Abstract
Ribonucleoside 2′,3′-cyclic phosphates (N>p’s) are generated by multiple prebiotically plausible processes and are credible building blocks for the assembly of early RNA oligomers. While N>p’s can be polymerized into short RNAs by non-enzymatic processes with variable efficiency and regioselectivity, no enzymatic route for RNA synthesis had been described. Here we report such a non-canonical 3′-5′ nucleotidyl transferase activity. We engineered a variant of the hairpin ribozyme to catalyze addition of all four N>p’s (2′,3′-cyclic A-, G-, U-, and CMP) to the 5′-hydroxyl termini of RNA strands with 5′ nucleotide addition enhanced in all cases by eutectic ice phase formation at −7 °C. We also observed 5′ addition of 2′,3′-cyclic phosphate-activated β-nicotinamide adenine dinucleotide (NAD>p) and ACA>p RNA trinucleotide, and multiple additions of GUCCA>p RNA pentamers. Our results establish a new mode of RNA 3′-5′ extension with implications for RNA oligomer synthesis from prebiotic nucleotide pools.
The conjecture of an “RNA world”, a primordial biology preceding our own in which RNA was the central biomolecule for both catalysis and information storage, is supported by growing evidence.1 A recent key advance has been the establishment of a prebiotically plausible synthesis for the pyrimidine nucleotides.2 Ribonucleoside 2′,3′-cyclic phosphates (N>p’s) are not only the main products of said synthesis but also of prebiotic nucleoside phosphorylation and iterative degradation of RNA through transesterification.3 Furthermore, continuous N>p regeneration from the 2′- and 3′-monophosphates (resulting from, e.g., N>p hydrolysis and/or RNA degradation) is possible by activation with simple prebiotically plausible electrophiles.3a,4 The sustainable and varied prebiotic supply routes thus point to N>p’s as plausible building blocks for early RNA oligomer synthesis (Chart S1). However, the 2′,3′-cyclic phosphate is a poor activating group, and polymerization of N>p’s under aqueous conditions is thermodynamically disfavored (Keq ≈ 1–4 M–1).5 Therefore, RNA oligomer formation requires high concentrations of N>p’s such as might have arisen by evaporation or eutectic freezing.6 Here, we sought to explore if small catalytic RNAs (ribozymes) could have promoted incorporation of >p-activated substrates into RNA under favorable conditions. We describe the engineering and characterization of variants of the small hairpin ribozyme (HPz) to catalyze RNA 5′ addition of >p-activated mono- and oligonucleotides.
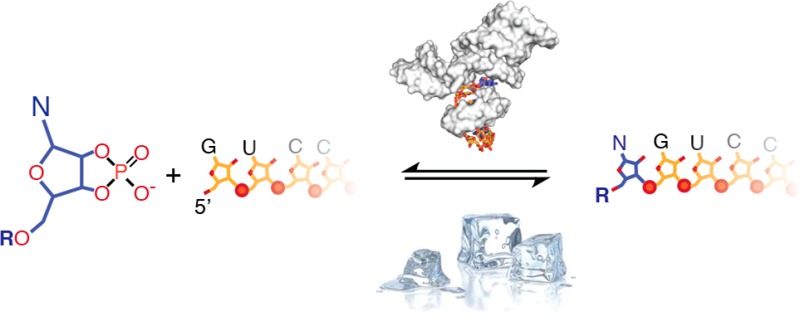
A minimal hairpin ribozyme–substrate complex comprises a two-way junction that forms a docking complex through RNA tertiary interactions (Figure 1a).7 Reversible cleavage of substrate RNA results from a general acid–base mechanism that catalyzes nucleophilic attack of the 2′-oxygen atom from position A-1 upon the scissile phosphorus between A-1 and G+1, leaving G+1 with a 5′-OH and A-1 with a 2′,3′-cyclic phosphate (Figure 1a).8 In the active complex, A-1 is positioned by a single hydrogen bond from A9 and a cross-strand base stack to G8.9 This binding mode is compatible with all four bases, and the HPz tolerates any base at position -1 during substrate cleavage.10 With the goal to create a rudimentary but general binding pocket for N>p’s, we connected the 5′ fragment of the pre-ligation substrate strand lacking A-1 with the unpaired A residue at the 3′-end of helix H3 by a short C5 linker (Figure 1b). We reasoned that the resulting ribozyme construct (5NTz) would now present a major groove cavity able to bind N>p’s in an orientation suitable for 5′-nucleotidyl transfer to the acceptor strand (AS, Figure 1c).
