Abstract
Nontypeable Haemophilus influenzae (NTHI) forms biofilms in the middle ear during human infection. The biofilm matrix of NTHI contains extracellular DNA. We show that NTHI possesses a potent nuclease, which is a homolog of the thermonuclease of Staphylococcus aureus. Using a biofilm dispersal assay, studies showed a biofilm dispersal pattern in the parent strain, no evidence of dispersal in the nuclease mutant, and a partial return of dispersion in the complemented mutant. Quantitative PCR of mRNA from biofilms from a 24-h continuous flow system demonstrated a significantly increased expression of the nuclease from planktonic organisms compared to those in the biofilm phase of growth (P < 0.042). Microscopic analysis of biofilms grown in vitro showed that in the nuclease mutant the nucleic acid matrix was increased compared to the wild-type and complemented strains. Organisms were typically found in large aggregates, unlike the wild-type and complement biofilms in which the organisms were evenly dispersed throughout the biofilm. At 48 h, the majority of the organisms in the mutant biofilm were dead. The nuclease mutant formed a biofilm in the chinchilla model of otitis media and demonstrated a propensity to also form similar large aggregates of organisms. These studies indicate that NTHI nuclease is involved in biofilm remodeling and organism dispersal.
INTRODUCTION
Nontypeable Haemophilus influenzae (NTHI) is frequently found as a component of the normal upper respiratory tract bacterial flora (1). This species is a cause of airway infections, including otitis media in children, sinusitis, and acute exacerbations of chronic bronchitis in adults (1). NTHI has been shown to be capable of forming biofilms both in vitro and in the upper and lower human respiratory tract during human disease (2–8). Bacterial biofilm matrices are an elaborate network of molecules, which can include pili, polysaccharides, extracellular DNA (eDNA), and bacterial and host-derived substances that help shape and secure the biofilm to an inanimate or host surface (9). The matrix protects the underlying bacteria from assault by the host immune response and antibiotic treatment, thus contributing to the recalcitrance of biofilm infections to antimicrobial treatment (10). The matrix of NTHI biofilms has been shown to contain double-stranded eDNA (11). Our laboratory has been interested in studying possible mechanisms controlling the matrix eDNA in an NTHI biofilm. Studies of the sequenced genome of H. influenzae strains KW20 Rd (HI1296) and 86-028NP (NTHI1828) indicated that an open reading frame (ORF) with high homology to the Staphylococcus aureus thermonuclease was present. A number of studies have shown that mechanisms are present in bacteria to degrade biofilm matrix and release organisms from the biofilm to planktonic phase of growth (12–14). Studies by Steichen et al. demonstrated that Neisseria gonorrhoeae expressed a thermonuclease, which was involved in biofilm eDNA matrix remodeling (15). In S. aureus, it has been shown that its thermonuclease is regulated by the SaeRS two-component system (16) and that this nuclease is involved in facilitating the escape of S. aureus from neutrophil extracellular traps (NETs) (17). We investigate in the present study whether NTHI expresses a nuclease and, if so, whether it plays a role in biofilm remodeling and organism dispersal.
MATERIALS AND METHODS
Bacteria and culture conditions.
Bacterial strains used in the study are shown in Table 1. Nontypeable H. influenzae 2019 (NTHI 2019) is a clinical isolate described in previous studies (18). NTHI 2019 was grown from frozen stock cultures at 37°C in 5% CO2 in brain heart infusion agar (Difco) supplemented with 10 μg of hemin/ml and 10 μg of NAD/ml (sBHI). Escherichia coli K-12 was grown in Luria-Bertani medium with or without agar and supplemented with antibiotics as needed.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Nontypeable H. influenzae strain or plasmid | Genotype, relevant characteristic(s), and/or sequence (5′-3′)a | Source or reference |
|---|---|---|
| NTHI strains | ||
| 2019 | Wild type | Our laboratory |
| 2019Δnuc | Δnuc; Kan | This study |
| 2019Δnuc::nuc | Δnuc::nuc; Kan Sp | This study |
| Plasmids | ||
| pGEM-T | Promega | |
| pET151/D-TOPO | Amp | Life Technologies |
| pUC18K3 | Kan | This laboratory |
| p601.1-Sp2 | Sp | 10 |
| pCEC#27 | pUC18K3::Δnuc::kan; Kan | This study |
| pCEC#29 | pET151::nuc; Amp | This study |
| pCEC#31 | p601.1Sp2::nuc; Sp | This study |
| Primers | ||
| DNase 1 FRET assay primer | Cy3-CCGCGAGAAACCAAGCACAGAGCACCGAAGA-BHQ_2 | |
| qRT-PCR primers | ||
| nuc primers | ||
| Forward | AGGGTATGGCATGGGCTTAT | |
| Reverse | GTGCGAGCCTGTTCATATTG | |
| OMP p6 primers | ||
| Forward | GGTATTCFTGGGTGGTTGCGTATT | |
| Reverse | AGATCAGCAACAGAGTAACCGCCA |
Kan, kanamycin; Sp, spectinomycin; Amp, ampicillin; BHQ, black hole quencher.
Construction of the nuclease (nuc) deletion mutant.
