Abstract
A 14-bp insertion/deletion (indel) within the 3' untranslated region (3’UTR) that affects HLA-G expression has been associated with HIV-1 mother-to-child transmission (MTCT). However, other 3’UTR single nucleotide polymorphisms (SNPs) that influence HLA-G mRNA stability have been described but not analyzed in the context of MTCT, and little is known about the role of HLA-G alleles. We examined HLA-G alleles and 3’UTR SNPs, including the 14-bp indel, in 216 mother-infant pairs from Johannesburg, South Africa. Mother-infant pairs were classified as HIV-1 non-transmitting (NT, n=144) or HIV-1 transmitting (TR, n=72) with either intrapartum (IP, n=29) or in utero (IU, n=19) infected infants. We found HLA-G allele, G*01:01:02 (in strong linkage disequilibrium with the 14-bp insertion) and +3187G SNP were significantly over-represented in IU-TR mothers compared to NT mothers (P=0.036, OR=2.26; P=0.011, OR=2.96, respectively). These findings suggest that maternal HLA-G alleles and/or SNPs that might alter expression of HLA-G potentially influence IU HIV-1 MTCT.
Keywords: Mother-to-child transmission (MTCT), HIV-1, HLA-G alleles, HLA-G 3’UTR haplotypes
1. Introduction
Prior to current prevention strategies, mother-to-child transmission (MTCT) or vertical transmission of HIV-1 occurred at an estimated rate of more than 30%, and still is the major cause of HIV/AIDS in children (Taha, 2011, da Silva et al., 2013). MTCT can occur during pregnancy (in utero, IU), at the time of delivery (intrapartum, IP), or postpartum (PP) through breast feeding (Duri et al., 2010, Kourtis et al., 2001). While the use of antiretroviral therapy (ART) during pregnancy has been shown to reduce the risk of MTCT, in the absence of ART, large proportions of infants remain HIV-1 uninfected and appear to have “natural protection”. Thus, studying the mechanisms of natural protection in HIV-1 exposed but uninfected (EU) infants may help to determine correlates of protection in both infants and adults. Several studies have suggested that host genetic factors, such as human leukocyte antigen (HLA) class I and II alleles (Matt and Roger, 2001, Kuhn et al., 2004, MacDonald et al., 1998, Polycarpou et al., 2002) and killer immunoglobulin-like receptors (KIR) (Hong et al., 2013, Paximadis et al., 2011) contribute to MTCT. Recently, there has been increased interest in the role of HLA-G in MTCT, because of its preferential expression at the maternal-foetal interface and its immunosuppressive properties. HLA-G can inhibit differentiation, proliferation, cytolysis, cytokine secretion and immunoglobulin production upon binding to their specific inhibitory receptors: immunoglobulin-like transcript (ILT)-2, ILT-4 and KIR2DL4 expressed by many immune cells, such as B and T lymphocytes as well as natural killer (NK) cells (Amiot et al., 2014).
Unlike the classical class I HLA molecules, the non-classical HLA-G molecule has limited allelic polymorphism in the coding region, with only 50 HLA-G alleles having been described to date (IMGT/HLA, version 3.16.0, 2014/04/14) (Robinson et al., 2013). These alleles encode 16 distinct transmembrane proteins (G*01:01, G*01:02, G*01:03, G*01:04, G*01:06, G*01:07, G*01:08, G*01:09, G*01:10, G*01:11, G*01:12, G*01:14, G*01:15, G*01:16, G*01:17, G*01:18) and two truncated proteins (G*01:05N and G*01:13N) (Robinson et al., 2013). Alternative splicing of the HLA-G primary transcript can generate seven alternative mRNAs that encode membrane-bound (HLA-G1, −G2, −G3, and −G4) and soluble (HLA-G5, −G6, and −G7) protein isoforms (Donadi et al., 2011). HLA-G1 can also be proteolytically cleaved from the membrane and released as soluble HLA-G1 (sHLA-G) allowing it to have systemic immunoregulatory effects in the absence of localized tissue expression (Solier et al., 2002).
In contrast to the coding region, the 3' untranslated region (UTR) of HLA-G (which exhibits several regulatory elements, including AU-rich motifs, a poly-A signal, as well as signals that regulate the spatial and temporal expression of an mRNA) presents a high degree of variation (Lynge Nilsson et al., 2014, Donadi et al., 2011). Amongst these 3’UTR polymorphisms (Figure 1), a 14-bp indel (rs66554220) has been the most studied and has been associated with the magnitude of HLA-G production, with modulation of HLA-G mRNA stability, and with binding for specific microRNAs (Castelli et al., 2014). In general, the presence of a 14-bp insertion (Ins) (5’-ATTTGTTCATGCCT-3’) introduces an alternative splicing site that generates a 92-bp deletion in the 3’UTR that influences mRNA stability; and individuals with Ins/Ins genotype have been associated with lower mRNA production compared to Ins/Del and Del/Del genotypes (Hviid et al., 2003, Castelli et al., 2010, Castelli et al., 2009). Furthermore, other 3’UTR SNPS have also been implicated in the regulation of HLA-G expression. For example, a SNP at position +3142C/G (rs1063320), the presence of a Guanine (G) increases the affinity of this region for microRNAs (miR-148a, miR-148b and miR-152), thereby decreasing HLA-G expression by mRNA degradation and translational suppression (Castelli et al., 2014, Tan et al., 2007, Veit and Chies, 2009). While polymorphisms at positions +3187A/G (rs9380142) and +3196C/G (rs1610696), located near an AU-rich motif in the HLA-G mRNA, have also been associated with mRNA stability and degradation. It was demonstrated that an Adenine (A) at +3187 was associated with decreased HLA-G expression due to the increased number of Adenines in this AU-rich motif (Yie et al., 2008). In addition, polymorphisms within the 3’UTR seem to be arranged in several haplotypes, each of them associated either with a single or a group of coding and/or promoter region polymorphisms (Alvarez et al., 2009, Castelli et al., 2010, Donadi et al., 2011).
Figure 1.
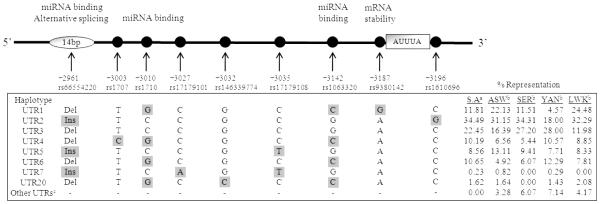
Polymorphisms in the 3’ untranslated region (UTR) of the HLA-G gene and the 3’UTR haplotypes. Polymorphic positions are relative to the first ATG codon at exon 1. UTR haplotypes were numbered according to (Sabbagh et al., 2014).
Del, Deletion; Ins, insertion.
a SA is represented by the total group of HIV-1 positive mothers in our MTCT cohort (n=216)
b Represents 3’UTR frequencies reported by (Sabbagh et al., 2014) which pooled some data from the 1000 genome project. ASW, people of African ancestry from the south western United States (n=61); SER, Serer from Niakhar, Senegal (n=239); YAN, Yansi from Bandundu, Democratic Republic of the Congo (N=175); and LWK, Luhya from Webuye, Kenya (n=96).
c Represents other 3’ UTR haplotypes not identified in SA
In the context MTCT of HIV-1, several reports have associated the 14-bp indel with the risk of MTCT (Segat et al., 2014, Sanches et al., 2013, Segat and Crovella, 2012, Segat et al., 2009, Fabris et al., 2009, Aikhionbare et al., 2006); but only one has associated HLA-G alleles with MTCT (Luo et al., 2013). However, some of these reports have been conflicting, for example: in Zambian infants, the Ins was associated with protection from IU and IP HIV-1 infection (Segat et al., 2014); yet in Brazilian children, the Del allele and Del/Del genotype associated with a protective effect from vertical transmission (Fabris et al., 2009).Whilst, in another study, HIV-1 transmitting mothers were found to have almost four times more placental sHLA-G compared to non-transmitting mothers (Moodley and Bobat, 2011). Similarly, while differing in the mode of HIV-1 transmission, higher levels of sHLA-G in the female genital tract were independently associated with both HIV-1 infection and bacterial vaginosis in sex workers from Benin (Thibodeau et al., 2011). Moreover G*01:03-alleles, which reportedly translate into lower plasma sHLA-G, were associated with a reduced risk of vertical transmission of HIV-1 (Luo et al., 2013). Whereas, in the context of sexual transmission, G*01:04:04, an allele with reportedly higher levels of plasma sHLA-G, was associated with susceptibility to HIV-1 infection (Turk et al., 2013). Of note, amongst all these studies, few have assessed the roles of HLA-G alleles and 3’UTR SNPs collectively.
In this study, we investigated the role of HLA-G in MTCT of HIV-1 in a Black South African cohort of 216 mother-infant pairs, particularly focusing on the coding region representing extracellular α1, α2 and α3 domains, as well as the 3’UTR, since polymorphisms within these regions might influence peptide binding as well as HLA-G expression, respectively. We found maternal possession of the G*01:01:02 allele, as well as the G allele at position +3187 of the 3’UTR (indicative of the UTR1 haplotype), independently increased the risk for IU transmission. Overall, these findings suggest that polymorphisms that alter HLA-G expression are likely to influence MTCT of HIV-1 via the IU route.
2. Materials and Methods
2.1. Study population
This study is a nested case-control analysis of data collected prospectively on 216 Black mother-infant pairs recruited as part of four mother-to-infant HIV-1 transmission studies in Johannesburg, South Africa, that took place from 1996 to 2005. The studies aimed to identify immunogenetic correlates of HIV-1 transmission and have been described previously (Kuhn et al., 2007).
All available HIV-1 transmitting mother-infant samples (TR, n=72) from the four transmission studies were selected as “cases” for the nested analysis. As controls, two HIV-1 non-transmitting mother-infant samples (NT, n=144) for each case were randomly selected from each of the four transmission studies giving rise to the cases. HIV-1 infected infants were further characterized according to timing of transmission as determined by an HIV-1 DNA PCR test (Roche Amplicor version 1.5) at birth and at 6 weeks of age. In total 19 were in utero (IU) infected (PCR positive at birth and at 6 weeks), 29 were intrapartum (IP) infected (PCR negative at birth but positive at 6 weeks), and the remaining 24 infants were found to be positive at 6 weeks but had no birth sample available (unknown whether IU or IP).
For analyses confined to mother-infant pairs who received maternal sdNVP, in addition to analysing associations by known timing of transmission IU or IP as described above, we also analysed associations for an IU-enriched group called IU2. In IU2 we combined the 19 known IU-infected infants with the 24 infected infants of unknown transmission on the basis that 79% (19/24) of these mothers received sdNVP (Table 1). The rationale for this is that maternal sdNVP administration given only at the onset of labour is known to have little to no effect on reducing IU infection. Thus we can infer that when infection does occur in an sdNVP-exposed infant, there is a greater likelihood that it is due to IU infection than due to IP infection. We confirmed this in our data. Among infected infants with a known timing of infection (Table 1), significantly fewer acquired infection during the IP period when born to mothers that received sdNVP compared to ART-naive mothers (10/29, 34% vs. 19/29, 65%, respectively, P=0.035).