Figure 1.
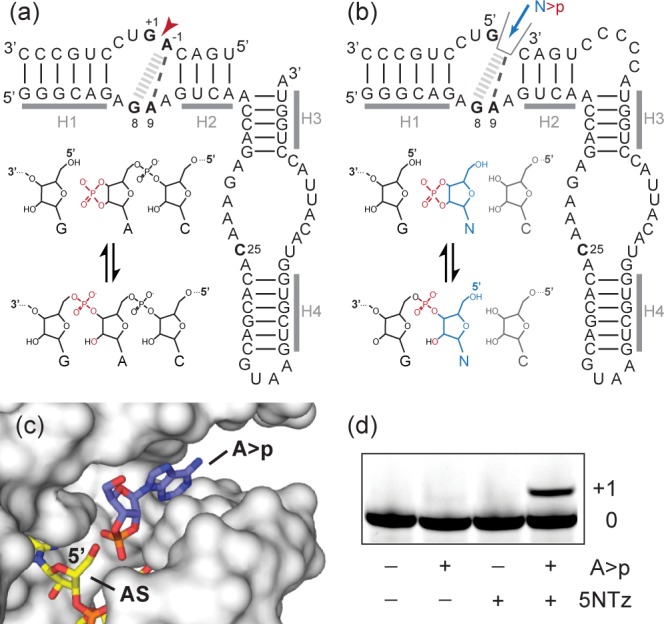
Design of a 5′-nucleotidyl transferase for N>p’s. (a) Two-way junction HPz, which catalyzes reversible RNA ligation using a 2′,3′-cyclic phosphate. (b) Redesign of HPz into 5NTz. (c) Structural model of the substrate-binding pocket of 5NTz (based on PDB 1M5V(9)). The ribozyme is shown as surface; A>p and the acceptor strand (AS) are shown in stick models. (d) 5NTz catalyzes 5′-adenylation in ice (2 mM A>p, 2 μM 5NTz, 1 μM 3′-FITC-labeled AS, 72 h in ice at −7 °C).
However, 5′-nucleotidyl transferase activity was only weak (although detectable) at ambient temperatures (Figure S1). We speculated that the entropically disfavored 5′ addition might be enhanced by the high substrate concentrations and low temperatures effected by eutectic ice-phase formation.11 Indeed, we found that 5′ addition of 2′,3′-cyclic adenosine monophosphate (A>p) proceeded with enhanced efficiency in ice (−7 °C) (Figure 1d), yielding >10% single-nucleotide addition. 5′ extension was 10-fold lower in unfrozen, supercooled samples and 20-fold lower at 17 °C (Figure S1), demonstrating that eutectic conditions promote N>p addition in spite of the unfavorable entropy term. No (or very weak) strand extension was observed with 5NTz variants lacking the N>p binding pocket or catalytic features of the parental HPz (Figure S2).