The whole gene, except for the first and the last codon of the nuc ORF, was replaced with a kanamycin resistance cassette. Approximately 500 bp upstream and downstream, the arms of nuc were PCR amplified. The upstream homology arm contained EcoRI at the 5′ end and KpnI at the 3′ end, and the downstream homology arm contained XbaI at the 5′ end and HindIII at the 3′ end. The homology arms were ligated into pUC18K3, flanking the kanamycin resistance cassette in the multiple cloning site. The resulting plasmid, pCEC#27, is shown in Table 1. The plasmid was transformed into NTHI 2019, and transformants were screened on sBHI plates containing 15 μg of ribostamycin/ml. The sequences of mutant transformants (NTHI 2019Δnuc) were confirmed by PCR and DNA sequencing.
Complementation of the nuc deletion.
Previous studies in our laboratory used p601.1-Sp2 plasmid for chromosomal complementation in NTHI 2019 (18). Appropriate primers to clone the entire nuc ORF were designed in which both the 5′ end of the forward primer and the 3′ end of the reverse primer contained SmaI restriction enzyme sites. The PCR-amplified product was cloned into p601.1-Sp2 using the SmaI restriction enzyme site. The final plasmid was pCEC#31 (Table 1). This plasmid was transformed into NTHI 2019Δnuc. The transformants were screened on BHI agar plate supplemented with 15 μg of ribostamycin/ml and 25 μg of spectinomycin/ml. The complemented strain was designated NTHI 2019Δnuc::nuc (Table 1). PCR and DNA sequencing were used to confirm the sequence fidelity and the correct orientation of the complementation.
Cloning, expression, and purification of Nuc.
Nuc, without the signal sequence, was expressed in pET151/D-TOPO (Life Technologies) with cleavable 6×His tag in BL21(DE3) E. coli cells and induced with 1.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Invitrogen) at an optical density of 0.6 at 18°C. Cell cultures were pelleted and frozen at −20°C in preparation for lysis. The frozen cells were thawed in lysis buffer (100 mM Tris [pH 9.1], 5 mM CaCl2, 200 mM NaCl, 50 mM imidazole) plus a Mini-Complete protease inhibitor cocktail tablet (Roche) and lysed with an Emulsiflex C3. Subsequently, His-tagged Nuc was added to Ni-nitrilotriacetic acid (Ni-NTA) affinity resin (Qiagen). The resin was then washed with the lysis buffer, and the protein was eluted with lysis buffer containing 250 mM imidazole. The eluted protein was concentrated and mixed 1:200 with Tobacco etch virus protease at 4°C for 16 h to cleave the His tag. The cleaved products were passed over a Ni-Sepharose column (HisTrap FF; GE Healthcare) and the flowthrough was subjected to gel filtration (Superdex 75 25/60; GE Healthcare) to separate Nuc from contaminants and to exchange it into the final purification and reaction buffer (100 mM Tris [pH 9.1], 5 mM CaCl2, 200 mM NaCl, 5 mM dithiothreitol [DTT]).
DNase assays.
Two different assays were used to assess the enzymatic activity of Nuc. The first assay involved combining either Nuc or DNase I (New England BioLabs, Inc.) with DNA template in DNase I buffer at 37°C. After 30 min, DNA running buffer was added to the samples, and they were immediately run on a 1% agarose gel. The gel was stained with 0.5 g of ethidium bromide/ml, destained with double-distilled H2O, and then viewed using UV light.
Nuclease enzyme activity was also measured by a fluorescence resonance energy transfer activity study (FRET) assay using a Tecan Infinite M200 Pro. The FRET substrate was a single-stranded 30-mer oligonucleotide (Table 1) with the 5′ end modified with Cy3 fluorophore and the 3′ end modified with Black Hole Quencher 2 (19). The substrate was incubated with either Nuc or DNase I (positive control) at 26°C, and then the absolute fluorescence was measured every 10 s for 5 min. Each study shown was performed a minimum of six times on multiple Nuc samples.
Biofilm dispersal studies.
Biofilm dispersal was analyzed according to the method of Kaplan and Fine (14), which takes advantage of the microcurrents caused by edge evaporation in an open 100-mm culture dish in which small numbers of organisms (102 to 103 CFU/ml) are incubated overnight at 37°C under 85% humidity and 5% CO2 for 24 h. After the incubation, the medium was cultured and carefully decanted, and the plate was stained with gentian violet in 20% ethanol for 2 min and then washed vigorously with distilled water. After 20 min, the plate was washed again with distilled water, dried, and viewed with a Nikon SMZ800 scope with a Diagnostic Instruments, Inc., digital camera module at ×5, ×20, and ×35. The images were then processed using SPOT software 5.1 (SPOT Imaging Solutions, Inc.). These experiments were performed in triplicate on separate occasions studying NTHI 2019, NTHI 2019Δnuc, and NTHI 2019Δnuc::nuc.
Biofilm growth over glass using continuous flow chambers.