Table 1.
Clinical characteristics and antiretroviral (ARV) administration in HIV-1 positive mothers who did/did not transmit (TR/NT) HIV-1 to their infants
|
HIV+ mothers
Median (IQR range) |
NT
(N=144) |
TR
(N=72) |
IP-TR
(N=29) |
IU-TR
(N=19) |
Unknown-TR
(N=24) |
|
| |||||
| Age (years) | 27 (22-30) | 29 (24-31) | 29 (22-31) | 29 (22-31) | 28 (25-30) |
| log10 VL (copies/ml) | 4.0 (3.2-4.6) | 4.8 (3.7-5.4) | 4.8 (3.9-5.3) | 4.9 (4.2-5.5) | 4.6 (2.6-5.4) |
| CD4 (cells/μ) | 449 (319-681) | 375 (253-575) | 378 (262-508) | 350 (182-647) | 437 (301-590) |
|
| |||||
|
ARV administration
N (%) |
NT
(N=144) |
TR
(N=72) |
IP-TR
(N=29) |
IU-TR
(N=19) |
Unknown-TR
(N=24) |
|
| |||||
| None | 60 (41.7%) | 29 (40.3%) | 19 (65.5%) | 7 (36.8%) | 3 (12.5%) |
| sdNVP | 80 (55.5%) | 41 (56.9%) | 10 (34.5%) | 12 (63.2%) | 19 (79.2%) |
| Other ARV | 4 (2.8%) | 2 (2.8%) | 0 (0%) | 0 (0%) | 2 (0%) |
IQR: interquartile range; NT: HIV-1 nontransmitting mother; TR: total group of HIV-1 transmitting mother; IP-TR: intrapartum HIV-1 transmitting mother; IU-TR: in utero HIV-1 transmitting mother; Unknown-TR: group of HIV-1 transmitting mothers where the infants’ mode of HIV-1 acquisition was unknown.
Maternal viral load (mVL) determinations were performed on samples collected at the time of delivery using the Roche Amplicor HIV-1 RNA Monitor assay version 1.5 (Roche Diagnostic Systems, Inc., Branchburg, NJ) and the CD4+ T cell counts (cells/μL) were quantified using the commercially available FACSCount System from Becton Dickinson (San Jose, CA).
This study was approved by the University of the Witwatersrand Committee for Research on Human Subjects and the Institutional Review Board of Columbia University. Written informed consent was obtained from all women in this study.
2.2. HLA-G genotyping
Genomic DNA was extracted from whole blood using a QIAamp DNA Blood Mini Kit (Qiagen, Dusseldorf, Germany) following manufacturer’s instructions. Exons 2, 3 and 4 of HLA-G were amplified and sequenced using previously published primers (Turk et al., 2013, Hviid et al., 1997). Briefly, a 994-bp fragment encompassing exons 2 and 3 was amplified using primers HLAG_2/3PCRF (5’-CGGCCCCTGCGCGGAGGAGGGAGGGG-3’) and HLAG_2/3PCRR (5’-TCAGGACCAGAGGGAGGGCGATATTC-3’); primers HLAG_4PCRF (5’-AGGTGCTG-CTGGAGTGTC-3’) and HLAG_4PCRR (5’-TCTGGGAAAGGAGGTGAAG-3’) were used to amplify a 463-bp fragment spanning exon 4 (Turk et al., 2013). Thermocycling conditions were: 94°C for 2 min followed by 35 cycles of 94°C for 30 sec, 61.5°C/60°C for 30 sec (for exons 2-3 and exon 4, respectively) and 72°C for 1 min followed by a final extension step of 72°C for 7 min. PCR amplicons were purified using the Invitek MSB Spin PCRapace cleanup kit (Berlin, Germany) and sequenced in both directions by capillary electrophoresis using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, California, USA) with the sequencing primers described by Turk et al. (2013): HLAG_2SEQR (5’-TCGTGATCTGCGCCCTG-3’); HLAG_3SEQF (5’-TGGGCGGGGCTGACCGAGGGGGTGGG-3’); HLAG_3SEQR (5’-TCAGGACCAGAG-GGAGGGCGATATTC-3’); HLAG_4SEQF (5’-GTGCTTGAATT-TTCTGACTCTT-3’) and HLAG_4SEQR (5’-TGCTTTCCCTAACAGACATGAT-3’). HLAG_2SEQF (5’-CTCCATGAGGTATTTCAGC[G]-3’) was an in-house primer designed using HLA-G intron 1 alignments and synthesised with a lock nucleic acid (LNA) modified 3’ end (indicated in square brackets). Sequence analysis and allele assignment were performed using Assign™ SBT version 4.7 software (Conexio Genomics, Fremantle, Western Australia) with the IMGT/HLA-G 2013 reference library compiled and supplied by Conexio Genomics (Fremantle, Western Australia) on personal request.
2.3 HLA-G 3’UTR genotyping
Nucleotide sequence variation of the HLA-G 3’UTR was evaluated by direct sequencing of a 343-bp fragment encompassing the genomic positions +2885 through to +3228, using PCR primers described by (Sizzano et al., 2012). Briefly, the 3’UTR region was amplified using HLAG_3UTRF (5’-TCACCCCTCACTGTGACTGA-3’) and HLAG_3UTRR (5’-CCCATCAA-TCTCTCTTGGAAA-3’) primers with the following thermocycling conditions: 95°C for 15 min followed by 30 cycles of 93°C for 1 min, 58°C for 1 min, 72°C for 1 min. A final extension step was carried out at 72°C for 10 min. Purified amplicons using the Invitek MSB Spin PCRapace cleanup kit (Berlin, Germany) were sequenced in both directions by capillary electrophoresis using an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, California, USA) using the same PCR primers. The chromatograms obtained were analysed using Sequencher version 4.10.1 (Gene Codes Corporation, Ann Arbor, Michigan, USA) and sequences were aligned with an available HLA-G 3’UTR reference sequence (GenBank Accession number NG_029039.1) to identify known polymorphic positions (Castelli et al., 2010) and any other polymorphism that had not been previously described.
2.4 HLA-G 3’UTR haplotypes
Arrangement of variations in the HLA-G 3’UTR into haplotypes was predicted both by visual examination of the genotypic data as well as using the Bayesian algorithm through the HAPLOTYPER software (Niu et al., 2002). Observed haplotypes were compared to published 3’UTR haplotypes and assigned the published nomenclature when the haplotypes corresponded in sequence/structure (Sabbagh et al., 2014). The frequencies of haplotypes were calculated by counting the number of alleles harbouring the haplotypes and dividing by the total number of alleles. Counting of the haplotypes was irrespective of the presence of additional SNPs not forming part of the haplotypes in question.
2.5 Classification of mother-infant HLA-G concordance
To assess the effect of concordance on MTCT of HIV-1, we directly counted the number of matching mother-infant pairs following the method reported by MacDonald et al. (1998). Since a child will inherit one allele from the mother and one from the father, it will match at least 50% of the mother’s HLA-G alleles, if the two alleles of the child matched two alleles of the mother, they were considered concordant. If the mother was homozygous at HLA-G and matched one of the child’s HLA-G alleles, she was also considered to be concordant since the child would not recognise any foreign antigens. At the 3’UTR, concordance was considered when both mother and infant had the same polymorphism and/or UTR haplotype.
2.6. Computational and statistical analysis
Allele frequencies at all polymorphic positions were determined by direct counting. Deviations from Hardy-Weinberg Equilibrium (HWE) were assessed using the conventional Monte Carlo exact test (Guo and Thompson, 1992) using the computer program TFPGA (Tools for Population Genetic Analyses version 1.3; 1997: author Mark. P. Miller). Linkage disequilibrium (LD) between HLA-G alleles and the polymorphic sites within the 3’UTR were evaluated using the likelihood ratio test as described by (Lewontin, 1964). The statistical significance of the LD between each of the SNP pairs was evaluated by the approximate chi-square described by (Liau et al., 1984). For associations of MTCT risk, comparisons of HLA-G allele and genotype frequencies were analyzed between NT and TR maternal groups, as well as EU and HIV-1 infected infants using the online Fisher exact test, VassarStats (http://www.vassarstats.net/odds2x2.html) which was also used to estimate the odds ratio (OR) and its 95% confidence interval (95%CI). Reported P-values are two-tailed and were considered significant when P<0.05. No adjustments were made for multiple comparisons, as adjustment for multiple comparisons correct for type 1 errors but increase the risk of type 2 errors. Given the complexity and multifactorial nature of maternal-infant HIV-1 transmission, we considered it more important to identify potential factors that may play a role in this route of infection rather than dismissing these leads as due to chance variations brought about by multiple comparisons. Unconditional logistic regression (Pezzullo, 2005) was used to adjust for the effects of the following variables: maternal viral load (mVL), maternal sdNVP, as well as infant possession of KIR2DS4-v which we have recently reported to impact on IU MTCT in the same cohort (Hong et al., 2013).
3 Results
3.1 HLA-G allele representation in Black South Africans
A total of 216 Black South African mother-infant pairs were genotyped, which included 144 NT/EU pairs and 72 TR/IP, IU or IU2 pairs. The clinical characteristics are shown in Table 1. Sixteen HLA-G alleles were identified in the study population (Table 2). These encoded six functional proteins (G*01:01, G*01:03, G*01:04, G*01:06, G*01:08 and G*01:11) and the truncated protein (G*01:05N). The three most common HLA-G alleles were G*01:01:01, G*01:01:02 and G*01:04:04 with frequencies greater than 15% in both mothers and infants, which corresponded with the most common genotypes G*01:01:01/G*01:01:01, G*01:01:01/G*01:01:02 and G*01:01:01/G*01:04:04 (Table 3). All HLA-G allele frequencies were all in Hardy–Weinberg equilibrium for both mother and infant groups (P=0.120 and P=0.769, respectively).
Table 2.
