Abstract
High-throughput screening has enabled the identification of small molecule modulators of important drug targets via well-established colorimetric or fluorimetric activity assays. However, existing methods to identify small molecule binders of nonenzymatic protein targets lack either the simplicity (e.g., require labeling one of the binding partners with a reporter) or throughput inherent in enzymatic assays widely used for HTS. Thus, there is intense interest in the development of high-throughput technologies for label-free detection of protein–small molecule interactions. Here we describe a novel self-referencing external cavity laser (ECL) biosensor approach that achieves high resolution and high sensitivity, while eliminating thermal noise with subpicometer wavelength accuracy. Using the self-referencing ECL biosensor, we demonstrate detection of binding between small molecules and a variety of immobilized protein targets, pairs that have binding affinities or inhibition constants ranging from subnanomolar to low micromolar. Finally, a “needle-in-the-haystack” screen for inhibitors against carbonic anhydrase isozyme II is performed, in which known inhibitors are clearly differentiated from inactive molecules within a compound library.
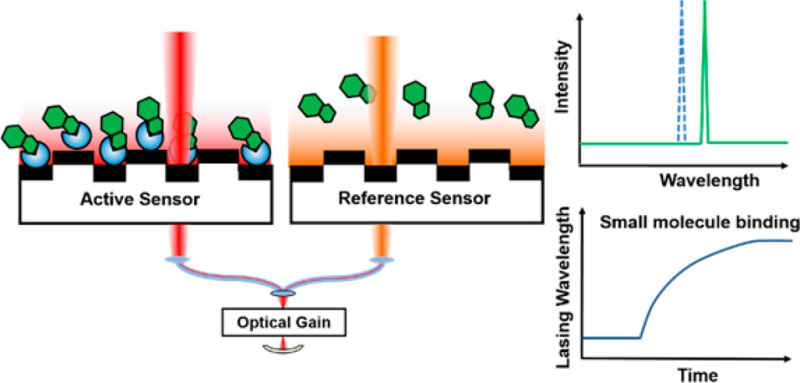
High-throughput screening (HTS) is a valuable tool for the identification of small molecule modulators of various macromolecular targets and is a critical early component of the pharmaceutical discovery process.1 Despite the utility of HTS in drug discovery, enzymatic protein targets with colorimetric or fluorimetric activity assay readouts are typically more amenable to this approach than nonenzymatic protein targets. Thus, there is an intense need for general binding assays that can be utilized to identify small molecule binders for proteins, not merely enzymatic inhibitors.2 Current assays utilized for direct detection of protein–small molecule binding include isothermal titration calorimetry (ITC),3 surface plasmon resonance (SPR),4,5 and small-molecule microarrays (SMMs).6 ITC and SPR are capable of quantitative detection of protein–small molecule interaction in low- or medium-throughput applications, while SMMs can identify small molecule probes that bind with specific protein targets using a fluorescence-based readout. As an alternative binding assay for high-throughput identification of small molecule binders, an external cavity laser (ECL) biosensor was recently reported.7,8 Utilizing a narrowband photonic crystal (PC) resonant reflector9 as optical feedback in a laser cavity, ECL achieves high spectral resolution through the stimulated emission. The ECL detects the adsorption of biomolecules to the PC biosensors with improved resolution when compared to simply measuring the passive resonant reflection spectrum10 and allows the measurement of small wavelength shifts.
Challenges confronting the detection with the ECL biosensor, as well as many types of optical biosensors, are the detection of noise signals that are indistinguishable from the signals generated by an actual binding event. For example, thermal fluctuations of the test sample, thermal expansion/contraction of sensor materials, and nonspecific binding can all lead to changes in the measured resonant wavelength that are of similar magnitude to wavelength changes generated by small molecule–protein interactions. Realizing that incorporation of accurate referencing is key to enable direct detection of small molecule binding to immobilized protein targets, herein we describe a self-referencing ECL biosensor detection instrument which is capable of lasing at two independent wavelengths. The introduction of two-wavelength lasing enables real-time self-referencing by designating one wavelength as a reference for the “active” biosensor. The active and reference resonators share all optical components and thermal environment and thus share common-mode sources of wavelength shift noise, including thermal drift. Utilizing the reference external cavity mode, the self-referencing ECL eliminates common-mode noise sources to achieve accurate referencing, while simultaneously maintaining high sensitivity and high spectral resolution. As demonstrated herein, using the newly developed instrument, it is possible to detect small molecule binding to immobilized protein targets with micromolar affinity and to perform HTS.