NTHI strains 2019, 2019Δnuc, and 2019Δnuc::nuc were grown in continuous flow chambers for in vitro biofilm growth by the method of Schwartz et al. (20). Chambers were made with the same dimension and material as previously described (20), except the outflow fitting material was changed from copper to plastic. Glass coverslips (22 mm by 50 mm) were placed in the chambers and were stabilized with silicone. The chambers with the coverslips and influent and effluent tubing were sterilized by autoclaving. NTHI strains were grown to mid-log phase in RPMI 1640 medium (Life Technologies) supplemented with protoporphyrin IX (1 μg/ml), hypoxanthine (0.1 mg/ml), uracil (0.1 mg/ml), β-NAD (10 μg/ml), sodium pyruvate (0.8 mM), and Neu5Ac (100 μm). The chambers with clamped effluent tubing on a flat surface were inoculated with 6-ml cultures diluted to an A600 of 0.25. The chambers were incubated at 37°C for 2 h to allow for bacteria to adhere to the coverslip. The influent tubing was aseptically connected to the flask containing supplemented RPMI medium, diluted 1:4 in phosphate-buffered saline (PBS), and then to the chambers via a 23-gauge needle (1 in.), which was aseptically inserted through the inlet stopper. Both inflow and outflow tubing were fed through the same pump to maintain constant volume in the chambers. The chambers, on a flat surface, were incubated at 37°C for 48 h at a flow rate of 100 μl/min. When indicated, biofilms were stained with Live/Dead stain according to the manufacturer's instructions (Life Technologies) prior to fixation. DRAQ-5 was included in the embedment in these experiments. In other experiments, at 48 h the medium was carefully replaced with 4% paraformaldehyde in PBS to fix the bacteria in situ. Fixed cells were labeled with monoclonal antibody (MAb) 6E4 (anti-KDO) and a fluorescein isothiocyanate-conjugated secondary antibody (GAM-IgG/FITC; Jackson Immunoresearch, West Grove, PA), taking care to avoid disruption of the biofilm on the coverslips during the procedures. The use of a mounting medium, Fluoro-Gel III with propidium iodide (Electron Microscopy Sciences), allowed labeling of the eDNA matrix in the specimens. All samples were viewed by confocal laser-scanning microscopy using a Nikon Digital Eclipse C1 laser scanning confocal microscope (Nikon Instruments, Inc.) at a magnification of ×20.
Chinchilla model of NTHI biofilm formation.
NTHI strains including NTHI 2019, NTHI 2019Δnuc and NTHI 2019Δnuc::nuc were studied for their ability to form biofilms in the middle ears of adult chinchillas (Chinchilla lanigera; mean weight, 400 to 600 g; Rauscher's Chinchilla Ranch, LaRue, OH). The animals were acclimated to the vivarium for a period of 7 to 10 days. Both middle ears were inoculated with 300 μl of sterile pyrogen-free saline containing 1,500 to 2,000 CFU of NTHI via transbullar inoculation as previously described (21, 22), with the actual inoculum received confirmed by plate count. Middle ears were then monitored daily for signs of otitis media via video otoscopy and tympanometry. Mature biofilms were formed by NTHI in the chinchilla middle ear 5 days after challenge (21, 22) and in the present study chinchillas were also sacrificed at this time point. The bullae were dissected away from the skull, opened to visualize the inferior bulla, and any fluid present was aseptically collected. Images were taken of all bullae. The mucosa, along with any biofilm present, were collected from the right bulla and placed in a preweighed tube. These were homogenized, serially diluted, and plated to determine CFU of NTHI strain/mg (wet weight) of tissue. The left bulla from each chinchilla was filled with optimal cutting temperature (OCT) compound and snap-frozen for histological analysis. Recovered effusions were serially diluted and plated for semiquantitative determination of CFU of NTHI/ml of middle ear fluid. All studies involving chinchillas were performed under an Institutional Animal Care and Use Committee-approved protocol in compliance with all relevant federal guidelines and institutional policies.
Embedding in OCT for cryosectioning.
To preserve the architecture of biofilms that had formed in vitro and in vivo, we embedded the biofilm sample in an OCT compound (Fisher Scientific, Pittsburgh, PA). The chinchilla biofilms were processed immediately after dissection for later cryosectioning. Briefly, the inferior and superior portions of iced, dissected bullae were separated, and any effusion present was retrieved by aspiration. The inferior bulla was then rinsed several times and drained via wicking onto absorbent paper. OCT was slowly added via 18-gauge needle. Bullae were then snap-frozen over liquid nitrogen and placed on a bed of dry ice. External bone was carefully chipped away, thereby eliminating the need for decalcification, as well as achieving our goal of leaving the middle ear mucosa and any attached biofilm intact. The resulting block was split in a plane perpendicular to the tympanic membrane. Serial sections (4-μm thickness) were cut on a Leica CM3050S cryotome (Leica Microsystems, Inc., Bannockburn, IL). Sections were placed on Superfrost slides (Fisher Scientific) and stored at −80°C. Prior to staining, sections were fixed in 4% (wt/vol) paraformaldehyde (in 0.1 M phosphate buffer [pH 7.4]). The in vitro-grown biofilms were embedded in OCT, cryosectioned, fixed in situ, and stained with MAb 6E4 and secondary antibody goat anti-mouse IgG conjugated to FITC (Jackson Immunoresearch). The DNA matrix in the specimens was stained with DAPI (4′,6′-diamidino-2-phenylindole).
COMSTAT analysis of confocal z-series.
Quantitative analysis of each z-series was performed using COMSTAT (23). COMSTAT is a mathematical script written for MATLAB 5.3 (The Mathworks, Inc., Natick, MA) that quantifies three-dimensional biofilm structures by evaluating confocal image stacks, so that pixels may be converted into relevant measurements of biofilm, including total biomass and average thickness. To complete COMSTAT analysis, an information file was created for each z-series to adjust for the pixel sizes of the x, y and z axes and number of images in each z-series. COMSTAT was then used to threshold the images to reduce background. Biomass and average and maximum thickness in each z-series were calculated by COMSTAT from the threshold images.
Quantitative real-time PCR.