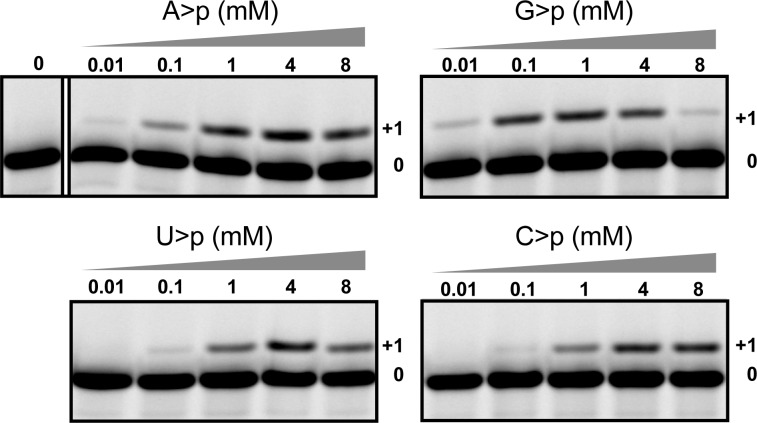
Next we investigated whether 5NTz could catalyze nucleotide transfer of the three other canonical N>p’s. Indeed, we observed ribozyme-dependent addition of G>p, U>p, and C>p (Figure 2). Strikingly, 5′ extension was already visible at an apparent N>p concentration as low as 10 μM for purine and 100 μM for pyrimidine nucleotides. The preference of 5NTz for purines may be due to their stronger stacking interactions with G8 (Figure 1b). A further increase in 5′-nucleotidyl transfer efficiency at high millimolar nucleotide concentrations was impeded by substrate inhibition, which was observed for all N>p’s except C>p. Inhibition was most severe for G>p (Figure 2), possibly due to competition of exogenous G>p with the internal G-1 base for pairing to C25 (Figure 1a), an interaction required for loop–loop docking during active-site assembly.7
Figure 2.
Concentration-dependent 5′ transfer of A>p, G>p, C>p, or U>p (−7 °C in ice, 72 h, 10 mM MgCl2).
In-ice 5′-adenlyation was nearly independent of bivalent metal ions, and we observed nucleotidyl transfer even in the absence of magnesium ions (Mg2+) (Figure S3a). As metal ions are also potent catalysts of RNA degradation,3d this would boost RNA synthesis efficiency by this route. Eutectic 5′ nucleotidyl transferase activity was also largely temperature independent (Figure S3b) above −28 °C, the eutectic freezing point, suggesting that the interstitial liquid brine is a prerequisite for ribozyme catalysis.11a,12
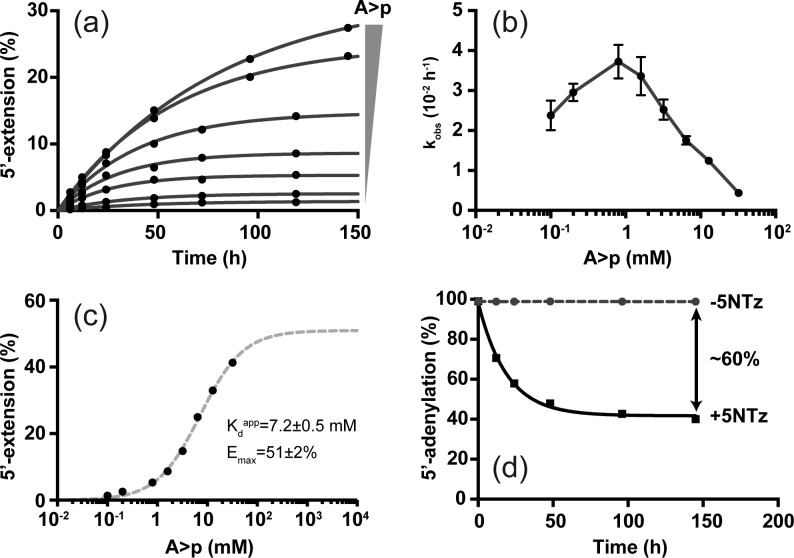
To further characterize 5NTz catalysis, we investigated the kinetics of 5′-adenylation under pseudo-first-order conditions using excess nucleotide (Figure 3). Time courses of 5′-adenylation were monophasic (Figure 3a and S4) but showed a biphasic concentration dependency (Figure 3b): The observed rate kobs increased from 2.4 × 10–2 h–1 (T1/2 = 29 h) at 0.1 mM A>p to 3.7 × 10–2 h–1 (T1/2 = 18.6 h) at 0.8 mM A>p but decreased at higher A>p concentrations. These rates are comparable to template-dependent 5′-3′ elongation rates of non-enzymatic (∼1 × 10–2 h–1 for adenosine 5′-monophosphoimidazole at −18.4 °C)13 and enzymatic (∼0.1 h–1 for 5′-triphosphates)14 primer extension reactions under eutectic conditions. The rate inhibition observed at high nucleotide concentrations complicates a detailed analysis of the reaction mechanism. However, from the fitted extension amplitudes, we estimate an apparent (aqueous) Kdapp = 7.2 mM of 5NTz for A>p at ∼25% extension and a maximal extension level (Emax) of ∼51% (Figure 3c). It is illustrative to compare these numbers with the average equilibrium constant for N>p addition by ribonucleases (Keq = 2.2 M–1 at 0 °C),5 which predicts that ∼150 mM N>p would be required for 25% 5′ extension under aqueous conditions. Similarly, the Kdapp of 5NTz for A>p increases to ∼170 mM at 0 °C (Figure S5). Altogether, these data imply that improved 5′ extension in ice is predominantly a result of the concentrating effect of eutectic phase formation rather than low temperature or product stabilization.