In order to achieve accurate referencing, two PC sensors in adjacent wells of a 384-well plate were utilized as wavelength selective elements in the ECL cavity at the same time. Each sensor selects its own resonant wavelength, so the ECL system can lase at two independent wavelengths simultaneously. A schematic diagram of the instrument is shown in Figure 1a, with the dual-mode lasing spectrum shown in Figure S1. Self-referencing was accomplished by designating one well as the “reference” well and the other as the “active” well, where both sensors were fabricated identically on the same substrate and were prepared identically with exception of the immobilized protein in the active well (Figure 1b). Using rigorous coupled-wave analysis computer simulations of the PC sensor with/without the presence of a 10 nm thick protein layer, we found that the sensitivity for the active and reference sensors is identical. No temperature or environmental control is applied to the detection instrument, sensor, or test samples. Due to the close physical proximity of the active and reference sensors, accurate referencing is achieved to compensate for thermal variations and nonspecific binding. Importantly, the active and reference laser cavities share the entire optical system, including the gain medium, optical fibers, and mechanical holding stages, thus any common-mode error that may cause the lasing wavelength to drift will occur to both devices in an identical fashion. Moreover, the side-by-side configuration of the active and reference sensors enables the use of a pulse-driven bistable shutter for alternate operation of the two lasing modes with a frequency of 0.5 Hz. This avoids competition between two simultaneously oscillating modes and enables stable operation of the ECL system. Kinetic monitoring of the lasing wavelength value (LWV) is achieved by directing a portion of the lasing emission energy with a beam splitter to a detection instrument such as a spectrometer or interferometer-based laser wavelength meter.
Figure 1.
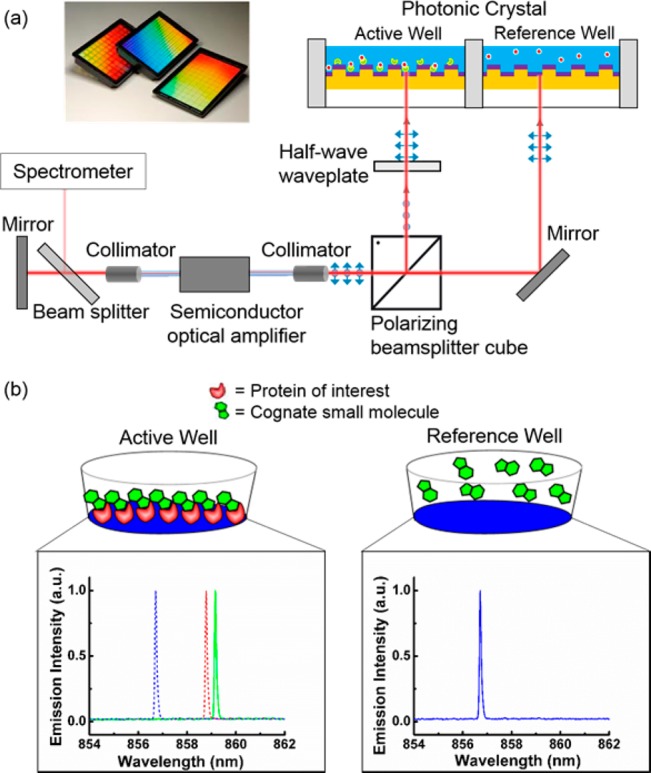
(a) Schematic diagram of the self-referencing ECL biosensor system. Insert: Images of PC biosensor films adhered to the bottom of standard microplates. (b) Biosensor assays used for detection of direct small molecule binding to immobilized protein targets with referencing. Active and reference sensors functionalized with glutaraldehyde are used. The immobilization of protein and the addition of cognate small molecule in the active well both result in shifts in LWV of the active sensor.
To validate the self-referencing technique, tests to demonstrate accurate compensation of environmental fluctuations were performed. By kinetically monitoring the LWVs of the alternating lasing modes, a self-referenced LWV shift was obtained by subtracting the LWV of the reference sensor from the active sensor, resulting in an effectively reduced noise level with a short-term standard deviation of σ = 0.8 pm over a 20 min time period (Figure S2). Moreover, the LWV variations of the active sensor have a fluctuation range of 15 pm (Figure S2), demonstrating the importance of the reference sensor. As a proof-of-concept, quantitative detection of biotin binding to an immobilized layer of streptavidin (SA) was performed. Upon establishment of a stable baseline, biotin was added to both the active and reference wells, and their kinetic LWVs were obtained simultaneously (Figure S3). By subtracting the signal of the reference sensor from the active sensor, a referenced LWV shift of 13 pm due to the binding of biotin to SA was observed (Figure S3). The LWV shift as a result of a binding interaction was considerably smaller than the noise introduced by thermal shift and would otherwise be undetectable without the incorporation of the referencing control.