SYBR green quantitative real-time PCR (qRT-PCR) was used to measure the expression of nuc in the 24-hour NTHI 2019 (24). For each target, qRT-PCR primer sets were selected using Primer Express software (Agilent Technologies) and obtained from Integrated DNA Technologies (Coralville, IA). RNA samples were converted into cDNA as follows: 2 μl of random hexamer primers (Invitrogen, Carlsbad, CA) was added to 2 μg of total RNA in a total volume of 12 μl, followed by incubation at room temperature for 10 min to allow primers to anneal, and then transitioned to 70°C to relax RNA secondary structure and finally cooled on ice for 2 min. Then, 2 μl of 0.1 M DTT, 4 μl of SuperScript II 10× reaction buffer, 1 μl of 10 mM deoxynucleoside triphosphates, and 1 μl of SuperScript II were added to each tube buffer (all reagents were obtained from Invitrogen), followed by incubation at 42°C for 4 h. At this point, RNA was degraded with 3.5 μl of 0.5 M EDTA at 65°C for 15 min, and reactions were neutralized with 5 μl of 1 M Tris and 21.5 μl of Tris-EDTA buffer (all reagents were obtained from Ambion/Applied Biosystems). cDNAs were purified using the Qiagen PCR cleanup kit, quantitated, and then diluted to 10 ng/μl and used as the templates for qRT-PCR. Relative RNA quantities were determined by standard curve (5-fold dilutions of purified genomic DNA [gDNA] ranging from 100 to 0.00032 ng/μl), and all values were normalized to the amount of outer membrane protein 6 (ompP6) RNA in each sample. OmpP6 is considered constitutively expressed in H. influenzae. Our data on more than 60 NTHI mRNA samples confirmed this fact. We performed 50-μl reactions in triplicate in 1× SYBR green master mix (Ambion/Applied Biosystems) with 3.5 mM magnesium chloride, 10 ng of template or gDNA standard, and 1 μM final concentrations of each primer. RT-PCR was performed on the ABI Prism 7000 sequence detection system (Quantum Analytics). Each RT-PCR assay was performed four times on three matched biofilm and planktonic samples, and genes were considered validated if results were consistent for all assays and the absolute fold change values were equivalent to or greater than the corresponding array fold change. The data were normalized against NTHI P6 (25) and standardized against NTHI 2019 wild-type genomic DNA.
Statistical analysis.
The statistical analyses for the qRT-PCR experiments were performed using the linear-mixed-modeling (LMM) framework. Estimation was performed using restricted maximum-likelihood (REML) estimation and the denominator degrees of freedom for tests of model coefficients were obtained using the containment method as implemented in SAS v9.3. For the qRT-PCR experiments, the outcome variable was the ratio of expression of HI1296 (nuc) and HI0501 (p6). A random intercept was included to adjust for the multiple measurements taken from each experiment. A model that included an intercept and an indicator for whether the observation came from the biofilm or planktonic phase growth was fit to the data. All significance tests based on LMM analyses were conducted at the α = 0.05 level and were performed using SAS v9.3. The CFU studies were analyzed using Prism 6.0 with analysis of variance using nonparametric tests (the Kruskal-Wallis test).
RESULTS
NTHI Nuc has homology to the S. aureus thermonuclease.
We provide here evidence that NTHI expresses a nuclease, which plays a role in biofilm remodeling and organism dispersal and appears to be under the control of quorum sensing. NTHI Nuc contains 154 amino acids, and the first 13 are a signal (SpII) peptidase sequence (Fig. 1). The nuclease has homology to the staphylococcal thermonuclease with conserved amino acids (35%) across 70% of the sequence and an E value of 4e–10. There are 24 NTHI genomes in the public databases which have high homology (E values of less than 10−38) to the NTHI 2019nuc amino acid sequence.
FIG 1.
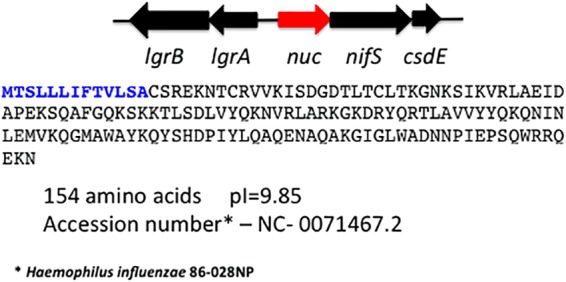
Genomic arrangement and sequence of the NTHI nuc (red) in the H. influenzae 2019 genome. The boldface blue letters indicate the 13-amino-acid signal sequence. The homologous sequence in the H. influenzae RD KW20 genome is HI1296.
NTHI Nuc has nuclease activity.
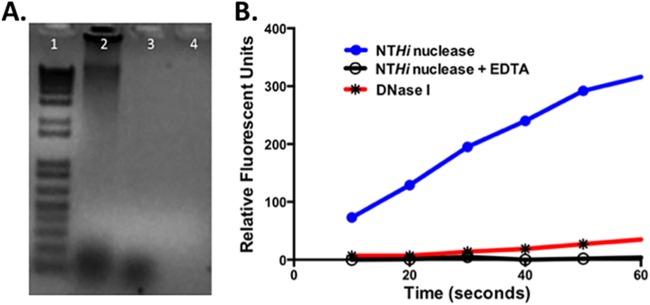
NTHI Nuc was expressed in E. coli without the signal sequence, purified by affinity and molecular sieve chromatography, and studied to determine whether it would digest DNA. Figure 2A shows the results of agarose gel studies demonstrating that NTHI Nuc can digest double-stranded DNA. Studies using FRET-labeled nucleotides demonstrated that NTHI Nuc is a nuclease that is dependent upon a divalent cation, calcium, for activity (Fig. 2B) and can also cleave single-stranded DNA. NTHI Nuc shows markedly increased activity compared to bovine DNase 1 at 180 times the concentration of NTHI Nuc.
