the cytoplasm (data not shown), further confirming the importance of TMHs in ElpA trafficking within mammalian cells. Further analysis showed that a portion of ElpA did not colocalize with the ER but concentrated in the perinuclear region (Fig. 3D), indicating that GFP-ElpA is not simply trapped in the ER following expression. Importantly, ElpA fused to mCherry also trafficked to the ER (data not shown), indicating that localization is directed by the effector.
FIG 3.
The N terminus of ElpA directs trafficking to the ER. ElpA (A), ElpA1–343 (B), and ElpA341–418 (C) were individually expressed in HeLa cells as GFP fusion proteins (green). At 18 h posttransfection, cells were processed for fluorescence microscopy. The ER was detected using antibody directed against calnexin (red) and DAPI-stained DNA (blue). Bar, 10 μm. Full-length ElpA and the N-terminal portion of ElpA traffic to the ER, as shown by fluorescence overlap (yellow) in merged panels, while ElpA341–418 is dispersed in the cytoplasm. (D) A population of GFP-ElpA does not colocalize with calnexin, as observed by partial fluorescence overlap in the perinuclear region.
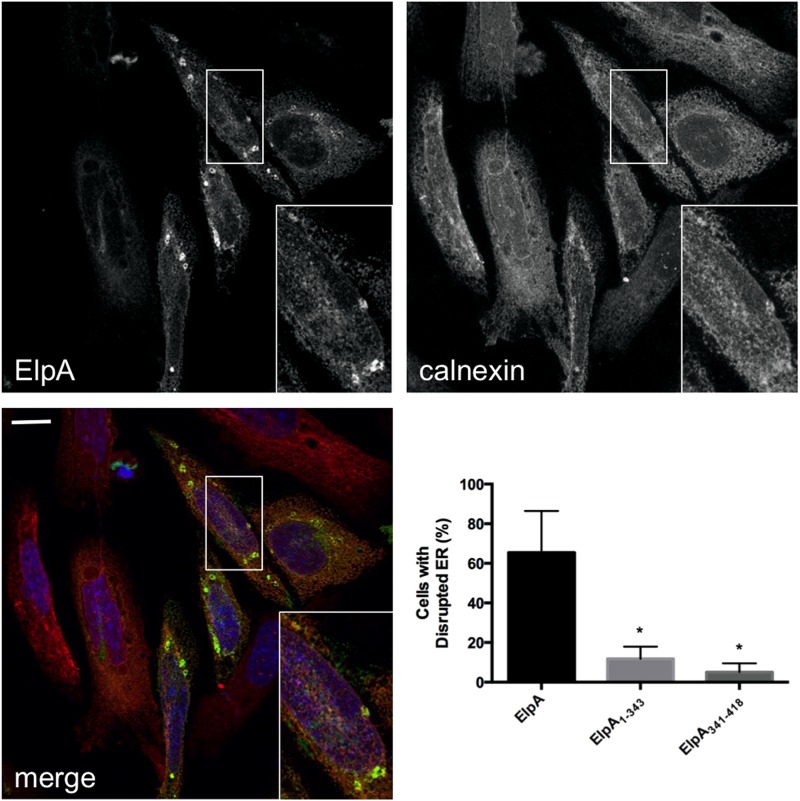
Interestingly, approximately 65% of ElpA-expressing cells contained severely disrupted ER structures, resulting in ElpA- and calnexin-rich compartments in the cytoplasm (Fig. 4). Full-length ElpA was required for ER disruption, as approximately 15% of ElpA1–343-expressing cells contained altered ER structures and ElpA341–418 did not significantly influence organelle structure regardless of the level of expression (<10% of cells contained disrupted ER). These results suggest that ElpA activity negatively impacts ER structure.
FIG 4.
ElpA triggers disruption of ER structure. Full-length GFP-ElpA, GFP-ElpA1–343, and GFP-ElpA341–418 were individually expressed in HeLa cells (green), and cells were processed at 18 h posttransfection for fluorescence microscopy. The ER was detected using antibody directed against calnexin (red) and DAPI-stained DNA (blue). Bar, 10 μm. The graph indicates the percentage of transfected cells containing disrupted ER structure under each condition. Error bars represent the standard deviations from the means, and data were collected from at least 100 cells/condition from experiments performed in triplicate. *, P < 0.0001 by Student's t test when truncations and full-length GFP-ElpA are compared. Ectopic expression of GFP-ElpA alters ER structure, leading to accumulation of calnexin-rich ElpA-containing compartments in the cytoplasm. In contrast, most cells expressing GFP-ElpA1–343 or GFP-ElpA341–418 contain a normal ER structure.
ElpA disrupts host secretory transport.