Next, the ECL biosensor instrument was used to study five well-characterized protein–small molecule binding interactions: CA II–dorzolamide (KD = 1.1 nM),11 NQO1–dicoumarol (KI = 1–10 nM),12 XIAP–SM-164 (KI = 0.56 nM),13 caspase-3–Q-VD-OPh (IC50 < 25 nM)14 and hLDH-A–N-hydroxyindole-1 (NHI-1) (KI = 10.8 μM).15 These protein–small molecule interactions have binding affinities or inhibition constants ranging from sub-nanomolar to low-micromolar, which are typical of most protein–small molecule interactions. The interaction between hLDH-A and NHI-1 (Figure 2a) can be readily detected (Figure 2b) despite the pair having the weakest inhibition constant among the tested pairs. As shown in Figure 2, the system can also detect the binding interaction of the other four protein–small molecule pairs: XIAP and SM-164 (Figure 2c), CA II and dorzolamide (Figure 2d), NQO1 and dicoumarol (Figure 2e), caspase-3 and Q-VD-OPh (Figure 2f). In addition, we were also able to detect the binding of SM-122 to immobilized XIAP (Ki = 180 nM)13 (Figure S4a), and ES936 to immobilized NQO1 (Ki = 450 nM)16 (Figure S4b) even though they have much weaker affinities to XIAP and NQO1 than SM-164 and dicoumarol. Since the binding interaction is measured in a microplate format, the binding rate between the small molecule and protein target is mainly limited by mass transport, prohibiting the determination of kinetic parameters associated with the binding event. Nonbinding signals of the other four noncognate small molecules can be readily distinguished from the positive binding signal of the cognate protein–small molecule pairs (Table 1), indicating that the observed LWV shift is not due to nonspecific aggregation to the sensor surface. In addition, the small LWV shift observed due to the noninteracting protein–small molecule pairs (Table 1) is within 3σ of the noise generated when only buffer solution was added to both the active and reference wells (Figure S2), demonstrating that the presence of the reference well aids in compensating the noise due to nonspecific binding. Lastly, using structurally similar but inactive analogues of NHI-1 (1b and 1g, Figure 2a),17 we were able to demonstrate the specific binding of NHI-1 to immobilized hLDH-A (Figure 2b).
Figure 2.
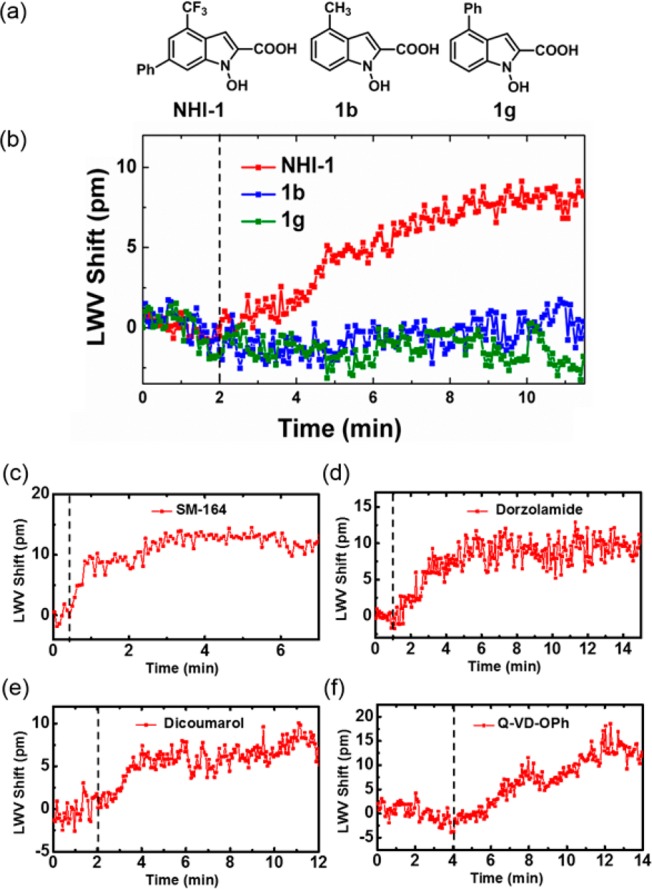
(a) Structure of NHI-1 and its inactive variants 1b and 1g. Observed LWV shift from the binding of 50 μM (b) NHI-1, 1b, or 1g to immobilized hLDH-A, (c) SM-164 to immobilized GST-XIAP, (d) dorzolamide to immobilized CA II, (e) dicoumarol to immobilized NQO1, and (f) Q-VD-OPh to immobilized caspase-3. The vertical dotted line indicates the addition of small molecules to both the active and reference wells.