FIG 2.
(A) Agarose gel demonstrating the enzymatic activity of Nuc against double-stranded genomic DNA after incubation. Lane 1, molecular weight markers; lane 2, herring sperm DNA alone; lane 3, DNase I (1 U) and herring sperm DNA; lane 4, Nuc (1 μg) and herring sperm DNA. (B) Results of digestion of 25 mM FRET-labeled nucleotide at 25°C with 0.035 nM nuclease (●) or 6.00 nM DNase 1 (✳). The differences in the velocity of the enzymes can be readily seen. The graph also demonstrates the loss of nuclease activity in the presence of 4 mM EDTA (○) due to chelation of the divalent cations Mg2+ and Ca2+.
Expression of nuc in biofilms and the planktonic phase of growth.
We measured the relative nuc expression and compared this to the constitutively expressed H. influenzae gene, ompP6. As shown in Table 2, the expression of nuc was significantly greater in the planktonic state than the biofilm state in NTHI 2019 (P < 0.042). In other studies we have performed, we have demonstrated that the NTHI Nuc has 1,200 to 1,400 times greater activity than DNase 1; thus, small changes in gene expression can substantially augment enzymatic activity (Fig. 2B and data not shown). Some idea of these differences in the velocity of NTHI Nuc and bovine DNase 1 can be seen in Fig. 2B. These qRT-PCR experiments suggest that the expression of nuc is regulated during biofilm formation and may play a role in the control of biofilm dispersal.
TABLE 2.
Results of qRT-PCR studies of nuc expression in biofilms and the planktonic phase of growtha
| State | n | Mean | SD | Range |
Fold difference | P | |
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | ||||||
| Planktonic phase | 12 | 407.2 | 116.1 | 252.5 | 648.17 | 1.52 | 0.042 |
| Biofilms | 12 | 267.3 | 93.3 | 141.0 | 430.5 | ||
Assays were derived from four separate biofilm/planktonic studies, with three assays on each sample. Mean, standard deviation, and range values are expressed as pg of Nuc/pg of P6.
Biofilm dispersal experiments.
In order to study the effect of Nuc on NTHI biofilm dispersal, we adapted the method of Kaplan to study NTHI 2019, NTHI 2019Δnuc, and NTHI 2019Δnuc::nuc. The results are shown in Fig. 3. In this assay, microturbulence in the plates causes a comet-like tail to form as organisms disperse from the nascent biofilm or microcolony (14). This can be seen by the comet tail of satellite colonies formed by NTHI 2019 grown for 24 h in this system (Fig. 3A). No evidence of this was found in the plates containing NTHI 2019Δnuc (Fig. 3B). The phenotype has been partially restored in NTHI 2019Δnuc::nuc. The nuc expression in NTHI 2019Δnuc::nuc is unregulated as the complementing gene is driven from the spectinomycin promoter. This results in smaller biofilms (microcolonies), but the comet-like plumes can still be seen in NTHI 2019Δnuc::nuc (Fig. 3C). These data strongly suggest that Nuc is involved in biofilm dispersal in NTHI.
FIG 3.
Evidence that the nuclease is the factor responsible for dispersal of NTHI biofilms. (A) “Comet tails” caused by the release of organisms from NTHI 2019 microcolonies/nascent biofilms as organisms transition from the biofilm to planktonic phase over a 24-h period. (B) Study performed on 2019Δnuc, which had no evidence of microcolony formation or dispersal of organisms. (C) Partial complementation in cis of 2019Δnuc because the expression of nuc is unregulated in the complemented strain. Small microcolonies were seen with the comet tail configurations similar to those seen in panel A.
Deletion of nuc affects biofilm formation.
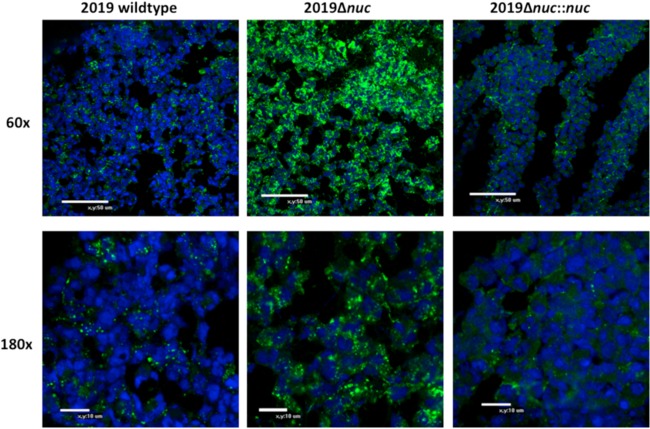
In order to determine the role of the nuclease in the NTHI biofilm, NTHI 2019, NTHI 2019Δnuc, and NTHI 2019Δnuc::nuc were studied in a continuous flow biofilm chamber (Fig. 4). Confocal microscopic analysis of the continuous flow biofilms indicated the organisms in the Δnuc biofilm were aggregated compared to the diffusely distributed organisms in the biofilm formed by the wild-type or Δnuc::nuc strains. The intensity of propidium iodine staining was also increased in the NTHI 2019Δnuc mutant, suggesting an increase in the amount eDNA in the biofilm matrix. The biofilm formed by NTHI 2019Δnuc::nuc restored the wild-type biofilm phenotypes. Frozen sections of a separate set of wild-type, mutant, and complemented mutant biofilms showed a similar clustering of organisms in the mutant compared to the wild type and the complemented mutant (Fig. 5). COMSTAT analysis indicated that the height and mass of the biofilms were not statistically different between the biofilms formed by the three strains. Our studies demonstrated that the majority of organisms in the NTHI 2019Δnuc biofilm were dead by 48 h (Fig. 6). All biofilm studies were performed in at least three separate experiments with similar results.