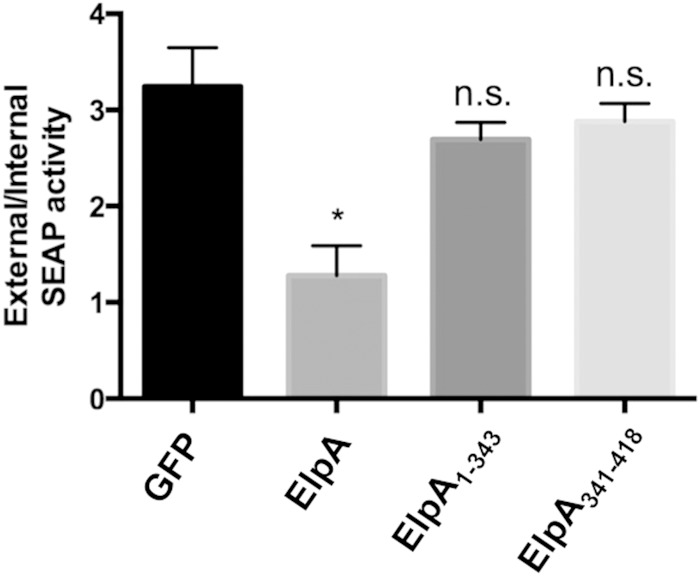
Bacterial effectors that traffic to the ER and/or alter organelle structure often disrupt secretory transport and are predicted to mediate ER-related processes during infection. Full-length ElpA traffics to the ER and triggers substantial rearrangement of the ER network (Fig. 3 and 4). Thus, we next assessed the impact of ElpA on host cell secretory transport. As determined by a standard secreted alkaline phosphatase (SEAP) assay (Fig. 5), GFP-ElpA expression was sufficient to prevent HEK293T cell secretion of alkaline phosphatase, similar to previously identified C. burnetii T4SS effectors (4). Supporting ER disruption results, only full-length ElpA altered secretion, as SEAP levels resulting from cells expressing ElpA1–343 or ElpA341–418 were similar to those obtained with cells expressing only GFP, suggesting that the full-length effector is needed to interact with host components that negatively influence secretion.
FIG 5.
ElpA alters host secretory transport. (A) HEK293T cells were cotransfected with a SEAP-expressing construct and GFP alone or one of the indicated GFP fusion proteins. SEAP levels were determined and are displayed as the ratio of intracellular to extracellular product. Experiments were performed in triplicate, and error bars represent the standard deviations from the means. *, P < 0.005; n.s., not significant (Student's t test). All GFP-ElpA samples were compared to GFP alone. The N- and C-terminal portions of ElpA alone do not significantly alter secretion, but full-length ElpA triggers approximately 60% less secretion than the GFP-only control, indicating that ElpA negatively influences host secretory transport.
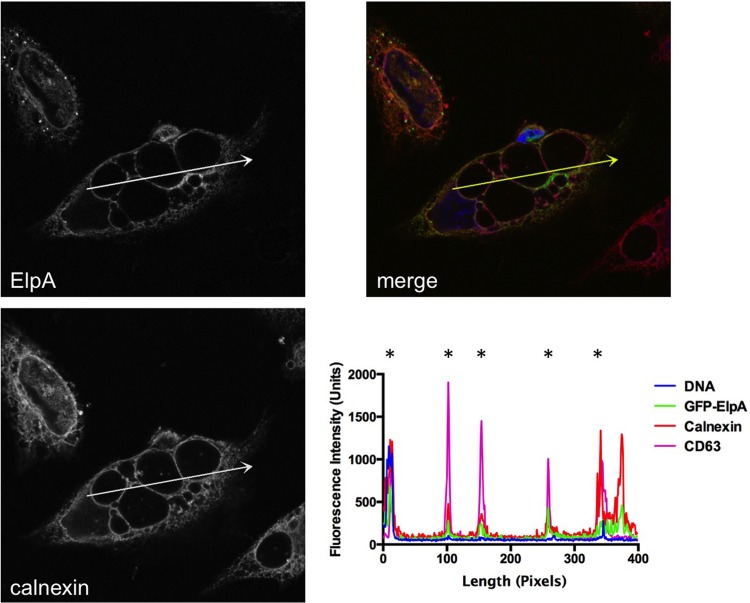
Calnexin and ElpA concentrate around the PV during C. burnetii intracellular growth.