Representation of HLA-G allele frequencies in HIV-1 positive mothers and their infants, N (%)
|
Maternal
HLA-G Alleles |
NT
(2n=288 ) |
IP-TR
(2n=58 ) |
IU-TR
(2n=38 ) |
IU2-
TR (2n=86 ) |
IP-TR vs. NT | IU-TR vs. NT | IU2-TR vs. NT | |||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||||
|
| ||||||||||
| G*01:01:01 | 85 (29.51) |
19 (32.76) |
15 (39.47) |
25 (29.07) |
1.16 (0.63- 2.12) |
0.63 9 |
1.55 (0.77- 3.13) |
0.26 1 |
0.97 (0.57- 1.66) |
1.00 0 |
| G*01:01:02 | 59 (20.49) |
11 (18.97) |
14 (36.84) |
26 (30.23) |
0.91 (0.44- 1.86) |
0.86 0 |
2.26 (1.10- 4.64) |
0.03
6 |
1.68 (0.97- 2.89) |
0.07
8 |
| G*01:01:08 | 8 (2.78) | 3 (5.17) |
1 (2.63) |
2 (2.33) |
1.91 (0.49- 7.42) |
0.40 3 |
0.94 (0.11- 7.77) |
1.00 0 |
0.83 (1.07- 3.99) |
1.00 0 |
| G*01:01:09 | 11 (3.82) |
2 (3.45) |
1 (2.63) |
1 (1.16) |
0.90 (0.19- 4.16) |
1.00 0 |
0.68 (0.08- 5.42) |
1.00 0 |
0.29 (0.04- 2.32) |
0.31 0 |
| G*01:01:15 | 1 (0.35) | 0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:01:17 | 1 (0.35) | 0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:01:19 | 8 (2.78) | 2 (3.45) |
0 (0.00) |
4 (4.65) |
1.25 (0.25- 6.04) |
1.00 0 |
- | 0.60 3 |
1.70 (0.50- 5.81) |
0.48 3 |
| G*01:01:20 | 1 (0.35) | 0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:03:01 | 23 (7.99) |
3 (5.17) |
3 (7.89) |
7 (8.14) |
0.63 (0.18- 2.16) |
0.59 2 |
0.98 (0.28- 3.45) |
1.00 0 |
1.02 (0.42- 2.46) |
1.00 0 |
| G*01:04:01 | 11 (3.82) |
5 (8.62) |
0 (0.00) |
1 (1.16) |
2.37 (0.79- 7.11) |
0.16 0 |
- | 0.37 5 |
0.29 (0.04- 2.32) |
0.31 0 |
| G*01:04:04 | 53 (18.40) |
10 (17.24) |
3 (7.89) |
15 (17.44) |
0.92 (0.44- 1.94) |
1.00 0 |
0.38 (0.11- 1.28) |
0.11 6 |
0.93 (0.50- 1.76) |
0.87 5 |
| G*01:05N | 27 (9.38) |
2 (3.45) |
0 (0.00) |
4 (4.65) |
0.34 (0.08- 1.49) |
0.19 4 |
- |
0.05
6 |
0.47 (0.16- 1.38) |
0.18 8 |
| G*01:06 | 0 (0.00) |
1 (1.72) |
0 (0.00) |
0 (0.00) |
- | 0.16 8 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:11 | 0 (0.00) | 1 (1.72) |
1 (2.63) |
1 (1.16) |
- | 1.00 0 |
- | 0.11 7 |
- | 0.23 0 |
|
| ||||||||||
|
Infant
HLA-G Alleles |
EU
(2n=288 ) |
IP
(2n=58 ) |
IU
(2n=38 ) |
IU2
(2n=86 ) |
IP vs. EU | IU vs. EU | IU2 vs. EU | |||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||||
|
| ||||||||||
| G*01:01:01 | 96 (33.33) |
22 (37.93) |
13 (34.21) |
21 (24.42) |
1.22 (0.68- 2.19) |
0.54 5 |
1.04 (0.51- 2.12) |
1.00 0 |
0.64 (0.37- 1.12) |
0.14 5 |
| G*01:01:02 | 59 (20.49) |
6 (10.34) |
9 (23.68) |
20 (23.26) |
0.45 (0.18- 1.09) |
0.09 6 |
1.20 (0.54- 2.68) |
0.67 2 |
1.17 (0.66- 2.09) |
0.65 2 |
| G*01:01:08 | 10 (3.47) |
4 (6.90) |
3 (7.89) |
4 (4.65) |
2.05 (0.62- 6.81) |
0.26 5 |
2.38 (0.62- 9.07) |
0.37 7 |
1.35 (0.41- 4.43) |
0.74 6 |
| G*01:01:09 | 7 (2.43) | 4 (6.90) |
1 (2.63) |
1 (1.16) |
2.97 (0.84- 10.5) |
0.09 4 |
1.08 (0.12- 9.06) |
1.00 0 |
0.47 (0.05- 3.89) |
0.68 8 |
| G*01:01:19 | 11 (3.82) |
2 (3.45) |
0 (0.00) |
5 (5.81) |
0.90 (0.19- 4.17) |
1.00 0 |
- | 0.37 5 |
1.55 (0.52- 4.60) |
0.54 2 |
| G*01:03:01 | 29 (10.07) |
4 (6.90) |
4 (10.53) |
8 (9.30) |
0.66 (0.22- 1.95) |
0.48 5 |
1.05 (0.35- 3.17) |
1.00 0 |
0.92 (0.40- 2.08) |
1.00 0 |
| G*01:04:01 | 11 (3.82) |
1 (1.72) |
1 (2.63) |
3 (3.49) |
0.44 (0.06- 3.49) |
0.49 7 |
0.68 (0.08- 5.42) |
1.00 0 |
0.91 (0.25- 3.33) |
1.00 0 |
| G*01:04:04 | 41 (14.24) |
11 (18.97) |
6 (15.79) |
19 (22.09) |
1.41 (0.67- 2.94) |
0.41 9 |
1.12 (0.44- 2.87) |
0.80 6 |
1.71 (0.93- 3.13) |
0.09 4 |
| G*01:04:05 | 1 (0.35) | 0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:05N | 21 (7.29) |
3 (5.17) |
1 (2.63) |
5 (5.81) |
0.69 (0.20- 2.41) |
0.60 0 |
0.34 (0.05- 2.63) |
0.34 3 |
0.78 (0.29- 2.15) |
0.81 0 |
| G*01:06 | 0 (0.00) | 1 (1.72) |
0 (0.00) |
0 (0.00) |
- | 0.16 8 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:08 | 1 (0.35) | 0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
| G*01:11 | 1 (0.35) | 0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
NT: HIV-1 nontransmitting mother; IP-TR: intrapartum HIV-1 transmitting mother; IU-TR: in utero HIV-1 transmitting mother; IU2-TR: enriched group of presumed in utero transmitting mothers; EU: HIV-1 exposed uninfected infant; INF; IP: intrapartum infected infant; IU: in utero infected infant; IU2: enriched group of presumed in utero infected infants. Highlighted and bold P values indicate significant differences (P<0.05) and bold P values indicate trends (P<0.090).
Table 3.
Representation of maternal HLA-G genotypes, N (%)
|
HLA-G
Genotypes* |
NT (n=14 4) |
IP- TR (n=2 9) |
IU- TR (n=19) |
IU- TR (n=4 3) |
IP-TR vs. NT | IU-TR vs. NT | IU2-TR vs. NT | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) |
P | OR (95% CI) |
P | OR (95% CI) |
P | |||||
| G*01:01:01/G*01
:01:01 |
17 (11.81 ) |
3 (10.3 4) |
5 (26.32 ) |
5 (11.6 3) |
0.86 (0.23- 3.16) |
1.00 0 |
2.66 (0.85- 8.34) |
0.14 3 |
0.98 (0.34- 2.84) |
1.00 0 |
| G*01:01:01/G*01
:01:02 |
15 (11.81 ) |
4 (13.7 9) |
3(15.7 9) |
4 (9.30 ) |
1.37 (0.42- 4.49) |
0.74 4 |
1.61 (0.42- 6.18) |
0.69 8 |
0.88 (0.27- 2.81) |
1.00 0 |
| G*01:01:01/G*01
:01:09 |
4 (2.78) |
0 (0.00 ) |
1 (5.26) |
1 (2.33 ) |
- | 0.60 8 |
1.94 (0.21- 18.3) |
1.00 0 |
0.83 (0.09- 7.65) |
1.00 0 |
| G*01:01:01/G*01
:03:01 |
4 (2.78) |
0 (0.00 ) |
1 (5.26) |
2 (4.65 ) |
- | 0.60 8 |
1.94 (0.21- 18.3) |
1.00 0 |
1.71 (0.30- 9.66) |
0.62 2 |
| G*01:01:01/G*01
:04:01 |
3 (2.08) |
2 (6.90 ) |
0 (0.00) |
0 (0.00 ) |
3.48 (0.56- 21.8) |
0.19 6 |
- | 1.00 0 |
- | 0.58 7 |
| G*01:01:01/G*01
:04:04 |
17 (11.81 ) |
4 (13.7 9) |
0 (0.00) |
6 (13.9 5) |
1.19 (0.37- 3.85) |
0.75 8 |
- | 0.22 4 |
1.21 (0.44- 3.29) |
0.79 2 |
| G*01:01:01/G*01
:05N |
7 (4.86) |
1 (3.45 ) |
0 (0.00) |
0 (0.00 ) |
0.70 (0.08- 5.91) |
1.00 0 |
- | 0.60 2 |
- | 0.20 5 |
| G*01:01:02/G*01
:01:02 |
6 (4.17) |
2 (6.90 ) |
3 (15.79 ) |
4 (9.30 ) |
1.70 (0.33- 8.89) |
0.62 3 |
4.31 (0.98- 18.9) |
0.07
2 |
2.35 (0.63- 8.77) |
0.24 2 |
| G*01:01:02/G*01
:01:08 |
3 (2.08) |
1 (3.45 ) |
0 (0.00) |
1 (2.33 ) |
1.67 (0.17- 16.7) |
1.00 0 |
- | 1.00 0 |
1.11 (0.11- 11.0) |
1.00 0 |
| G*01:01:02/G*01
:01:19 |
4 (2.78) |
0 (0.00 ) |
0 (0.00) |
0 (0.00 ) |
- | 0.60 8 |
- | 1.00 0 |
- | 0.57 5 |
| G*01:01:02/G*01
:03:01 |
4 (2.78) |
0 (0.00 ) |
2 (10.53 ) |
5 (11.6 3) |
- | 0.60 8 |
4.11 (0.70- 24.1) |
0.14 5 |
4.61 (1.17- 17.9) |
0.03
1 |
| G*01:01:02/G*01
:04:01 |
3 (2.08) |
0 (0.00 ) |
0 (0.00) |
1 (2.33 ) |
- | 1.00 0 |
- | 1.00 0 |
1.11 (0.11- 11.0) |
1.00 0 |
| G*01:01:02/G*01
:04:04 |
10 (6.94) |
2 (6.90 ) |
2 (10.53 ) |
4 (9.30 ) |
1.00 (0.21- 4.78) |
1.00 0 |
1.57 (0.32- 7.81) |
0.63 4 |
1.37 (0.41- 4.62) |
0.74 1 |
| G*01:01:02/G*01
:05N |
6 (4.17) |
0 (0.00 ) |
0 (0.00) |
2 (4.65 ) |
- | 0.38 7 |
- | 0.61 5 |
1.12 (0.22- 5.77) |
1.00 0 |
| G*01:01:08/G*01
:04:04 |
3 (2.08) |
0 (0.00 ) |
1 (5.26) |
1 (2.33 ) |
- | 1.00 0 |
2.61 (0.26- 26.5) |
0.39 4 |
1.11 (0.11- 11.0) |
1.00 0 |
| G*01:01:09/G*01
:05N |
3 (2.08) |
0 (0.0 0) |
0 (0.00) |
0 (0.00 ) |
- | 1.00 0 |
- | 1.00 0 |
- | 0.58 7 |
| G*01:03:01/G*01
:04:04 |
3 (2.08) |
1 (3.45 ) |
0 (0.00) |
0 (0.00 ) |
1.67 (0.17- 16.7) |
1.00 0 |
- | 1.00 0 |
- | 0.58 7 |
| G*01:03:01/G*01
:05N |
4 (2.78) |
0 (0.00 ) |
0 (0.00) |
0 (0.00 ) |
- | 0.60 8 |
- | 1.00 0 |
- | 0.57 5 |
| G*01:04:04/G*01
:04:04 |
6 (4.17) |
0 (0.00 ) |
0 (0.00) |
0 (0.00 ) |
- | 0.38 7 |
- | 0.61 5 |
- | 0.33 9 |
| G*01:04:04/G*01
:05N |
6 (4.17) |
1 (3.45 ) |
0 (0.00) |
2 (4.65 ) |
0.82 (0.09- 7.09) |
1.00 0 |
- | 0.61 5 |
1.12 (0.22- 5.77) |
1.00 0 |
indicates HLA-G genotype frequencies > 2% in the NT group. NT: HIV-1 nontransmitting mother; IP-TR: intrapartum HIV-1 transmitting mother; IU-TR: in utero HIV-1 transmitting mother; IU2-TR: enriched group of presumed in utero transmitting mothers. Highlighted and bold P values indicate significant differences (P<0.05) and bold P values indicate trends (P<0.090).