FIG 4.
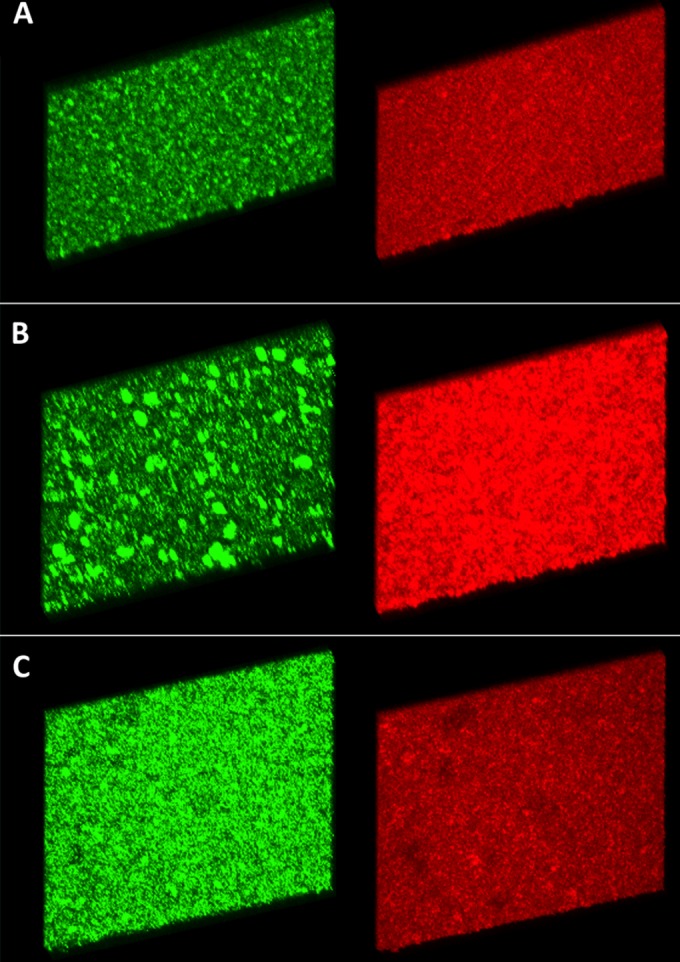
A 50-image stacked z-series at ×20 magnification of 24-h biofilms grown in continuous flow chambers. The samples were stained with propidium iodide (red) and MAb 6E4 (green) prior to visualization. At 24 h, the NTHI 2019Δnuc biofilm contains increased amounts of eDNA and large aggregates of organisms (B). This compares to lesser amounts of eDNA and diffuse arrangement of organisms within the biofilm of NTHI 2019 (A) and NTHI 2019Δnuc::nuc (C).
FIG 5.
Cryo sections through 24-h biofilms from NTHI 2019 wild type (A), NTHI 2019Δnuc (B), and NTHI 2019Δnuc::nuc (C). The DNA matrix is stained with DAPI, and the NTHI strains are stained with MAb 6E4. Scale bar, 20 μm. The organisms in the parent strain and in the complemented mutant are clearly dispersed throughout the biofilm, whereas they are clustered in the Δnuc mutant.
FIG 6.
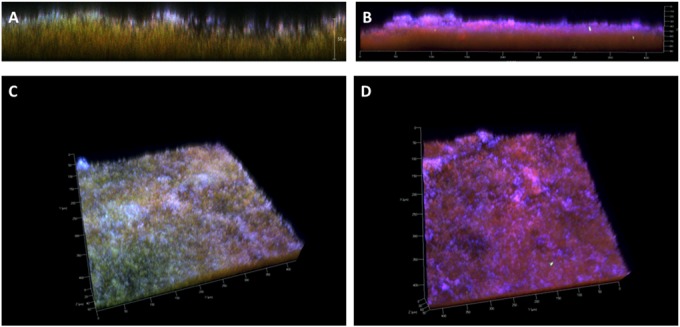
Lateral views and stacked z-series of 48-h biofilms stained with Live/Dead stain (green-red) and DRAQ-5 (blue). (A and C) Biofilm images of NTHI 2019 demonstrating a predominance of live organisms (green), while the images of NTHI 2019Δnuc biofilm (B and D) demonstrate that the majority of organisms were dead (red) by 48 h.
Chinchilla middle ear infection studies.