The C. burnetii PV has not been predicted to closely associate with the ER similarly to other intracellular pathogens. However, due to the negative impact of ElpA activity on ER structure and secretion, we assessed ER accumulation at the PV membrane. As shown in Fig. 6, PVs containing replicating C. burnetii were decorated heavily with the ER-resident protein calnexin. Fluorescence intensity analysis showed overlap of GFP-ElpA, calnexin, and the PV marker CD63 at the membrane of approximately 94% of vacuoles containing C. burnetii (Fig. 6 and data not shown). Additionally, no significant difference was observed in calnexin labeling of Dugway and Nine Mile I PV, with >90% of both isolate PVs associating with the ER (data not shown). Similar results for Nine Mile and Dugway suggest that ER recruitment to the PV is likely not ElpA dependent. Although the mechanism of ER association with, or active recruitment to, the PV has not been established, small PVs (<10 μM diameter) are also labeled with calnexin, suggesting that vacuole size alone is not responsible for the presence of ER components at the membrane. These results indicate that the PV closely associates with the ER network during infection, providing a platform for effector interactions with ER proteins after translocation across the PV membrane.
FIG 6.
The PV is decorated with ElpA and calnexin during C. burnetii intracellular growth. Full-length GFP-ElpA (green) was ectopically expressed in HeLa cells that were infected for 30 h with the Dugway isolate. At 18 h posttransfection, cells were processed for fluorescence microscopy. The ER was detected using antibody against calnexin (red) and DAPI-stained DNA (blue). *, PV membrane. Fluorescence intensity analysis (arrows) shows that GFP-ElpA and calnexin are enriched around the PV membrane, as indicated by overlap with the PV marker CD63.
DISCUSSION
In this study, we discovered a novel C. burnetii pathotype-specific Dot/Icm substrate termed ElpA that contains a predicted ER structural motif, traffics to the host ER, and disrupts secretory transport. ElpA is found in all C. burnetii isolates except the Nine Mile acute disease isolate, suggesting that Nine Mile has adapted to survive in the absence of the effector, possibly through the use of proteins with similar functions. Nine Mile encodes two other Dot/Icm substrates, CBU0372 and CBU1576, with TMHs and a CCD that localize to the ER, but neither effector disrupts secretory transport (31). Interestingly, only Nine Mile and Dugway contain CBU0372 and CBU1576, while other isolates exclusively encode ElpA. However, Dugway encodes all three proteins, suggesting that this pathotype may encode multiple Dot/Icm substrates with similar functions, as previously suggested by genomic comparison studies (8). Although CBUA0019, CBU0635, CBU1556, and CBU1825 do not traffic to the ER, each effector negatively impacts host cell secretory transport (4, 31). The presence of multiple effectors that traffic to the ER and/or disrupt secretory transport support the importance of interactions with this organelle for productive C. burnetii infection.
Disruption of host cell secretory transport is an emerging theme among intracellular bacterial pathogens. L. pneumophila and Brucella spp. produce multiple T4SS substrates that interact with ER proteins and prevent typical secretory transport (9, 32–34). Two L. pneumophila effectors, LegC2/YlfB and LegC7/YlfA, trigger substantial ER disruption similar to that observed in ElpA-expressing cells in the current study, suggesting similar intracellular functions. Additionally, L. pneumophila AnkX disrupts host Golgi architecture, ultimately preventing secretory transport (4, 6). Brucella type IV-secreted VceC localizes to the ER and triggers organelle rearrangement but not gross disruption, suggesting a function distinct from that of ElpA. However, VceC triggers ER stress by activating the unfolded protein response and altering Brucella-induced inflammation. These studies also used fluorescent fusion proteins similar to the GFP tag used in the current study, indicating that effector localization is not an artifact of the fluorescent protein. Thus, it is clear that intracellular pathogens target host cell secretory processes from multiple angles using type IV-secreted molecules. The ultimate impact of ElpA activity on C. burnetii infection is still under investigation, but our data indicate that the effector negatively impacts host protein secretion.
Two recent reports support a major role for C. burnetii proteins that influence secretory transport. From early stages of infection (6 h) onward, the PV is decorated with mammalian Rab1, a small GTPase involved in anterograde transport that functions at the ER-Golgi interface (35). GTPase-defective Rab1 does not support development of prototypical PV, and treatment of C. burnetii-infected cells with brefeldin A disrupts normal PV expansion, indicating that the pathogen requires a functional host secretory pathway for efficient intracellular growth. Additionally, Carey et al. (4) showed that type IV-secreted CBU0635 disrupts secretion of alkaline phosphatase using the SEAP assay that was used in the current study. Moreover, three additional T4SS effectors, CBUA0019, CBU1556, and CBU1825, prevent host secretion (31), indicating that C. burnetii encodes multiple effectors that influence ER and/or Golgi apparatus function.