Table 1. LWV Shift Values of Protein–Small Molecule Binding Interaction Detected on the ECL Biosensora.
| CA IIb | NQO1c | XIAPd | hLDH-Ae | Casp-3f | |
|---|---|---|---|---|---|
| dorzolamide | 12.5 ± (1.1) | –2.4 | 1.5 | 2.6 | 0.3 |
| dicoumarol | 1.2 | 11.2 ± (1.0) | –1.1 | –1.5 | –0.5 |
| SM-164 | –0.9 | 0.7 | 11.5 ± (0.5) | –0.2 | 1.4 |
| NHI-1 | 1.8 | 2.0 | 1.8 | 14.0 ± (1.0) | 1.2 |
| QVD-OPh | –1.5 | 1.1 | –0.2 | 0.4 | 19.3 ± (1.9) |
LWV shift values are reported in pm. Values in bold represent the mean of at least 3 independent measurements for the five cognate protein–small molecule pairs with positive binding signal. Values in parentheses indicate standard error of mean. Binding tests were performed using 50 μM of small molecule solution with 40 μg of immobilized protein at RT.
CA II, carbonic anhydrase isozyme II.
NQO1, human NAD(P)H dehydrogenase quinone 1.
XIAP, X-linked inhibitor of apoptosis protein.
hLDH-A, human lactate dehydrogenase-A.
Casp-3, caspase-3.
To confirm that the ECL biosensor instrument is sufficiently robust for screening protein–small molecule interactions, detection of warfarin binding to immobilized human serum albumin (HSA) (KD = 1.2 μM)18 was tested. When the active and reference sensors were exposed to a cocktail of 5 nonbinding compounds (Figure 3a), no binding signal was observed in the absence of warfarin, while the addition of warfarin in the cocktail resulted in a LWV shift of 14 pm (Figure 3b) due to its binding to immobilized HSA. Importantly, it was determined that the observed shift of 14 pm correlates well with the concentration of warfarin available to bind with HSA, based on further dose-response measurements (Figure S5). This set of data demonstrates the specificity of the ECL biosensor for detection of protein–small molecule binding interactions.
Figure 3.
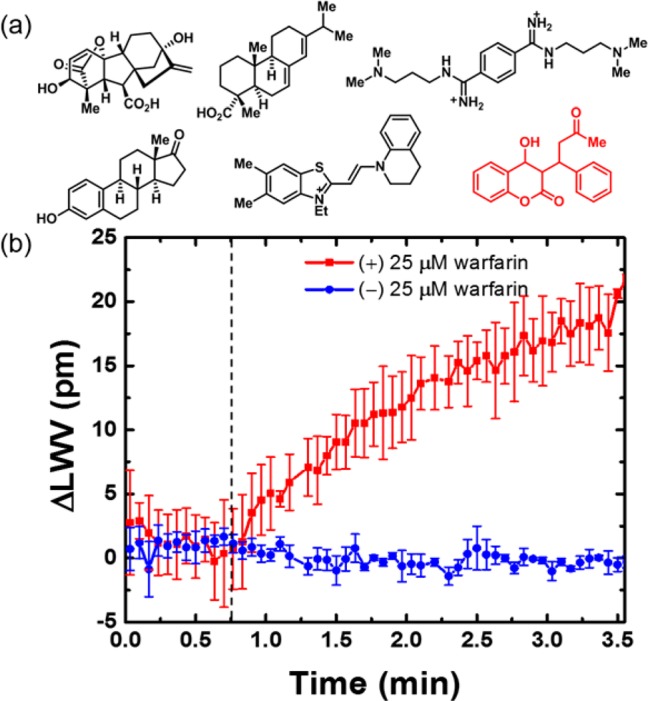
Specific detection of HSA-warfarin interaction. (a) Mixture of small molecules used in the test. Warfarin is highlighted in red. (b) LWV shift due to the binding of warfarin to HSA in the compound mixture. The dotted line indicates the addition of the compound mixture to both the active and reference wells. Values shown are the mean of at least three independent measurements; error bars represent standard error of mean.