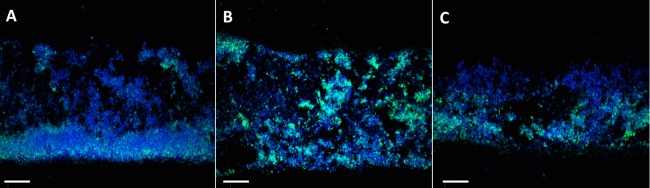
To study the impact of the nuc mutation during in vivo infection, NTHI 2019, NTHI 2019Δnuc, and NTHI 2019Δnuc::nuc were studied in a chinchilla model of experimental otitis media over a 5-day period (21). Three animals were studied for each strain. Both ears were infected and, at sacrifice, one bulla was used for microscopy and the second bulla was used for microbiological studies. Gross examination of the middle ears at sacrifice indicated that each of the strains appeared to induce biofilms of similar size based on the wet weight of the biofilms. Moreover, semiquantitative cultures from nasopharyngeal lavage fluids and middle ear fluids showed no significant differences in relative CFU of NTHI/ml; however, despite variability among tissue samples recovered from these outbred animals, there was a notable trend to increased NTHI/mg of middle ear mucosa (including associated mucosal biofilm) in animals challenged with NTHI 2019Δnuc at sacrifice (see Fig. S1 in the supplemental material). Confocal analysis of 10-μm-thick cryosections of the biofilm stained with DRAQ5 and MAb 6E4 showed evidence of inflammatory cell infiltration, eDNA, and organisms (Fig. 7) in biofilms from each of the animals. However, in the cohort of animals challenged with NTHI 2019Δnuc, there was increased density of NTHI with aggregation of organisms in the NTHI 2019Δnuc biofilm compared to the biofilms induced by either the wild-type strain or its the complemented mutant (Fig. 7). These data are similar to the aggregation seen in the in vitro studies conducted with NTHI 2019Δnuc and suggest that the dispersal of organisms from the NTHI 2019Δnuc mutant was also diminished in vivo.
FIG 7.
Confocal microscopy analysis of 10-μm-thick sections of biofilm formation at day 5 in the chinchilla middle ear after infection with NTHI 2019, NTHI 2019Δnuc, and NTHI 2019Δnuc::nuc. The NTHI are stained with MAb 6E4 (green), and DNA is stained with DRAQ5 (blue). There is an aggregation of organisms staining with MAb 6E4 in NTHI 2019Δnuc (B) compared to the more diffuse display of organisms in the wild type and the complemented mutant (A and C).
DISCUSSION
A number of bacterial species responsible for causing human disease, including Neisseria gonorrhoeae, Campylobacter jejuni, Enterococcus faecalis, and Bacillus anthracis, encode a protein homologous to the S. aureus thermonuclease. The nuclease is present among many NTHI strains since 24 NTHI genomes contain a gene with high homology (<e10−38) to NTHI 2019nuc. The functional properties and structure of the S. aureus thermonuclease have been studied for over 50 years (26). Recently, studies have shown that S. aureus and N. gonorrhoeae encode a thermonuclease, which plays a role in remodeling the eDNA matrix of their biofilms (16, 27). We provide evidence that the NTHI Nuc is a nuclease capable of digesting single- and double-stranded nucleic acids. It is an extremely potent thermonuclease and has ∼1,400-fold greater activity than DNase I on a molar basis (unpublished data). NTHI makes a biofilm whose matrix is also comprised primarily of eDNA (11), and in the present study, we show that the nuclease produced by NTHI is involved in the remodeling of the biofilm eDNA matrix structure, as well as organism dispersal, and its absence results in aggregation of organism within biofilms in vitro and in vivo.
At the functional level, like the S. aureus and N. gonorrhoeae thermonucleases, the NTHI nuclease plays a role in remodeling the eDNA matrix of the biofilm (27, 28). The biofilm formed by the NTHI 2019Δnuc mutant shows aggregation of organisms within the biofilm and an increase in eDNA. Live/Dead studies of in vitro grown biofilms in flow chambers showed decreased viability of organisms within the 2019Δnuc biofilm (Fig. 6). This is most probably due to loss of remodeling of the matrix, failure to disperse organisms, and aberrant water channel formation, leading to nutrient limitation. Studies of the NTHI 2019, 2019Δnuc, and 2019Δnuc::nuc biofilms in vitro and in the chinchilla model of otitis media showed that loss of NTHI Nuc activity resulted in a substantial increase in the aggregation of organisms within the biofilm compared to the wild-type strain and the complemented mutant strain. Quantitative cultures of fluid from the middle ear chamber and mucosa/biofilm of the mutant strain showed wide variability but overall similar numbers of viable organisms compared to the wild-type and complemented mutant strains (see Fig. S1 in the supplemental material). The differences were not significant. Juneau et al. showed that NTHI induced polymorphonuclear neutrophil (PMN) NETs in murine bone marrow-derived macrophages (29). These NETs had no impact on NTHI viability; thus, it is doubtful that the increased bacterial population seen in the NTHI2019 Δnuc biofilm in the chinchilla was due to loss of the effects of the NTHI nuclease on PMN nets. We would speculate that NTHI in biofilm in the middle ear environment are trapped within the biofilm, and it is possible that in this in vivo environment adequate nutrients are available to allow them to survive. Based on these results, we would surmise that the absence of the nuclease results in an increase in eDNA matrix and retention of organisms within restricted areas of the biofilm, supporting the hypothesis that the nuclease is involved in biofilm matrix remodeling and organism release, as has been described for S. aureus thermonuclease (28). This is supported by our dispersion studies, which showed no evidence of sustained organism dispersion in the NTHI 2019Δnuc mutant.