3.2 HLA-G alleles and mother-to-child transmission of HIV-1
To investigate the influence of HLA-G alleles and genotypes on MTCT of HIV-1 we compared HIV-1 non-transmitting mothers (NT) to HIV-1 transmitting mothers (TR) and their respective infants: exposed uninfected (EU), in utero infected (IU and IU2) and intrapartum infected (IP). Maternal possession of the G*01:01:02 allele was associated with increased risk for IU HIV-1 transmission (Table 2). Representation of G*01:01:02 was higher in IU-TR (P=0.036, OR=2.26) and IU2-TR (P=0.078, OR=1.68) mothers compared to NT mothers. While G*01:01:02/G*01:01:02 genotype showed a strong trend (P=0.072, OR=4.31) towards increased representation in IU-TR mothers compared to NT mothers (Table 3). In addition, the combination of G*01:01:02 with G*01:03:01 (G*01:01:02/G*01:03:01) was significantly higher in IU2-TR mothers compared to NT mothers (P=0.031, OR=4.61). All of these allele associations were significant post adjustments for mVL but only G*01:01:02 homozygosity maintained significance post adjustment for maternal sdNVP and infant KIR2DS4-v (Table 7). Infant HLA-G alleles (Table 2) and HLA-G genotypes (Table S1) showed no significant associations with regards to acquisition of HIV-1.
Table 7.
Logistic regression analysis, adjustments made for maternal factors that influence MTCT of HIV-1
| Allele/genotype | Association | Unadjusted | Adjusted mVL | Adjusted mNVP | Adjusted Infant KIR2DS4-v |
||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) |
P | OR (95% CI) |
P | OR (95% CI) |
P | OR (95% CI) |
P | ||
| G*01:01:02 | IU-TR vs. NT |
2.26 (1.10- 4.64) |
0.0
36 |
3.22 (1.03- 10.1) |
0.0
43 |
2.02 (0.74- 5.52) |
0.1 68 |
2.13 (0.79- 5.78) |
0.1 34 |
| G*01:01:02 | IU2-TR vs. NT |
1.68 (0.97- 2.89) |
0.0
78 |
2.39 (1.10- 5.20) |
0.0
27 |
1.54 (0.73- 3.22) |
0.2 52 |
1.79 (0.88- 3.65) |
0.1 05 |
|
G*01:01:02/G*01
:01:02 |
IU-TR vs. NT |
4.31 (0.98- 18.9) |
0.0
72 |
8.79 (1.46- 52.8) |
0.0
17 |
4.81 (1.04- 22.3) |
0.0
44 |
5.08 (1.12- 22.9) |
0.0
35 |
|
G*01:01:02/G*01
:03:01 |
IU2-TR vs. NT |
4.61 (1.17- 17.9) |
0.0
31 |
10.0 (2.15- 46.8) |
0.0
03 |
3.14 (0.73- 13.5) |
0.1 22 |
3.81 (0.89- 16.3) |
0.0 71 |
| 3187G | IU-TR vs. NT |
2.96 (1.31- 6.67) |
0.0
11 |
2.17 (0.67- 6.99) |
0.1 92 |
2.56 (0.90- 7.29) |
0.0 77 |
2.42 (0.86- 6.83) |
0.0 94 |
| 3187GG | IU-TR vs. NT |
14.4 (2.18- 94.7) |
0.0
11 |
29.6 (2.36- 371.0) |
0.0
09 |
- | - | - | - |
| 3187GG | IU2-TR vs. NT | 5.39 (0.86- 33.7) |
0.0
80 |
6.69 (0.91- 49.4) |
0.0
62 |
10.9 (1.06- 113.6) |
0.0
44 |
6.51 (1.04- 40.7) |
0.0
45 |
| UTR1 | IU-TR vs. NT |
2.96 (1.31- 6.67) |
0.0
11 |
2.17 (0.67- 6.99) |
0.1 92 |
2.56 (0.90- 7.29 |
0.0 77 |
2.42 (0.86- 6.83) |
0.0 94 |
| UTR1/UTR1 | IU-TR vs. NT |
13.3 (2.07- 85.7) |
0.0
12 |
29.6 (2.36- 371.0) |
0.0
09 |
- | - | - | - |
| UTR1/UTR2 | IU2-TR vs. NT |
3.66 (1.01- 13.3) |
0.0
52 |
5.44 (1.32- 22.4) |
0.0
19 |
3.43 (0.91- 12.8) |
0.0 67 |
3.57 (0.95- 13.3) |
0.0 58 |
| UTR3/UTR4 | IP-TR vs. NT |
8.19 (1.30- 51.5) |
0.0
34 |
6.17 (0.91- 41.4) |
0.0 61 |
4.24 (0.63- 28.5) |
0.1 37 |
N/A | N/ A |
|
| |||||||||
| Combined effect | |||||||||
|
| |||||||||
|
G*01:01:02 &
3187G |
IU-TR vs. NT |
4.12 (0.70- 24.2) |
0.1 45 |
6.62 (0.82- 52.9) |
0.0 74 |
3.81 (0.64- 22.5) |
0.1 40 |
4.19 (0.70- 24.9) |
0.1 16 |
|
G*01:01:02 or 3187G |
IU-TR vs. NT |
4.51 (1.25- 16.1) |
0.0
14 |
4.93 (1.24- 19.5) |
0.0
23 |
4.14 (1.14- 15.1) |
0.0
31 |
4.12 (1.13- 14.9) |
0.0
31 |
mVL: maternal viral load; mNVP: maternal single-dose Nevirapine; KIR2DS4-v: non-functional KIR2DS4; N/A: not an applicable adjustment, since infant possession of KIR2DS4-v has been association with IU HIV-1 transmission and not IP HIV-1 transmission. Highlighted and bold P values indicate significant differences (P<0.05).
3.3 HLA-G 3’UTR allele, genotype and haplotype frequencies in MTCT of HIV-1
Sequencing analysis of HLA-G 3’UTR revealed the presence of 11 polymorphic sites in our cohort. Nine of these have been previously described, namely: +2961Ins/Del (rs66554220), +3003T/C (rs1707), +3010C/G (rs1710), +3027C/A (rs17179101), +3032G/C (rs146339774), +3035C/T (rs17179108), +3142G/C (rs1063320), +3187A/G (rs9380142), and +3196C/G (rs1610696). Two new SNPs, +3076C/T and +3114G/A, were found in EU infants, but had frequencies below 1% and were not considered in further analyses. Observed genotype frequencies at all variation sites were in adherence with HWE. Using HAPLOTYPER software and in combination with published 3’UTR haplotypes (Sabbagh et al., 2014), we found eight distinct 3’UTR haplotypes (UTR1-UTR7 and UTR20) within our cohort (Figure 1). UTR1, UTR2 and UTR3 were the most frequent haplotypes, which together accounted for over 65% of the haplotypes in our South African study population, which was most comparable to the Senegalese population (SER, n=239) reported by Sabbagh et al. (2014).
When comparing the nine polymorphic sites within the 3’UTR with risk of MTCT, only one position (+3187A/G) showed an association with increased risk for IU MTCT (Table 4). Maternal frequencies of the G allele as well as G/G homozygosity at position +3187 were significantly over-represented in IU-TR mothers compared to NT mothers (P=0.011, OR=2.96 and P=0.011, OR=14.4, respectively). However, only G/G homozygosity maintained significance post adjustment for mVL (P=0.009, OR=29.6), but the confidence interval (CI) was considerably large (2.36-371.0), most likely due to the small number (Table 7). Comparison of haplotype associations in MTCT however confirmed the UTR1 haplotype (the only haplotype to have a G at position +3187), as well as the UTR1/UTR1genotype (Table 5), were both significantly over-represented in IU-TR mothers compared to NT mothers (P=0.011, OR=2.96 and P=0.012, OR=13.3, respectively). The genotype UTR1/UTR2 also showed a strong trend towards increased representation in IU2-TR mothers compared to NT mothers (P=0.052, OR=3.66). Both UTR1/UTR1 and UTR1/UTR2 genotypes maintained significance after adjustment for mVL (P=0.009, OR=29.6 and P=0.019, OR=5.44, respectively) but not after adjustment for maternal sdNVP or KIR2DS4-v (Table 7). Taken together, a Guanine at +3187 and/or the UTR1 haplotype, showed a significant association with increased risk for IU transmission. In contrast, IP-TR mothers had a significantly higher representation of the UTR3/UTR4 genotype compared to NT mothers (P=0.034, OR=8.19), but this association was not significant post adjustment for mVL or maternal sdNVP (Table 7). Comparison between genotypes UTR1/UTR1 (associated with IU transmission) and UTR3/UTR4 (associated with IP transmission) showed differences at four positions. The UTR3/UTR4 combination is heterozygous at positions +3003T/C, +3010C/G, +3142G/C, and has an Adenine at position +3187; while UTR1/UTR1 is homozygous at positions +3003T, +3010G, +3142C, and has a Guanine at position +3187.
Table 4.