Figure 3.
Kinetic characterization of eutectic A>p transfer by 5NTz. (a) Kinetics of 5′ extension at increasing A>p concentrations. Solid circles are experimental data, while solid lines represent monoexponential fits. (b) Replot of observed rate constants with their approximate standard errors. (c) Replot of amplitudes (black circles). The dashed gray line represents the best fit of a one-site binding model. (d) Quasi-irreversible cleavage of 5′-adenylated substrate strand by 5NTz indicates that only ∼60% of the ribozyme population is active.
We hypothesized that Emax of 5NTz may be limited by the presence of an inactive ribozyme fraction (as with the parental hairpin ribozyme).12,15 Indeed, only ∼60% of a fully adenylated 5′-RNA was deadenylated in presence of 5NTz under quasi-irreversible conditions (ice, 0 μM A>p) (Figure 3d). Additionally, the end points of 5′ extension diverged by ∼35% when the reaction in presence of 3 mM A>p was started from either the substrate or product side (Figure S6), suggesting that 35–40% of ribozyme/AS complexes reside in an unproductive conformation.
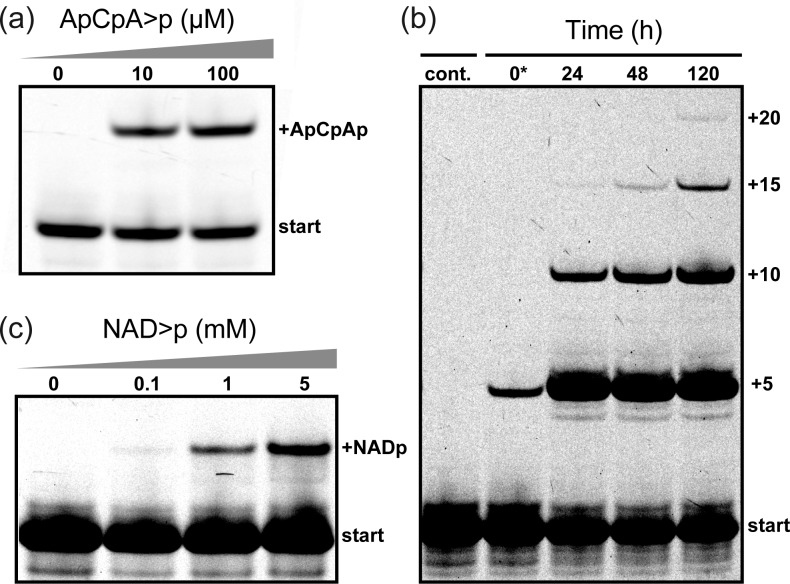
We anticipated that additional interactions such as Watson–Crick base-pairing would allow 5′ additions with enhanced efficiencies and at lower substrate concentrations compared to N>p’s. We found that a truncated variant of 5NTz that lacks C-2 and A-3 (5NTzΔ2,Figure S7a) was able to catalyze 5′ transfer of an ACA>p trinucleotide with near-maximal yields even at low micromolar concentrations (Figure 4a). This trinucleotide can form two base pairs (in place of C-2 and A-3) as part of helix H2, resulting in an increase in affinity by ∼3 orders of magnitude compared to mononucleotides. Thus, trinucleotides (as well as presumably di-, tetra-, pentanucleotides, etc.) resulting from polymerization or degradation of longer RNA oligomers are potential substrates for 5′ addition by 5NTz. Indeed, we were able to redesign HPz (Figure S7b,c) for iterative additions of GUCCA>p pentamers (Figure 4b).