Finally, to demonstrate the HTS capability of the ECL biosensor assay, a “needle-in-the-haystack” screen for inhibitors against CA II was performed. The Z′-factor of the assay was determined to be 0.52 (Figure S6), indicating that it is sufficiently robust to discern binders from nonbinders.19 A 47-member compound collection obtained from an in-house screening library, plus dorzolamide, was screened at 50 μM (1 compound per well) with CA II immobilized to the sensor surface. Each of the 48 compounds was added individually into both the active and reference wells and the LWV shift was measured. As shown in Figure 4, dorzolamide is clearly differentiated from the other nonbinding compounds, demonstrating the HTS capability of the assay in identifying protein–small molecule binders. To validate that the “hit” obtained from the screen was real, a range of concentrations of dorzolamide was tested against CA II to obtain dose–response data (Figure S7).
Figure 4.
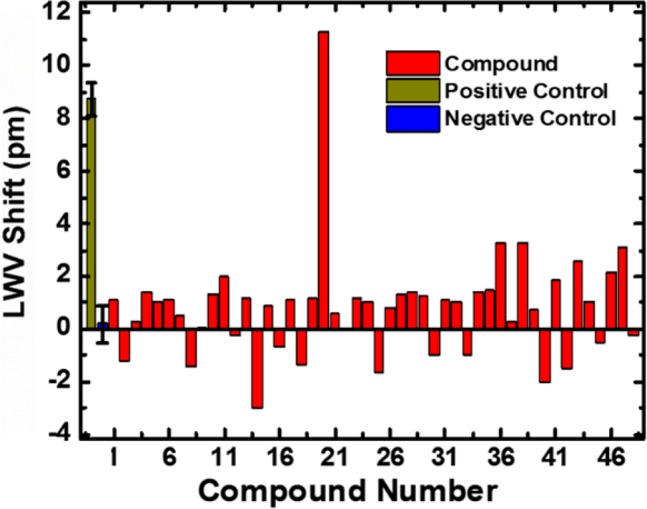
Binding of dorzolamide (compound 20) to immobilized CA II can be detected in the HTS. LWV shift data for all 48 compounds screened at 50 μM with 40 μg of immobilized CA II. 50 μM acetazolamide and 5% DMSO were the positive and negative control, respectively. The values shown for the positive and negative controls are mean of at least three independent measurements; error bars represent standard error of mean.
A self-referencing ECL biosensor system was developed to detect protein–small molecule binding. This optical biosensor system achieves high detection sensitivity and high spectral resolution (Table S1) while being capable of eliminating thermal noise through accurate referencing. The detection instrument and PC biosensor enable the quantitative detection of small molecule binding to immobilized protein targets with various binding affinities. In addition, highly specific detection of protein–small molecule binding was achieved using this robust label-free assay system. Finally, as an initial demonstration of the HTS capability of this technology, a “needle-in-the-haystack” screen with CA II was performed in serial, and dorzolamide was easily identified among the screening compounds. As demonstrated in this work, protein–small molecule binding can be accurately detected in this robust detection system with no temperature control of the reagents and the sensor, in an HTS-compatible microplate format that minimizes reagent consumption, and with “flat” low-density surface chemistry. In light of previous optical biodetection approaches, and SPR biosensing in particular, the ECL-based optical biosensing approach represents substantial technical advance through its ability to use the narrow bandwidth output of the laser to achieve high resolution for detection of small wavelength shifts that occur for small molecule binding interactions with large immobilized proteins (Table S1).
In the future, we plan to incorporate highly optimized surface chemistry and functionalization techniques to increase the binding response of small molecule binders to the immobilized protein targets. To further enrich the capability of the self-referencing ECL beside identification of small molecule binders, active transport (such as a microplate stirring system) can be implemented to mitigate mass transport limitations of the current setup for measurement of kinetic binding constants. Lastly, with this initial demonstration of the technology, it is our intention to improve the throughput of the current setup to facilitate future HTS endeavors through engineering of the instrument and test method. Higher throughput of the setup can be achieved either via an end point reading method or increasing the number of semiconductor optical amplifiers operating in parallel within the instrument (refer to Section 2, Supporting Information). With the capability of performing higher throughput assays, this newly developed system offers a sensing platform that can be useful for the community of chemical biologists and medicinal chemists for the identification and validation of small molecule binders to protein targets in a broad range of biologically significant applications.