Representation 3’UTR polymorphisms of HLA-G in HIV-1 positive mothers, N (%)
| Polymorphism | NT (n=144) |
IP-TR (n=29) |
IU-TR (n=19) |
IU2-TR (n=43) |
IP-TR vs. NT | IU-TR vs. NT | IU2-TR vs. NT | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) |
P | OR (95% CI) |
P | OR (95% CI) |
P | ||||||
|
+2961
(Indel) |
D | 166 (57.64) |
36 (62.07) |
19 (50.00) |
43 (50.00) |
1.20 (0.67- 2.14) |
0.562 | 0.73 (0.37- 1.45) |
0.388 | 0.73 (0.45- 1.19) |
0.218 |
| I | 122 (42.36) |
22 (37.93) |
19 (50.00) |
43 (50.00) |
1 | 1 | 1 | ||||
| DD | 53 (36.81) |
11 (37.93) |
6 (31.58) |
12 (27.91) |
1.61 (0.47- 5.48) |
0.564 | 0.58 (0.17- 1.97) |
0.527 | 0.58 (0.23- 1.45) |
0.344 | |
| ID | 60 (41.67) |
14 (48.28) |
7 (36.84) |
19 (44.19) |
1.81 (0.55- 5.96) |
0.414 | 0.52 (0.16- 1.69) |
0.348 | 0.81 (0.35- 1.90) |
0.668 | |
| II | 3 (21.53) |
4 (13.79) |
6 (31.58) |
12 (27.91) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3003.T/C | C | 31 (10.76) |
9 (15.52) |
1 (2.63) | 4 (4.65) | 1.52 (0.68- 3.40) |
0.366 | 0.22 (0.03- 1.69) |
0.149 | 0.40 (0.14- 1.18) |
0.095 |
| T | 257 (89.24) |
49 (84.48) |
37 (97.37) |
82 (95.35) |
1 | 1 | 1 | ||||
| CC | 4 (2.78) | 1 (3.45) | 0 (0.00) | 0 (0.00) | 1.39 (0.15- 13.1) |
1.000 | - | 1.000 | - | 0.573 | |
| TC | 23 (15.97) |
7 (24.14) |
1 (5.26) | 4 (9.30) | 1.69 (0.36- 4.45) |
0.418 | 0.28 (0.03- 2.22) |
0.311 | 0.52 (0.17- 1.60) |
0.329 | |
| TT | 117 (81.25) |
21 (72.41) |
18 (94.74) |
39 (90.70) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3010.C/G | G | 100 (34.72) |
22 (37.93) |
15 (39.47) |
26 (30.23) |
1.14 (0.64- 2.06) |
0.653 | 1.23 (0.61- 2.45) |
0.590 | 0.81 (0.48- 1.37) |
0.516 |
| C | 188 (65.28) |
36 (62.07) |
23 (60.53) |
60 (69.77) |
1 | 1 | 1 | ||||
| GG | 23 (15.97) |
4 (13.79) |
5 (26.32) |
5 (11.63) | 1.06 (0.31- 3.66) |
1.000 | 1.61 (0.49- 5.32) |
0.518 | 0.66 (0.22- 1.95) |
0.609 | |
| CG | 54 (37.50) |
14 (48.28 |
5 (26.32) |
16 (37.21) |
1.58 (0.66- 3.76) |
0.379 | 0.69 (0.22- 2.18) |
0.582 | 0.90 (0.43- 1.89) |
0.853 | |
| CC | 67 (46.53) |
11 (37.93) |
9 (47.37) |
22 (51.16) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3027.C/A | A | 1 (0.35 | 0 (0.00) | 0 (0.00) | 0 (0.00) | - | 1.000 | - | 1.000 | - | 1.000 |
| C | 287 (99.65) |
58 (100.00) |
38 (100.00) |
86 (100.00) |
1 | 1 | 1 | ||||
| AA | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | - | 1.000 | - | 1.000 | - | 1.000 | |
| CA | 1 (0.69) | 0 (0.00) | 0 (0.00) | 0 (0.00) | - | 1.000 | - | 1.000 | - | 1.000 | |
| CC | 143 (99.31) |
29 (100.00) |
19 (100.00) |
43 (100.00) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3032.G/C | C | 4 (1.39) | 1 (1.72) | 1 (2.63) | 2 (2.33) | 1.00 (0.11- 8.66) |
1.000 | 1.52 (0.17- 13. 5) |
1.000 | 1.34 (0.26- 7.07) |
1.000 |
| G | 284 (98.61) |
57 (98.28) |
37 (97.37) |
84 (97.67) |
1 | 1 | 1 | ||||
| CC | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | - | 1.000 | - | 1.000 | - | 1.000 | |
| GC | 4 (2.78 | 1 (3.45) | 1 (5.26) | 2 (4.65) | 1.67 (0.16- 16.6) |
1.000 | 2.59 (0.26- 26.4) |
0.396 | 2.28 (0.37- 14.1) |
0.591 | |
| GG | 140 (97.22) |
28 (96.55) |
18 (94.74) |
41 (95.35) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3035.C/T | C | 263 (91.32 |
53 (91.38) |
34 (89.47) |
78 (90.70 |
1.01 (0.37- 2.75) |
1.000 | 0.81 (0.26- 2.46) |
0.760 | 0.93 (0.40- 2.14) |
1.000 |
| T | 25 (8.68) |
5 (8.62) | 4 (10.53) |
8 (9.30) | 1 | 1 | 1 | ||||
| CC | 121 (84.03) |
24 (82.76) |
15 (78.95 |
35 (81.40) |
- | 1.000 | - | 1.000 | - | 1.000 | |
| CT | 21 (14.58) |
5 (17.24) |
4 (21.05) |
8 (18.60) |
- | 1.000 | - | 1.000 | - | 1.000 | |
| TT | 2 (1.39) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3142.G/C | C | 100 (34.72) |
22 (37.93) |
15 (39.47) |
26 (30.23) |
1.14 (0.64- 2.06) |
0.653 | 1.23 (0.61- 2.45) |
0.590 | 0.81 (0.48- 1.37) |
0.516 |
| G | 188 (65.28) |
36 (62.07) |
23 (60.53) |
60 (69.77) |
1 | 1 | 1 | ||||
| CC | 23 (15.97) |
4 (13.79) |
5 (26.32) |
5 (11.63) |
1.06 (0.31- 3.65) |
1.000 | 1.61 (0.49- 5.33) |
0.518 | 0.66 (0.22- 1.95) |
0.609 | |
| GC | 54 (37.50) |
14 (48.28) |
5 (26.32) |
16 (37.21) |
1.58 (0.66- 3.76) |
0.379 | 0.68 (0.21- 2.18) |
0.582 | 0.90 (0.43- 1.89) |
0.853 | |
| GG | 67 (46.53) |
11 (37.93) |
9 (47.37) |
22 (51.16) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3187.A/G | G | 31 (10.76) |
6 (10.34) |
10 (26.32) |
14 (16.28) |
0.96 (0.37- 2.41) |
1.000 | 2.96 (1.31- 6.67) |
0.011 | 1.61 (0.81- 3.19) |
0.187 |
| A | 257 (89.24) |
52 (89.66) |
28 (73.68) |
72 (83.72) |
1 | 1 | 1 | ||||
| GG | 2 (1.39) | 0 (0.00) | 3 (15.79) |
3 (6.98) | - | 1.000 | 14.4 (2.18- 94.7) |
0.011 | 5.39 (0.86- 33.7) |
0.080 | |
| AG | 27 (18.75) |
6 (20.69) |
4 (21.05) |
8 (18.60) |
1.11 (0.41- 2.99) |
1.000 | 1.42 (0.42- 4.74) |
0.740 | 1.06 (0.44- 2.57) |
1.000 | |
| AA | 115 (79.86) |
23 (79.31) |
12 (63.16) |
32 (74.42) |
1 | 1 | 1 | ||||
|
| |||||||||||
| +3196.C/G | G | 97 (33.68) |
17 (29.31) |
15 (39.47) |
35 (40.70) |
0.82 (0.44- 1.51) |
0.545 | 1.28 (0.64- 2.57) |
0.586 | 1.35 (0.82- 2.21) |
0.249 |
| C | 191 (66.32) |
41 (70.69) |
23 (60.53) |
51 (59.30) |
1 | 1 | 1 | ||||
| GG | 18 (12.50) |
2 (6.90) | 4 (21.05) |
7 (16.28) |
0.52 (0.11- 2.48) |
0.514 | 1.81 (0.48- 6.68) |
0.464 | 1.68 (0.60- 4.76) |
0.399 | |
| CG | 61 (42.36) |
13 (44.83) |
7 (36.84) |
21 (48.84) |
0.98 (0.43- 2.27) |
1.000 | 0.93 (0.32- 2.73) |
1.000 | 1.49 (0.71- 3.16) |
0.347 | |
| CC | 65 (45.14) | 14 (48.28) | 8 (42.11) | 15 (34.88) | 1 | 1 | 1 | ||||
NT: HIV-1 nontransmitting mother; IP-TR: intrapartum HIV-1 transmitting mother; IU-TR: in utero HIV-1 transmitting mother; IU2-TR: enriched group of presumed in utero transmitting mothers. Highlighted and bold P values indicate significant differences (P<0.05) and bold P values indicate trends (P<0.090).
Table 5.