Figure 4.
Eutectic 5′ addition of other substrates activated by 2′,3′-cyclic phosphates. (a) Transfer of the trinucleotide ApCpA>p (72 h, −7 °C). (b) Time series of multiple GpUpCpCpA>p (25 μM) 5′-additions by a HPz derivative. Small amounts of transfer did already occur during sample preparation (*). The left lane shows a primer-only control. (c) Transfer of NAD>p at different dinucleotide concentrations (96 h, −7 °C).
This prompted us to further explore 5′ addition of other prebiotically plausible building blocks. 5NTz was able to charge the 5′-end of RNA with β-nicotinamide adenine dinucleotide 2′,3′-cyclic phosphate (NAD>p) (Figure 4c). This ubiquitous cofactor of modern enzymes is widely considered a relic of early metabolism16 and has been shown to expand the catalytic repertoire of ribozymes.17 Additionally, the β-nicotinamide mononucleotide moiety has been shown to be an effective leaving group for ribozyme-catalyzed RNA ligation.18 Thus, simple 5′ transferase ribozymes as described here might not only extend but also activate RNA oligomers for further ligation and extension cycles.
Our results have implications for a better understanding of the emergence of the RNA world from prebiotic chemistry. N>p’s are generated (and regenerated) by a multitude of prebiotically plausible pathways and therefore deserve consideration as substrates for early RNA oligomer synthesis.3a,4 The ease with which a simple ribozyme could be repurposed as a 3′-5′ mono- and oligonucleotidyl transferase suggests that even in absence of more highly activated nucleotide substrates, multiple routes for the extension and elaboration of RNA oligomers were available.
The hairpin ribozyme is one of the simplest RNA catalysts and likely would have been among the first ribozymes to emerge from pools of short random RNA oligomers.11c,19 It has been estimated that one micromole of random RNA 50-mers may contain >2 × 107 active hairpin ribozymes.20 Even this is a likely underestimate since such calculation does not take into account the powerful concentrating effect of eutectic phase formation, which not only enhances nucleotide addition catalysis but also enables assembly of active hairpin ribozymes from RNA oligomer fragments.12 Furthermore, many other small catalytic RNAs such as the hammerhead, hepatitis delta virus, and twister ribozyme families realize an analogous transesterification reaction in unrelated sequence motifs,21 which may likewise be amenable to reconfiguration for 5′ addition. Random cleavage and ligation catalyzed by such ribozymes has been proposed to allow bootstrapping of more complex activities20,22 and, indeed, HPz self-replication from prefabricated oligonucleotides and self-processing into new topologies has been demonstrated.23 Our success in engineering HPz for multiple 5′ addition suggests a route by which larger RNAs might have been assembled from the short oligonucleotides provided by non-enzymatic processes.
In conclusion, we describe a strategy for 5′ addition of ribonucleotide substrates to RNA oligomers. Such 3′-5′ nucleotidyl transfer reactions are unusual in modern biology with tRNAHis guanylyl transferases the only known example.24 However, this mode of 3′-5′ addition from prebiotically plausible building blocks, together with others, such as chemoselective acetylation,25 may have aided expansion of primitive RNA oligomer pools in both length and complexity. Early RNA oligomers may thus have been able to grow “from both ends” both by canonical 3′ extension (by addition of 5′-activated monomers) as well as by non-canonical 5′ extension utilizing both non-enzymatic and enzymatic routes.
Acknowledgments
This work was supported by a FEBS long-term fellowship (H.M.) and by the Medical Research Council (program no. U105178804).