Nuc expression studies comparing 24-h biofilm mRNA with planktonic mRNA suggested that the NTHI nuc expression is significantly reduced in the organisms in the biofilm phase compared to those in the planktonic phase of growth. The NTHI nuclease is extremely potent and, while the differences in expression are not great (1.5-fold), they are significant between NTHI 2019 planktonic bacteria and biofilm (Table 2) samples. These studies implicate a potential role for an autoinducer in control of expression of nuclease activity in the biofilm but further studies are necessary. Due to potentially deleterious effects of the nuclease on the NTHI DNA and RNA, it is reasonable that there would be tight control of the NTHI nuc expression, and it would appear that Nuc is important to ensure the organism's survival and dispersal in the biofilm environment.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIAID grant AI024616 (M.A.A.) and by NIDCD grant DC003915 (L.O.B.). A Howard Hughes Medical Student Research Fellowship and The University of Iowa Carver College of Medicine Physician Scientist Training Pathway supported C.C.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02601-14.
REFERENCES
- 1.Murphy TF, Apicella MA. 1987. Nontypeable Haemophilus influenzae: a review of clinical aspects, surface antigens and the human response to infection. Rev Infect Dis 9:1–15. doi: 10.1093/clinids/9.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Chin CL, Manzel LJ, Lehman EE, Humlicek AL, Shi L, Starner TD, Denning GM, Murphy TF, Sethi S, Look DC. 2005. Haemophilus influenzae from patients with chronic obstructive pulmonary disease exacerbation induce more inflammation than colonizers. Am J Respir Crit Care Med 172:85–91. doi: 10.1164/rccm.200412-1687OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Post JC. 2001. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 111:2083–2094. doi: 10.1097/00005537-200112000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ, Daigle BJ, Ehrlich MD, Post JC. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287:1710–1715. doi: 10.1001/jama.287.13.1710. [DOI] [PubMed] [Google Scholar]
- 5.Starner TD, Zhang N, Kim G, Apicella MA, McCray PB Jr. 2006. Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med 174:213–220. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swords WE, Moore ML, Godzicki L, Bukofzer G, Mitten MJ, VonCannon J. 2004. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect Immun 72:106–113. doi: 10.1128/IAI.72.1.106-113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greiner LL, Watanabe H, Phillips NJ, Shao J, Morgan A, Zaleski A, Gibson BW, Apicella MA. 2004. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect Immun 72:4249–4260. doi: 10.1128/IAI.72.7.4249-4260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. 2005. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun 73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 10.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 11.Jurcisek JA, Bakaletz LO. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol 189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolodkin-Gal I, Cao S, Chai L, Bottcher T, Kolter R, Clardy J, Losick R. 2012. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell 149:684–692. doi: 10.1016/j.cell.2012.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan JB, Fine DH. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol 68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steichen CT, Cho C, Shao JQ, Apicella MA. 2011. The Neisseria gonorrhoeae biofilm matrix contains DNA, and an endogenous nuclease controls its incorporation. Infect Immun 79:1504–1511. doi: 10.1128/IAI.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. 2013. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun 81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Kockritz-Blickwede M. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun 2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, Gibson BW, Apicella MA, Munson RS Jr. 2007. Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol Microbiol 66:26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee SP, Porter D, Chirikjian JG, Knutson JR, Han MK. 1994. A fluorometric assay for DNA cleavage reactions characterized with BamHI restriction endonuclease. Anal Biochem 220:377–383. doi: 10.1006/abio.1994.1353. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz K, Stephenson R, Hernandez M, Jambang N, Boles BR. 2010. The use of drip flow and rotating disk reactors for Staphylococcus aureus biofilm analysis. J Vis Exp pii:2470. doi: 10.3791/2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakaletz LO. 2009. Chinchilla as a robust, reproducible and polymicrobial model of otitis media and its prevention. Expert Rev Vaccines 8:1063–1082. doi: 10.1586/erv.09.63. [DOI] [PubMed] [Google Scholar]
- 22.Bakaletz LO, Tallan BM, Hoepf T, DeMaria TF, Birck HG, Lim DJ. 1988. Frequency of fimbriation of nontypeable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect Immun 56:331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10):2395–2407. [DOI] [PubMed] [Google Scholar]
- 24.Falsetta ML, Bair TB, Ku SC, Vanden Hoven RN, Steichen CT, McEwan AG, Jennings MP, Apicella MA. 2009. Transcriptional profiling identifies the metabolic phenotype of gonococcal biofilms. Infect Immun 77:3522–3532. doi: 10.1128/IAI.00036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson MB, Apicella MA, Murphy TF, Vankeulen H, Spotila LD, Rekosh D. 1988. Cloning and sequencing of Haemophilus influenzae outer membrane protein P6. Infect Immun 56:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham L. 1958. Micrococcal nuclease and some products of its action. Ann N Y Acad Sci 3:27–39. [DOI] [PubMed] [Google Scholar]
- 27.Steichen CT, Cho C, Hunt J, Shao J, Apicella MA. 2011. The Neisseria gonorrhoeae biofilm matrix contains DNA and an endogenous nuclease controls its incorporation. Infect Immun 79:1504–1511. doi: 10.1128/IAI.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juneau RA, Pang B, Weimer KE, Armbruster CE, Swords WE. 2011. Nontypeable Haemophilus influenzae initiates formation of neutrophil extracellular traps. Infect Immun 79:431–438. doi: 10.1128/IAI.00660-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.