Representation 3’UTR haplotype and genotype frequencies of HLA-G in HIV-1 positive mothers, N (%)
| 3’ UTR | NT (n=144 ) |
IP- TR (n=29 ) |
IU- TR (n=19 ) |
IU2- TR (n=43 ) |
IP-TR vs. NT | IU-TR vs. NT | IU2-TR vs. NT | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||||
| UTR1 | 31 (10.76) |
6 (10.34 ) |
10 (26.32 ) |
14 (16.28 ) |
0.96 (0.38- 2.41) |
1.00 0 |
2.96 (1.31- 6.67) |
0.01
1 |
1.61 (0.81- 3.19) |
0.18 7 |
| UTR2 | 97 (33.68) |
17 (29.31 ) |
15 (39.47 ) |
35 (40.70 ) |
0.82 (0.44- 1.51) |
0.54 5 |
1.28 (0.64- 2.57) |
0.58 6 |
1.44 (0.87- 2.37) |
0.15 5 |
| UTR3 | 66 (22.92) |
14 (24.14 ) |
4 (10.53 ) |
17 (19.77 ) |
1.07 (0.55- 2.07) |
0.86 5 |
0.39 (0.13- 1.16) |
0.09 4 |
0.82 (0.46- 1.51) |
0.55 9 |
| UTR4 | 31 (10.76) |
9 (15.52 ) |
1 (2.63) |
4 (4.65) |
1.52 (0.68- 3.39) |
0.36 6 |
0.22 (0.03- 1.69) |
0.14 9 |
0.40 (0.14- 1.18) |
0.09 5 |
| UTR5 | 24 (8.33) |
5 (8.62) |
4 (10.53 ) |
8 (9.30) |
1.03 (0.39- 2.84) |
1.00 0 |
1.29 (0.42- 3.96) |
0.75 6 |
1.13 (0.49- 2.61) |
0.82 6 |
| UTR6 | 34 (11.81) |
6 (10.34 ) |
3 (7.89) |
6 (6.98) |
0.86 (0.34- 2.16) |
0.82 7 |
0.64 (0.18- 2.19) |
0.59 6 |
0.56 (0.23- 1.38) |
0.23 8 |
| UTR7 | 1 (0.35) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
| UTR20 | 4 (1.39) |
1 (1.72) |
1 (2.63) |
2 (2.33) |
1.24 (0.14- 11.3) |
1.00 0 |
1.91 (0.21- 17.6) |
0.46 4 |
1.69 (0.30- 9.39) |
0.62 4 |
|
| ||||||||||
|
UTR1/UTR
1 |
2 (1.39) |
0 (0.00) |
3 (15.79 ) |
3 (6.98) |
- | 1.00 0 |
13.3 (2.07- 85.7) |
0.01
2 |
5.32 (0.86- 32.9) |
0.08
1 |
|
UTR1/UTR
2 |
5 (3.47) |
3 (10.34 ) |
2 (10.53 ) |
5 (11.63 ) |
3.21 (0.72- 14.3) |
0.13 2 |
3.27 (0.59- 18.2) |
0.19 0 |
3.66 (1.01- 13.3) |
0.05
2 |
|
UTR1/UTR
3 |
12 (8.33) |
1 (3.45) |
0 (0.00) |
0 (0.00) |
0.39 (0.05- 3.14) |
0.47 5 |
- | 0.36 3 |
- |
0.07
1 |
|
UTR1/UTR
4 |
5 (3.47) |
1 (3.45) |
0 (0.00) |
0 (0.00) |
0.99 (0.11- 8.83) |
1.00 0 |
- | 1.00 0 |
- | 0.34 7 |
|
UTR1/UTR
5 |
2 (1.39) |
0 (0.00) |
1 (5.26) |
2 (4.65) |
- | 1.00 0 |
3.94 (0.34- 45.7) |
0.31 2 |
3.46 (0.47- 25.4) |
0.22 7 |
|
UTR1/UTR
6 |
3 (2.08) |
0 (0.00) |
1 (5.26) |
1 (2.33) |
- | 1.00 0 |
2.61 (0.26- 26.5) |
0.39 4 |
1.12 (0.11- 11.0) |
1.00 0 |
|
UTR1/UTR
20 |
0 (0.00) |
1 (3.45) |
0 (0.00) |
0 (0.00) |
- | 0.16 8 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR2/UTR
2 |
18 (12.50) |
2 (6.90) |
4 (21.05 ) |
7 (16.28 ) |
0.52 (0.11- 2.37) |
0.53 4 |
1.87 (0.56- 6.25) |
0.47 4 |
1.36 (0.53- 3.51) |
0.61 0 |
|
UTR2/UTR
3 |
23 (15.97) |
4 (13.79 ) |
3 (15.79 ) |
10 (23.26 ) |
0.84 (0.27- 2.64) |
1.00 0 |
0.99 (0.26- 3.66) |
1.00 0 |
1.59 (0.69- 3.68) |
0.36 1 |
|
UTR2/UTR
4 |
12 (8.33) |
3 (10.34 ) |
0 (0.00) |
1 (2.33) |
1.27 (0.33- 4.81) |
0.71 9 |
- | 0.36 3 |
0.26 (0.03- 2.07) |
0.30 4 |
|
UTR2/UTR
5 |
10 (6.94) |
2 (6.90) |
2 (10.53 ) |
5 (11.63 ) |
0.99 (0.21- 4.78) |
1.00 0 |
1.58 (0.32- 7.81) |
0.63 4 |
1.76 (0.57- 5.47) |
0.34 2 |
|
UTR2/UTR
6 |
9 (6.25) |
1 (3.45) |
0 (0.00) |
0 (0.00) |
0.54 (0.06- 4.40) |
0.69 9 |
- | 0.39 0 |
- | 0.12 1 |
|
UTR2/UTR
7 |
1 (0.69) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR2/UTR
20 |
1 (0.69) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR3/UTR
3 |
8 (5.56) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 0.35 5 |
- | 0.59 8 |
- | 0.20 1 |
|
UTR3/UTR
4 |
2 (1.39) |
3 (10.34 ) |
0 (0.00) |
2 (4.65) |
8.19 (1.30- 51.5) |
0.03
4 |
- | 1.00 0 |
3.46 (0.47- 25.4) |
0.22 7 |
|
UTR3/UTR
5 |
5 (3.47) |
3 (10.34 ) |
0 (0.00) |
0 (0.00) |
3.21 (0.72- 14.3) |
0.13 2 |
- | 1.00 0 |
- | 0.34 7 |
| UTR3/UTR 6 | 7 (4.86) |
3 (10.34 ) |
1 (5.26) |
4 (9.30) |
2.25 (0.55- 9.31) |
0.37 4 |
1.09 (0.12- 9.35) |
1.00 0 |
2.01 (0.56- 7.21) |
0.46 1 |
|
UTR3/UTR
20 |
1 (0.69) |
0 (0.00) |
0 (0.00) |
1 (2.33) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR4/UTR
4 |
4 (2.78) |
1 (3.45) |
0 (0.00) |
0 (0.00) |
1.25 (0.13- 11.6) |
1.00 0 |
- | 1.00 0 |
- | 0.57 5 |
|
UTR4/UTR
5 |
1 (0.69) |
0 (0.00) |
1 (5.26) |
1 (2.33) |
- | 1.00 0 |
7.94 (0.48- 132.6) |
0.22 0 |
3.40 (0.21- 55.6) |
0.40 8 |
|
UTR4/UTR
6 |
2 (1.39) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR4/UTR
20 |
1 (0.69) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
3.40 (0.21- 55.6) |
0.40 8 |
|
UTR5/UTR
5 |
2 (1.39) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR5/UTR
6 |
2 (1.39) |
0 (0.00) |
0 (0.00) |
0 (0.00) |
- | 1.00 0 |
- | 1.00 0 |
- | 1.00 0 |
|
UTR6/UTR
6 |
5 (3.47) |
1 (3.45) |
0 (0.00) |
0 (0.00) |
0.99 (0.11- 8.83) |
1.00 0 |
- | 1.00 0 |
- | 0.34 7 |
|
UTR6/UTR
20 |
1 (0.69) |
0 (0.00) |
1 (5.26) |
1 (2.33) |
- | 1.00 0 |
7.94 (0.48- 132.6) |
0.22 0 |
3.40 (0.21- 55.6) |
0.40 8 |
NT: HIV-1 nontransmitting mother; IP-TR: intrapartum HIV-1 transmitting mother; IU-TR: in utero HIV-1 transmitting mother; IU2-TR: enriched group of presumed in utero transmitting mothers. Highlighted and bold P values indicate significant differences (P<0.05) and bold P values indicate trends (P<0.090).
Comparing the 3’UTR polymorphisms, as well as the haplotypes, in the infant group revealed no significant associations or trends (data not shown); suggesting that in our cohort, polymorphisms within 3’UTR did not play a role in infant acquisition of HIV-1.
3.4 Linkage disequilibrium (LD) between HLA-G alleles and the 3’UTR
In order to further investigate the relationships between HLA-G alleles and the variants within the 3’UTR, pairwise LD between the HLA-G alleles and the 14-bp indel, as well as the other SNPs and haplotypes was calculated. Table 6 shows LD results only for significant pairwise combinations. Seven of the HLA-G alleles were in strong LD with the 14-bp indel: G*01:01:01, G*01:04:01 and G*01:04:04 were significantly linked with the Del; while, G*01:01:02, G*01:03:01, G*01:05N and G*01:01:19 were significantly linked with the Ins. As expected, comparison of G*01:01:01, G*01:04:01 and G*01:04:04 with the 3’UTR haplotypes containing the 14-bp Del showed G*01:01:01 was in strong LD with UTR1, UTR6 and UTR20, while G*01:04:04 and G*01:04:04 were in strong LD with UTR3. Alleles associated with the Ins, G*01:01:02, G*01:03:01, G*01:05N and G*01:01:19, were found to be in significant LD with UTR2, UTR5 and UTR7.
Table 6.
Significant linkage disequilibrium (LD) between HLA-G alleles, the 14-bp indel, 3’UTR SNPs and haplotypes in HIV-1 positive mothers (n=216)
| LD comparisons | N | (%) | D' | X2 | P-value | |
|---|---|---|---|---|---|---|
| G*01:01:01 | Del | 126 | 29.17 | 0.95 | 45.19 | <0.001 |
| UTR1 | 44 | 10.19 | 0.80 | 37.70 | <0.001 | |
| UTR4 | 33 | 7.64 | 0.64 | 19.97 | <0.001 | |
| UTR6 | 45 | 10.42 | 0.94 | 47.47 | <0.001 | |
| UTR20 | 6 | 1.39 | 1.00 | 5.93 | <0.05 | |
|
| ||||||
| G*01:01:02 | Ins | 96 | 22.22 | 1.00 | 66.88 | <0.001 |
| UTR2 | 96 | 22.22 | 1.00 | 98.49 | <0.001 | |
|
| ||||||
| G*01:01:19 | Ins | 14 | 3.24 | 1.00 | 6.98 | <0.01 |
| UTR2 | 14 | 3.24 | 1.00 | 10.27 | <0.01 | |
|
| ||||||
| G*01:03:01 | Ins | 33 | 7.64 | 1.00 | 17.64 | <0.001 |
| UTR5 | 33 | 7.64 | 1.00 | 189.72 | <0.001 | |
|
| ||||||
| G*01:04:01 | Del | 17 | 3.94 | 1.00 | 4.16 | <0.05 |
| UTR3 | 17 | 3.94 | 1.00 | 27.24 | <0.001 | |
|
| ||||||
| G*01:04:04 | Del | 78 | 18.06 | 1.00 | 24.37 | <0.001 |
| UTR3 | 78 | 18.06 | 1.00 | 159.40 | <0.001 | |
|
| ||||||
| G*01:05N | Ins | 33 | 7.64 | 1.00 | 17.64 | <0.001 |
| UTR2 | 33 | 7.64 | 1.00 | 25.98 | <0.001 | |
| UTR7 | 1 | 0.23 | 1.00 | 5.83 | <0.05 | |
|
| ||||||
| Del | +3003 C | 44 | 10.19 | 1.00 | 11.94 | <0.01 |
| +3010 G | 148 | 34.26 | 1.00 | 65.44 | <0.001 | |
| +3035 C | 245 | 56.71 | 1.00 | 7.30 | <0.05 | |
| +3142 C | 148 | 34.26 | 1.00 | 65.44 | <0.001 | |
| +3187 G | 51 | 11.81 | 1.00 | 14.23 | <0.001 | |
| +3196 C | 245 | 56.71 | 1.00 | 126.48 | <0.001 | |
|
| ||||||
| Ins | +3003 T | 187 | 43.29 | 1.00 | 4.78 | <0.05 |
| +3010 C | 187 | 43.29 | 1.00 | 60.58 | <0.001 | |
| +3035 T | 38 | 8.80 | 1.00 | 20.71 | <0.001 | |
| +3142 G | 187 | 43.29 | 1.00 | 60.58 | <0.001 | |
| +3187 A | 187 | 43.29 | 1.00 | 6.44 | <0.01 | |
| +3196 G | 149 | 34.49 | 1.00 | 136.23 | <0.001 | |
N: the number of individuals; D’: measure of linkage disequilibrium; X2: Chi-Square value from which the P-value was determined significant at P<0.05.