Supporting Information Available
Experimental procedures and supplemental Chart S1, Figures S1–S7, and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Atkins J. F.; Cech T.; Gesteland R. F.. RNA worlds: from life’s origins to diversity in gene regulation; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2011. [Google Scholar]
- Powner M. W.; Gerland B.; Sutherland J. D. Nature 2009, 459, 239. [DOI] [PubMed] [Google Scholar]
- a Lohrmann R.; Orgel L. E. Science 1968, 161, 64. [DOI] [PubMed] [Google Scholar]; b Lohrmann R.; Orgel L. E. Science 1971, 171, 490. [DOI] [PubMed] [Google Scholar]; c Schoffstall A. M.; Barto R. J.; Ramos D. L. Orig. Life 1982, 12, 143. [DOI] [PubMed] [Google Scholar]; d Li Y.; Breaker R. R. J. Am. Chem. Soc. 1999, 121, 5364. [Google Scholar]
- Crowe M. A.; Sutherland J. D. Chembiochem 2006, 7, 951. [DOI] [PubMed] [Google Scholar]
- Mohr S. C.; Thach R. E. J. Biol. Chem. 1969, 244, 6566. [PubMed] [Google Scholar]
- a Renz M.; Lohrmann R.; Orgel L. E. Biochim. Biophys. Acta 1971, 240, 463. [DOI] [PubMed] [Google Scholar]; b Verlander M. S.; Orgel L. E. J. Mol. Evol. 1974, 3, 115. [DOI] [PubMed] [Google Scholar]
- Muller S.; Appel B.; Krellenberg T.; Petkovic S. IUBMB Life 2012, 64, 36. [DOI] [PubMed] [Google Scholar]
- Kath-Schorr S.; Wilson T. J.; Li N. S.; Lu J.; Piccirilli J. A.; Lilley D. M. J. Am. Chem. Soc. 2012, 134, 16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert P. B.; Massey A. P.; Sigurdsson S. T.; Ferre-D’Amare A. R. Science 2002, 298, 1421. [DOI] [PubMed] [Google Scholar]
- Chowrira B. M.; Berzal-Herranz A.; Burke J. M. Nature 1991, 354, 320. [DOI] [PubMed] [Google Scholar]
- a Attwater J.; Wochner A.; Pinheiro V. B.; Coulson A.; Holliger P. Nat. Commun. 2010, 1, 76. [DOI] [PubMed] [Google Scholar]; b Menor-Salvan C.; Marin-Yaseli M. R. Chem. Soc. Rev. 2012, 41, 5404. [DOI] [PubMed] [Google Scholar]; c Monnard P. A.; Kanavarioti A.; Deamer D. W. J. Am. Chem. Soc. 2003, 125, 13734. [DOI] [PubMed] [Google Scholar]
- Vlassov A. V.; Johnston B. H.; Landweber L. F.; Kazakov S. A. Nucleic Acids Res. 2004, 32, 2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard P. A.; Szostak J. W. J. Inorg. Biochem. 2008, 102, 1104. [DOI] [PubMed] [Google Scholar]
- Attwater J.; Wochner A.; Holliger P. Nat. Chem. 2013, 5, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban J. A.; Walter N. G.; Kotzorek G.; Heckman J. E.; Burke J. M. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Li N.; Ellington A. D. Chem. Biodivers. 2007, 4, 633. [DOI] [PubMed] [Google Scholar]
- Tsukiji S.; Pattnaik S. B.; Suga H. J. Am. Chem. Soc. 2004, 126, 5044. [DOI] [PubMed] [Google Scholar]
- Fujita Y.; Furuta H.; Ikawa Y. RNA 2009, 15, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris J. P.; Hill A. R. Jr.; Liu R.; Orgel L. E. Nature 1996, 381, 59. [DOI] [PubMed] [Google Scholar]
- Briones C.; Stich M.; Manrubia S. C. RNA 2009, 15, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ferre-D’Amare A. R.; Scott W. G. Cold Spring Harbor Perspect. Biol. 2010, 2, a003574. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Roth A.; Weinberg Z.; Chen A. G. Y.; Kim P. B.; Ames T. D.; Breaker R. R. Nat. Chem. Biol. 2014, 10, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrubia S. C.; Briones C. RNA 2007, 13, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gwiazda S.; Salomon K.; Appel B.; Muller S. Biochimie 2012, 94, 1457. [DOI] [PubMed] [Google Scholar]; b Petkovic S.; Muller S. FEBS Lett. 2013, 587, 2435. [DOI] [PubMed] [Google Scholar]
- Abad M. G.; Rao B. S.; Jackman J. E. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler F. R.; Chan C. K.; Duffy C. D.; Gerland B.; Islam S.; Powner M. W.; Sutherland J. D.; Xu J. Nat. Chem. 2013, 5, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.