Abstract
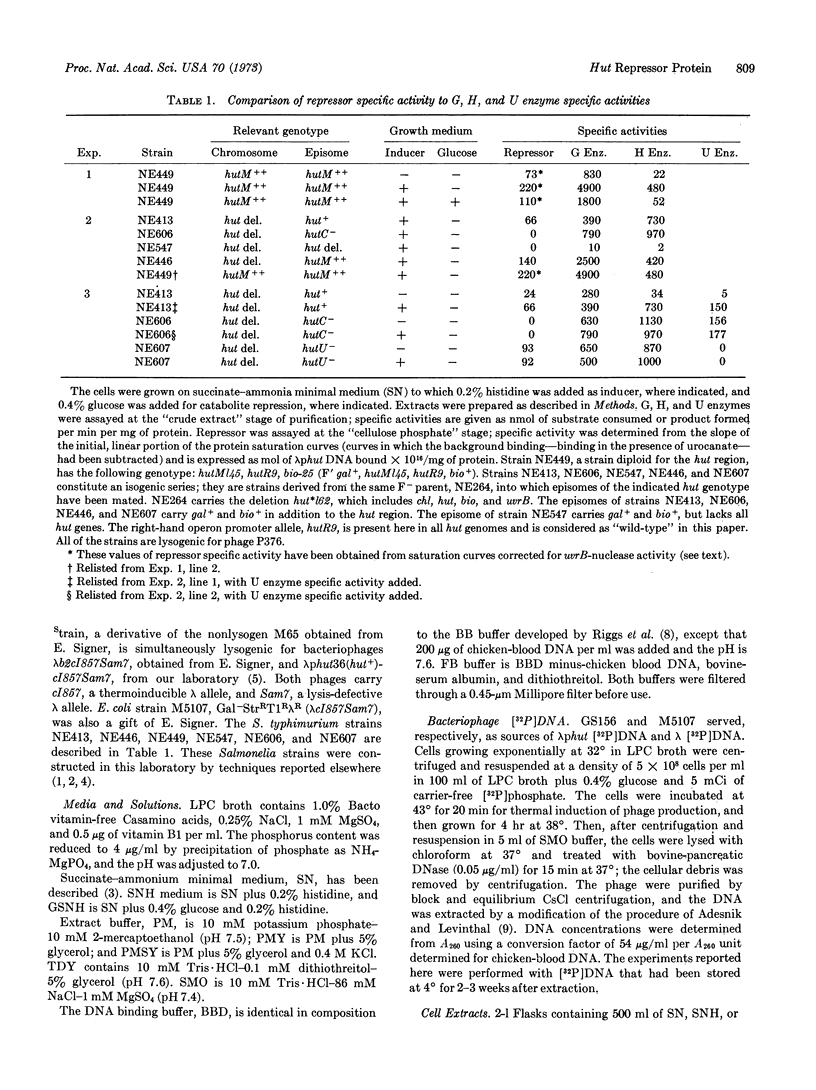
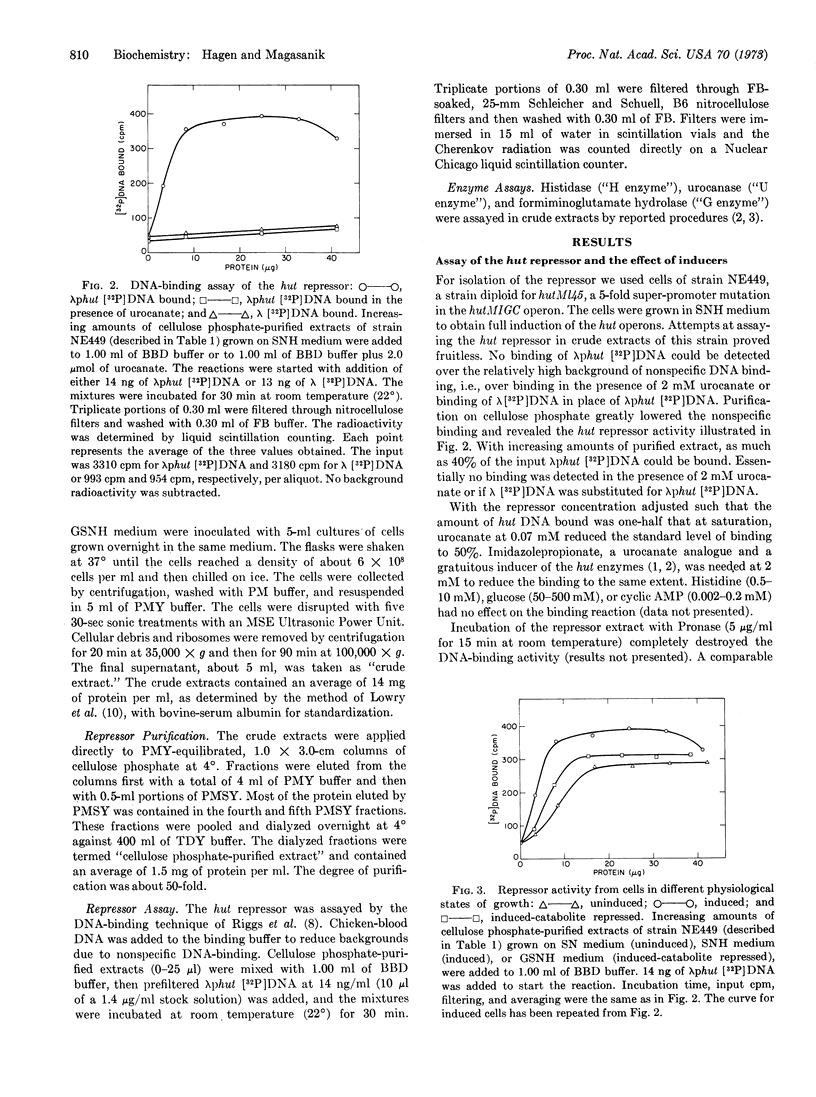
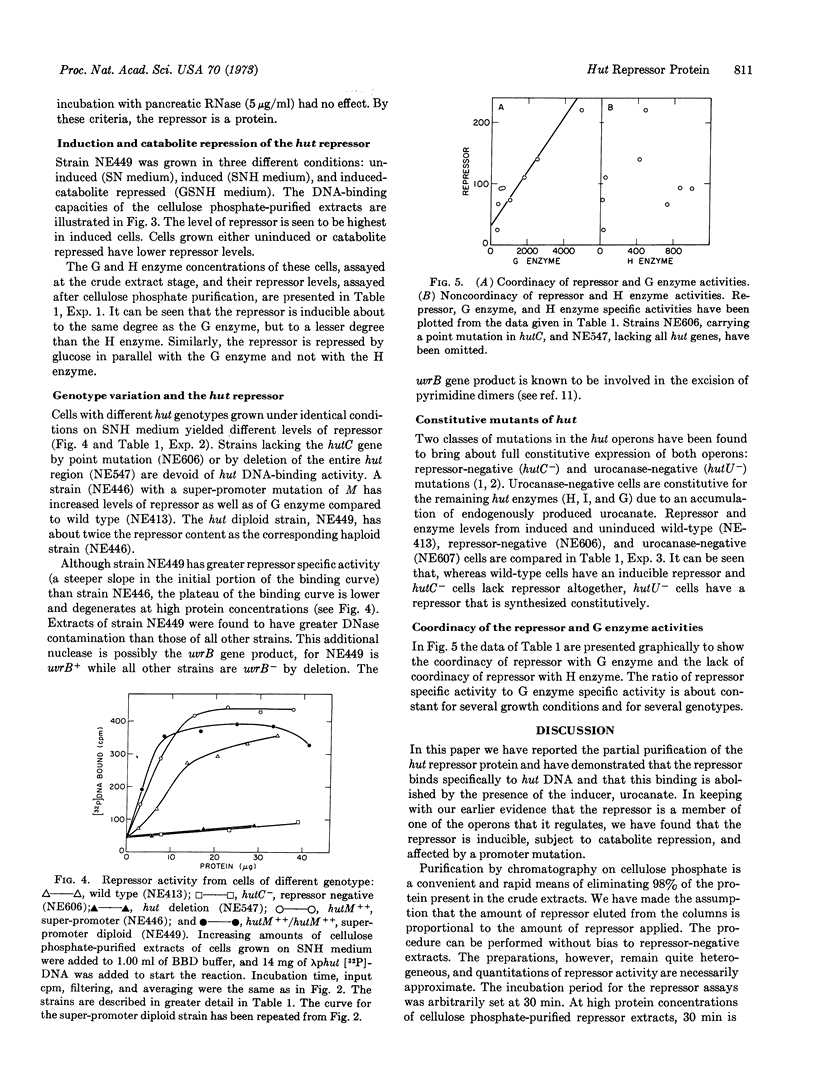
In Salmonella typhimurium the structural genes of the enzymes responsible for histidine utilization (hut) are clustered in two adjacent operons. A single repressor regulates both operons. The repressor itself is a member of one of the hut operons and, thus, regulates its own synthesis. We have assayed the hut repressor by its ability to bind radioactive DNA to nitrocellulose filters. The binding is specific for DNA bearing the hut operons, and the binding is abolished by the inducer, urocanate. As a member of one of the hut operons, the repressor is inducible, subject to catabolite repression, and affected by a promoter mutation.
Keywords: histidine utilization, DNA-binding, urocanate, λphut
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brill W. J., Magasanik B. Genetic and metabolic control of histidase and urocanase in Salmonella typhimurium, strain 15-59. J Biol Chem. 1969 Oct 10;244(19):5392–5402. [PubMed] [Google Scholar]
- Howard-Flanders P. DNA repair. Annu Rev Biochem. 1968;37:175–200. doi: 10.1146/annurev.bi.37.070168.001135. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Meiss H. K., Brill W. J., Magasanik B. Genetic control of histidine degradation in Salmonella typhimurium, strain LT-2. J Biol Chem. 1969 Oct 10;244(19):5382–5391. [PubMed] [Google Scholar]
- Pirrotta V., Chadwick P., Ptashne M. Active form of two coliphage repressors. Nature. 1970 Jul 4;227(5253):41–44. doi: 10.1038/227041a0. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Newby R. F., Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968 Jul 14;34(2):365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Halpern Y. S., Magasanik B. Genetic and metabolic control of enzymes responsible for histidine degradation in Salmonella typhimurium. 4-imidazolone-5-propionate amidohydrolase and N-formimino-L-glutamate formiminohydrolase. J Biol Chem. 1971 May 25;246(10):3320–3329. [PubMed] [Google Scholar]
- Smith G. R., Magasanik B. Nature and self-regulated synthesis of the repressor of the hut operons in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1493–1497. doi: 10.1073/pnas.68.7.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Magasanik B. The two operons of the histidine utilization system in Salmonella typhimurium. J Biol Chem. 1971 May 25;246(10):3330–3341. [PubMed] [Google Scholar]
- Smith G. R. Specialized transduction of the Salmonella hut operons by coliphage lambda: deletion analysis of the hut operons employing lambda-phut. Virology. 1971 Jul;45(1):208–223. doi: 10.1016/0042-6822(71)90128-0. [DOI] [PubMed] [Google Scholar]