Thus since the G*01:01:02 allele and +3187G (UTR1) are not in LD (D’=-0.12; X2=0.09, P>0.1), the effects of these two genotypic variations on IU transmission are independent of each other. Thus the comparison of IU-TR mothers vs. NT mothers having both G*01:01:02 and +3187G (UTR1) did not show a significant additive association, likely due to the small number of individuals carrying both these variants (Table 7). However, comparison of IU-TR mothers with EU mothers and representation of G*01:01:02 or +3187G (UTR1), was more significant (P=0.014; OR=4.51) than possession of G*01:01:02 alone, but not more significant than having +3187G (UTR1) alone (Table 7). The significantly greater representation of having either G*01:01:02 or +3187G (UTR1) was the only association maintained throughout all adjustments (Table 7).
3.5 Mother-infant concordance and MTCT
HLA concordance amongst the classical class I molecules (HLA-A, −B and −C) (MacDonald et al., 1998), as well as a synonymous SNP in exon 2 of HLA-G (Aikhionbare et al., 2001), have been described as a risk factors for MTCT; thus, we wanted to determine if concordance for HLA-G allele and 3’UTR polymorphisms between mother and infant had a similar association. Out of the 216 mother-infant pairs, concordance for HLA-G alleles did not significantly differ between NT and TR mother-infant pairs (Table S2). Similarly, mother-infant concordance for the 14-bp Ins/Ins, Ins/Del or Del/Deland other 3’ UTR haplotypes showed no significant associations. However, mother-infant concordance at SNP +3187A was found to be significantly lower in IU-TR/IU mother-infant pairs compared to NT/EU pairs (47% vs. 76%, P=0.014, OR=0.29).
4. Discussion
The role of HLA-G has become increasingly evident in many infectious diseases, primarily due to its inhibitory effect on the innate and adaptive immune system. By directly binding to the inhibitory receptors KIR2DL4, ILT-2 and ILT-4 that are present on NK and CD8+ T cells, HLA-G can also inhibit differentiation, proliferation, cytolysis, cytokine secretion, and immunoglobulin production (Amiot et al., 2014). Therefore, certain HLA-G alleles and/or 3’UTR polymorphisms that alter HLA-G expression may influence the function of NK and CD8+ T cells and ultimately influence the susceptibility to HIV-1 transmission. Several studies have found associations between certain HLA-G alleles, as well as a 14-bp indel, with an altered risk for vertical transmission of HIV-1 (Aikhionbare et al., 2006, Lajoie et al., 2009, Luo et al., 2013, Moodley and Bobat, 2011). However, these studies have largely focused on each of these parameters individually and, where studies have been comparable, the results have often not shown consensus. In addition, other SNPs within the 3’UTR have not been extensively analyzed for their clinical relevance in MTCT. In this study, we report on the combined analysis of HLA-G alleles as well as the 3’UTR of HLA-G in a HIV-1 MTCT cohort of 216 Black South African mother-infant pairs.
We found G*01:01:01, G*01:01:02 and G*01:04:04 were the most common alleles in our cohort, which corresponded with the most frequent genotypes G*01:01:01/G*01:01:01, G*01:01:01/G*01:01:02, and G*01:01:01/G*01:01:04. With regards to MTCT, maternal possession of G*01:01:02 allele, a Guanine at the +3187A/G 3’UTR SNP and the UTR1 haplotype, were significantly associated with increased risk for IU transmission (Tables 2, 4 and 5). Furthermore, strong LD between G*01:01:02 and the 14-bp Ins, as well as UTR1 with the 14-bp Del, suggested that differences in HLA-G expression altered the risk of MTCT.
Several studies have associated HLA-G with heterosexual transmission of HIV-1. In one study G*01:01:01 was significantly enriched in HIV-1 resistant sex workers, whereas G*01:04:04 was significantly associated with susceptibility to HIV-1 infection (Turk et al., 2013). However in Zimbabwean women, G*01:01:08 was associated with an increased risk of HIV-1 infection, while G*01:05N offered protection from heterosexual HIV-1 infection (Matte et al., 2004). In both these studies, HLA-G alleles that had higher HLA-G expression were associated with increased risk of HIV-1 transmission. It was further reported that individuals with G*01:04-had higher sHLA-G levels than individuals with the more frequent G*01:01:01; whereas, G*01:01:01 individuals had higher sHLA-G levels compared to individuals with G*01:01:03 and G*01:05N (Rebmann et al., 2001). Similarly, the 3’UTR haplotypes have also been associated with differences in sHLA-G expression levels, for example, UTR1 was associated with higher expression of sHLA-G, whereas UTR5 and UTR7 with lower expression and other UTRs (UTR2, 3, 4 and 6) exhibited intermediate levels of sHLA-G (Martelli-Palomino et al., 2013). What is unique about the UTR1 haplotype (14-bp Del, +3003T, +3010G, +3027C, +3032G, +3035C, +3142C, +3187G, +3196C) is that it carries the majority of the polymorphisms that associate with increased expression of HLA-G, namely: the 14-bp Del that is associated with highly soluble HLA-G expression, Cytosine at +3142 that is less sensitive to specific miRNAs (miR-148a, miR-148b and miR-152) and Guanine at +3187 that increases mRNA stability (Castelli et al., 2014).
Therefore in our study, UTR1 and its strong LD with G*01:01:01, may share a similar mechanism to that of heterosexual HIV-1 transmission, through increased expression of HLA-G being a risk factor for IU transmission. It has been postulated that HLA-G alleles that have higher HLA-G expression would exert greater NK and CD8+ T cell inhibition, which could result in the decreased ability of these cells to target viral-infected cells and thus increasing the risk for HIV-1 transmission (da Silva et al., 2014, Luo et al., 2013, Matte et al., 2004, Moodley and Bobat, 2011). In agreement with this hypothesis, placental expression of HLA-G was 3.9 times more up-regulated in transmitting mothers compared to non-transmitting mothers (Moodley and Bobat, 2011).
However, while certain alleles and 3’UTR polymorphisms would imply either increased/decreased expression of HLA-G, this is not absolute. Le Discorde et al. (2005) reported that even the null allele, G*0105N, had HLA-G protein expression. G*01:05N is characterized by a single cytosine deletion in exon 3 that presents a stop codon in exon 4 blocking the translation of HLA-G1 and HLA-G5 isoforms; it does however, encode both membrane-bound and soluble functional HLA-G proteins that are able to inhibit NK-cell cytolysis (Le Discorde et al., 2005). It is possible that even low expressing HLA-G alleles, such as G*01:01:02 and its strong LD with the 14-bp Ins, might have other mechanisms influencing HLA-G expression. Indeed, a fraction of HLA-G mRNA transcripts presenting the 14-bp Ins can be further processed (alternatively spliced) by the removal of 92 bases from the mature HLA-G mRNA, which yields smaller HLA-G transcripts, reported to be more stable than the complete mRNA forms (Donadi et al., 2011, Rousseau et al., 2003).
Interestingly in another study, G*01:01:02, in the heterozygous and homozygous state, was associated with increased expression of genital sHLA-G in HIV-1-infected sex workers compared with those in both the HIV-1-uninfected sex workers (P= 0.051) and non-sex workers (P=0.002) groups (Thibodeau et al., 2011). Additionally in a Canadian Human Papillomavirus (HPV) study, one of the most common sexually transmitted infections, both G*01:01:02 and G*01:03-alleles (also in high LD with the 14-bp Ins) were associated with increased risk for HPV-16 infection and persistent infections with HPV types from the alpha species (Ferguson et al., 2011). It is possible that polymorphisms within the HLA-G promoter region (5’URR, the upstream regulatory region) may influence HLA-G expression. For example, in a study of recurrent pregnancy loss, the tri-allelic polymorphism −725C/G/T (rs1233334) within the 5’URR was evaluated in relation to plasma sHLAG concentration, and the CC genotype was found to have significantly lower levels of sHLAG than the CG and CT genotypes (Jassem et al., 2012). Additionally, it was suggested that the presence of Guanine in position −725 may alter the methylation profile of CpG dinucleotides resulting in a modification of gene expression, influencing the binding of regulatory factors as IRF-1 (Interferon response factor-1) to ISRE (interferon specific regulation element) (Costa et al., 2012). Indeed, G*01:01:02 alleles were reported in 5’URR haplotypes having the −725C allele (Costa et al., 2012). It is also important to note that haplotype structures often differ between different ethnic groups and thus analysis of the 5’URR together with HLA-G alleles and the 3’UTR, and the respective haplotypes across this entire region with expression levels of HLA-G would be very informative in trying to determine the role of this molecule in HIV-1 MTCT.
Of note, while these two HLA-G variations [G*01:01:02 and +3187G (UTR1)] independently associated with increased risk for IU transmission, the effect of having either G*01:01:02 or +3187G (UTR1) was significantly over-represented in IU-TR mothers compared to NT mothers and was maintained throughout all adjustments. Interestingly, this association was stronger than the effect of G*01:01:02 alone, but weaker than that of UTR1 alone.
With regards to mother-infant concordance, we and others (Luo et al., 2013, Rousseau et al., 2003, Segat and Crovella, 2012) found no association for HLA-G alleles and the 14-bp indel with risk of vertical transmission. Only one study reported an association between HLA-G discordance at codon 57 in exon 2 in HIV-1 non-transmitting mother-infant pairs, but the sample size was small (n=34) and the study did not consider different routes of transmission (Aikhionbare et al., 2001). In keeping with this limitation, we acknowledge that in our study, we too may be biased in our analysis of the IU enriched group, IU2, as IU transmission is assumed based on maternal sdNVP treatment. Nevertheless, “true” IU infected infants were analysed on their own account and in most cases, associations within the IU enriched group (IU2) followed similar trends with the IU group.
In conclusion, we show that certain maternal HLA-G alleles (G*01:01:02) and 3’UTR polymorphisms (+3187G and UTR1) associated with an increased risk for IU MTCT, and that this is likely though modulation of maternal HLA-G expression at the maternal-foetal interface. However, further investigations in larger cohorts are necessary to confirm these associations and future studies should collectively assess HLA-G alleles as well as the 5’URR and the 3’UTR, as polymorphisms in all three regions might impact HLA-G expression.