Acknowledgments
The authors gratefully acknowledge the National Science Foundation (CBET 1132225) and National Institutes of Health (R21EB009695 and R01GM90220) for providing financial support for this work. We thank Prof. Shaomeng Wang (U. Michigan) for SM-122 and SM-164, Prof. Colin Duckett (U. Michigan) for pGEX-XIAP, Prof. Filippo Minutolo (U. Pisa) for NHI-1, 1b and 1g, Prof. David Boothman (UTSW) for dicoumarol, and Rachel Botham for the purification of caspase-3. We would also like to thank Chun Ge and Meng Lu for the help in setting up the self-referencing ECL system.
Supporting Information Available
Experimental details and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kodadek T. Nat. Chem. Biol. 2010, 6, 162. [DOI] [PubMed] [Google Scholar]
- Makley L. N.; Gestwicki J. E. Chem. Biol. Drug Des. 2013, 81, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt S.; Freire E. Curr. Opin. Struct. Biol. 2001, 11, 560. [DOI] [PubMed] [Google Scholar]
- Karlsson R. J. Mol. Recogn. 2004, 17, 151. [DOI] [PubMed] [Google Scholar]
- Myszka D. G. Anal. Biochem. 2004, 329, 316. [DOI] [PubMed] [Google Scholar]
- Vegas A. J.; Fuller J. H.; Koehler A. N. Chem. Soc. Rev. 2008, 37, 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C.; Lu M.; George S.; Flood T. A.; Wagner C.; Zheng J.; Pokhriyal A.; Eden J. G.; Hergenrother P. J.; Cunningham B. T. Lab Chip 2013, 13, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Ge C.; Lu M.; Zhang Z.; Cunningham B. T. Appl. Phys. Lett. 2013, 102, 213701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. T.; Li P.; Schulz S.; Lin B.; Baird C.; Gerstenmaier J.; Genick C.; Wang F.; Fine E.; Laing L. J. Biomol. Screen. 2004, 9, 481. [DOI] [PubMed] [Google Scholar]
- Heeres J. T.; Kim S.-H.; Leslie B. J.; Lidstone E. A.; Cunningham B. T.; Hergenrother P. J. J. Am. Chem. Soc. 2009, 131, 18202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T.; Hara K.; Hata S. Drug Metab. Dispos. 1994, 22, 377. [PubMed] [Google Scholar]
- Asher G.; Dym O.; Tsvetkov P.; Adler J.; Shaul Y. Biochemistry 2006, 45, 6372. [DOI] [PubMed] [Google Scholar]
- Lu J.; Bai L.; Sun H.; Nikolovska-Coleska Z.; McEachern D.; Qiu S.; Miller R. S.; Yi H.; Shangary S.; Sun Y.; Meagher J. L.; Stuckey J. A.; Wang S. Cancer Res. 2008, 68, 9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta T. M.; Smith A. N.; Gultice A. D.; Reedy M. A.; Brown T. L. Apoptosis 2003, 8, 345. [DOI] [PubMed] [Google Scholar]
- Calvaresi E. C.; Granchi C.; Tuccinardi T.; Di Bussolo V.; Huigens R. W.; Lee H. Y.; Palchaudhuri R.; Macchia M.; Martinelli A.; Minutolo F.; Hergenrother P. J. ChemBioChem. 2013, 14, 2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winski S. L.; Faig M.; Bianchet M. A.; Siegel D.; Swann E.; Fung K.; Duncan M. W.; Moody C. J.; Amzel L. M.; Ross D. Biochemistry 2001, 40, 15135. [DOI] [PubMed] [Google Scholar]
- Granchi C.; Roy S.; Giacomelli C.; Macchia M.; Tuccinardi T.; Martinelli A.; Lanza M.; Betti L.; Giannaccini G.; Lucacchini A.; Funel N.; LeÒn L. G.; Giovannetti E.; Peters G. J.; Palchaudhuri R.; Calvaresi E. C.; Hergenrother P. J.; Minutolo F. J. Med. Chem. 2011, 54, 1599. [DOI] [PubMed] [Google Scholar]
- Day Y. S. N.; Myszka D. G. J. Pharm. Sci. 2003, 92, 333. [DOI] [PubMed] [Google Scholar]
- Zhang J.-H.; Chung T. D. Y.; Oldenburg K. R. J. Biomol. Screen. 1999, 4, 67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