Although related pathogens, including L. pneumophila and Brucella abortus, replicate within an ER-derived vacuole, formation of the C. burnetii PV is not predicted to require ER interactions due to vast supplies of membrane obtained from lysosomes, autophagosomes, and endosomes. However, ElpA and the ER protein calnexin concentrate around the PV, supporting a potential role for PV-ER interactions during infection. This finding adds ElpA to a growing list of effectors that traffic to the C. burnetii PV membrane after translocation. Previous studies showed that CBU0077, AnkN, AnkO, CvpA, CpeB, and CpeL colocalize with lysosomal and/or autophagosomal proteins when ectopically expressed in mammalian cells, suggesting that they are involved in PV development through recruitment of vacuolar compartments (4, 5, 7, 12, 18). Future dissection of the role of ElpA at the PV-ER interface will shed light on an underappreciated aspect of C. burnetii-host cell interactions.
The current study presents a novel pathotype-specific Dot/Icm substrate that localizes to the PV membrane and associates with the host ER. The absence of ElpA in the Nine Mile isolate provides a natural mutant for future comparative studies to dissect the effector's mechanism of action. Additionally, structural determination and subsequent mutational analysis will provide a better understanding of the role of ElpA thioredoxin folds and protein-protein interaction domains in C. burnetii parasitism of human cells. Importantly, the current study further supports the presence of interactions between C. burnetii and the ER, which is a new area of investigation into the unique C. burnetii-host cell interaction.
ACKNOWLEDGMENTS
This research was supported by funding to D.E.V. from the NIH/NIAID (R01AI087669), the Arkansas Biosciences Institute, and the Center for Microbial Pathogenesis and Host Inflammatory Responses (NIH/NIGMS P20GM103625). J.G.G. was supported by a supplement to R01AI087669 to promote diversity in research.
We thank J. Craig Forrest for providing HEK293T cells for SEAP analysis.
REFERENCES
- 1.Maurin M, Raoult D. 1999. Q fever. Clin Microbiol Rev 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voth DE, Heinzen RA. 2007. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 3.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175–11. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey KL, Newton HJ, Luhrmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maturana P, Graham JG, Sharma UM, Voth DE. 2013. Refining the plasmid-encoded type IV secretion system substrate repertoire of Coxiella burnetii. J Bacteriol 195:3269–3276. doi: 10.1128/JB.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beare PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, Williams KP, Sobral BW, Kupko JJ III, Porcella SF, Samuel JE, Heinzen RA. 2009. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 77:642–656. doi: 10.1128/IAI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myeni S, Child R, Ng TW, Kupko JJ III, Wehrly TD, Porcella SF, Knodler LA, Celli J. 2013. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog 9:e1003556. doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klingenbeck L, Eckart RA, Berens C, Luhrmann A. 2013. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol 15:675–687. doi: 10.1111/cmi.12066. [DOI] [PubMed] [Google Scholar]
- 11.Luhrmann A, Nogueira CV, Carey KL, Roy CR. 2010. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A 110:E4770–E4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winchell CG, Graham JG, Kurten RC, Voth DE. 2014. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun 82:2229–2238. doi: 10.1128/IAI.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. 2014. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, Porcella SF, Heinzen RA. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voth DE, Heinzen RA. 2009. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun 77:205–213. doi: 10.1128/IAI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voth DE, Howe D, Heinzen RA. 2007. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun 75:4263–4271. doi: 10.1128/IAI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL. 2011. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS One 6:e27724. doi: 10.1371/journal.pone.0027724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res 32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotkiewicz P. 2007. iMol molecular visualization program. [Google Scholar]
- 24.Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. 2011. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A 102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A 110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y. 2009. I-TASSER: fully automated protein structure prediction in CASP8. Proteins 77(Suppl 9):S100–S113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ushioda R, Hoseki J, Nagata K. 2013. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol Biol Cell 24:3155–3163. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. 2008. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol 70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog 4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jong MF, Starr T, Winter MG, den Hartigh AB, Child R, Knodler LA, van Dijl JM, Celli J, Tsolis RM. 2013. Sensing of bacterial type IV secretion via the unfolded protein response. mBio 4:e00418–12. doi: 10.1128/mBio.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campoy EM, Zoppino FC, Colombo MI. 2011. The early secretory pathway contributes to the growth of the Coxiella-replicative niche. Infect Immun 79:402–413. doi: 10.1128/IAI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoenner HG, Lackman DB. 1960. The biologic properties of Coxiella burnetii isolated from rodents collected in Utah. Am J Hyg (Lond) (71):45–51. [DOI] [PubMed] [Google Scholar]
- 37.Beare PA, Samuel JE, Howe D, Virtaneva K, Porcella SF, Heinzen RA. 2006. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J Bacteriol 188:2309–2324. doi: 10.1128/JB.188.7.2309-2324.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadosky AB, Wiater LA, Shuman HA. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61:5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bardill JP, Miller JL, Vogel JP. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]