Supplementary Material
Highlights.
HLA-G is an immunosuppressive molecule expressed at the maternal-foetal interface
We assessed both HLA-G alleles and the 3’UTR in HIV-1 mother-to child transmission
Two maternal HLA-G variants independently increased in utero (IU) HIV-1 transmission
IU mothers had increased G*01:01:02 and a SNP within the UTR1 haplotype, +3187G
Findings suggest that maternal expression of HLA-G can influence HIV-1 transmission
Acknowledgements
The authors would like to thank the participants and clinical and laboratory staff at the NICD. HH is the recipient of the National Research Foundation, Poliomyelitis Research Foundation and University of Witwatersrand Postgraduate Merit Award. This work is based on the research supported by grants from NICHD 42402, the Elizabeth Glaser Paediatric AIDS Foundation, and the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- AIKHIONBARE FO, HODGE T, KUHN L, BULTERYS M, ABRAMS EJ, BOND VC. Mother-to-child discordance in HLA-G exon 2 is associated with a reduced risk of perinatal HIV-1 transmission. AIDS. 2001;15:2196–8. doi: 10.1097/00002030-200111090-00019. [DOI] [PubMed] [Google Scholar]
- AIKHIONBARE FO, KUMARESAN K, SHAMSA F, BOND VC. HLA-G DNA sequence variants and risk of perinatal HIV-1 transmission. AIDS Res Ther. 2006;3:28. doi: 10.1186/1742-6405-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALVAREZ M, PIEDADE J, BALSEIRO S, RIBAS G, REGATEIRO F. HLA-G 3'-UTR SNP and 14-bp deletion polymorphisms in Portuguese and Guinea-Bissau populations. Int J Immunogenet. 2009;36:361–6. doi: 10.1111/j.1744-313X.2009.00875.x. [DOI] [PubMed] [Google Scholar]
- AMIOT L, VU N, SAMSON M. Immunomodulatory Properties of HLA-G in Infectious Diseases. Journal of Immunology Research. 20142014:14. doi: 10.1155/2014/298569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASTELLI EC, MENDES-JUNIOR CT, DEGHAIDE NH, DE ALBUQUERQUE RS, MUNIZ YC, SIMOES RT, CAROSELLA ED, MOREAU P, DONADI EA. The genetic structure of 3'untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun. 2010;11:134–41. doi: 10.1038/gene.2009.74. [DOI] [PubMed] [Google Scholar]
- CASTELLI EC, MOREAU P, OYA E CHIROMATZO A, MENDES-JUNIOR CT, VEIGA-CASTELLI LC, YAGHI L, GIULIATTI S, CAROSELLA ED, DONADI EA. In silico analysis of microRNAS targeting the HLA-G 3' untranslated region alleles and haplotypes. Hum Immunol. 2009;70:1020–5. doi: 10.1016/j.humimm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- CASTELLI EC, VEIGA-CASTELLI LC, YAGHI L, MOREAU P, DONADI EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res. 20142014:734068. doi: 10.1155/2014/734068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSTA CH, GELMINI GF, WOWK PF, MATTAR SB, VARGAS RG, ROXO VM, SCHUFFNER A, BICALHO MDA G. HLA-G regulatory haplotypes and implantation outcome in couples who underwent assisted reproduction treatment. Hum Immunol. 2012;73:891–7. doi: 10.1016/j.humimm.2012.06.002. [DOI] [PubMed] [Google Scholar]
- DA SILVA GK, VIANNA P, VEIT TD, CROVELLA S, CATAMO E, CORDERO EA, MATTEVI VS, LAZZARETTI RK, SPRINZ E, KUHMMER R, CHIES JA. Influence of HLA-G polymorphisms in human immunodeficiency virus infection and hepatitis C virus co-infection in Brazilian and Italian individuals. Infect Genet Evol. 2014;21:418–23. doi: 10.1016/j.meegid.2013.12.013. [DOI] [PubMed] [Google Scholar]
- DA SILVA RC, BEDIN E, MANGANO A, AULICINO P, PONTILLO A, BRANDAO L, GUIMARAES R, ARRAES LC, SEN L, CROVELLA S. HIV mother-to-child transmission: a complex genetic puzzle tackled by Brazil and Argentina research teams. Infect Genet Evol. 2013;19:312–22. doi: 10.1016/j.meegid.2013.03.005. [DOI] [PubMed] [Google Scholar]
- DONADI EA, CASTELLI EC, ARNAIZ-VILLENA A, ROGER M, REY D, MOREAU P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. 2011;68:369–95. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DURI K, GUMBO FZ, KRISTIANSEN KI, KUREWA NE, MAPINGURE MP, RUSAKANIKO S, CHIRENJE MZ, MULLER F, STRAY-PEDERSEN B. Antenatal HIV-1 RNA load and timing of mother to child transmission; a nested case-control study in a resource poor setting. Virol J. 2010;7:176. doi: 10.1186/1743-422X-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FABRIS A, CATAMO E, SEGAT L, MORGUTTI M, ARRAES LC, DE LIMA-FILHO JL, CROVELLA S. Association between HLA-G 3'UTR 14-bp polymorphism and HIV vertical transmission in Brazilian children. AIDS. 2009;23:177–82. doi: 10.1097/QAD.0b013e32832027bf. [DOI] [PubMed] [Google Scholar]
- FERGUSON R, RAMANAKUMAR AV, RICHARDSON H, TELLIER PP, COUTLEE F, FRANCO EL, ROGER M. Human leukocyte antigen (HLA)-E and HLA-G polymorphisms in human papillomavirus infection susceptibility and persistence. Hum Immunol. 2011;72:337–41. doi: 10.1016/j.humimm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- GUO SW, THOMPSON EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–72. [PubMed] [Google Scholar]
- HONG HA, PAXIMADIS M, GRAY GE, KUHN L, TIEMESSEN CT. KIR2DS4 allelic variants: Differential effects on in utero and intrapartum HIV-1 mother-to-child transmission. Clin Immunol. 2013;149:498–508. doi: 10.1016/j.clim.2013.09.005. [DOI] [PubMed] [Google Scholar]
- HVIID TV, HYLENIUS S, RORBYE C, NIELSEN LG. HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics. 2003;55:63–79. doi: 10.1007/s00251-003-0547-z. [DOI] [PubMed] [Google Scholar]
- HVIID TV, MELDGAARD M, SORENSEN S, MORLING N. Polymorphism of exon 3 of the HLA-G gene. J Reprod Immunol. 1997;35:31–42. doi: 10.1016/s0165-0378(97)00051-x. [DOI] [PubMed] [Google Scholar]
- JASSEM RM, SHANI WS, LOISEL DA, SHARIEF M, BILLSTRAND C, OBER C. HLA-G polymorphisms and soluble HLA-G protein levels in women with recurrent pregnancy loss from Basrah province in Iraq. Hum Immunol. 2012;73:811–7. doi: 10.1016/j.humimm.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOURTIS AP, BULTERYS M, NESHEIM SR, LEE FK. Understanding the timing of HIV transmission from mother to infant. JAMA. 2001;285:709–12. doi: 10.1001/jama.285.6.709. [DOI] [PubMed] [Google Scholar]
- KUHN L, ABRAMS EJ, PALUMBO P, BULTERYS M, AGA R, LOUIE L, HODGE T, PERINATAL ACTS. Maternal versus paternal inheritance of HLA class I alleles among HIV-infected children: consequences for clinical disease progression. AIDS. 2004;18:1281–9. doi: 10.1097/00002030-200406180-00006. [DOI] [PubMed] [Google Scholar]
- KUHN L, SCHRAMM DB, DONNINGER S, MEDDOWS-TAYLOR S, COOVADIA AH, SHERMAN GG, GRAY GE, TIEMESSEN CT. African infants' CCL3 gene copies influence perinatal HIV transmission in the absence of maternal nevirapine. AIDS. 2007;21:1753–61. doi: 10.1097/QAD.0b013e3282ba553a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAJOIE J, FONTAINE J, TREMBLAY C, ROUTY JP, POUDRIER J, ROGER M. Persistence of high levels of blood soluble human leukocyte antigen-G is associated with rapid progression of HIV infection. AIDS. 2009;23:1437–40. doi: 10.1097/QAD.0b013e32832d0825. [DOI] [PubMed] [Google Scholar]
- LE DISCORDE M, LE DANFF C, MOREAU P, ROUAS-FREISS N, CAROSELLA ED. HLA-G*0105N null allele encodes functional HLA-G isoforms. Biol Reprod. 2005;73:280–8. doi: 10.1095/biolreprod.104.037986. [DOI] [PubMed] [Google Scholar]
- LEWONTIN RC. The Interaction of Selection and Linkage. Ii. Optimum Models. Genetics. 1964;50:757–82. doi: 10.1093/genetics/50.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAU SW, MICKEY R, ROMANO P, LEE TD. Study of the HLA system in the Haitian population. Tissue Antigens. 1984;23:308–13. doi: 10.1111/j.1399-0039.1984.tb00050.x. [DOI] [PubMed] [Google Scholar]
- LUO M, CZARNECKI C, RAMDAHIN S, EMBREE J, PLUMMER FA. HLA-G and mother-child perinatal HIV transmission. Hum Immunol. 2013;74:459–63. doi: 10.1016/j.humimm.2012.11.023. [DOI] [PubMed] [Google Scholar]
- LYNGE NILSSON L, DJURISIC S, HVIID TV. Controlling the Immunological Crosstalk during Conception and Pregnancy: HLA-G in Reproduction. Front Immunol. 2014;5:198. doi: 10.3389/fimmu.2014.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACDONALD KS, EMBREE J, NJENGA S, NAGELKERKE NJ, NGATIA I, MOHAMMED Z, BARBER BH, NDINYA-ACHOLA J, BWAYO J, PLUMMER FA. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J Infect Dis. 1998;177:551–6. doi: 10.1086/514243. [DOI] [PubMed] [Google Scholar]
- MARTELLI-PALOMINO G, PANCOTTO JA, MUNIZ YC, MENDES-JUNIOR CT, CASTELLI EC, MASSARO JD, KRAWICE-RADANNE I, PORAS I, REBMANN V, CAROSELLA ED, ROUAS-FREISS N, MOREAU P, DONADI EA. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PLoS One. 2013;8:e71742. doi: 10.1371/journal.pone.0071742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATT C, ROGER M. Genetic determinants of pediatric HIV-1 infection: vertical transmission and disease progression among children. Mol Med. 2001;7:583–9. [PMC free article] [PubMed] [Google Scholar]
- MATTE C, LAJOIE J, LACAILLE J, ZIJENAH LS, WARD BJ, ROGER M. Functionally active HLA-G polymorphisms are associated with the risk of heterosexual HIV-1 infection in African women. AIDS. 2004;18:427–31. doi: 10.1097/00002030-200402200-00008. [DOI] [PubMed] [Google Scholar]
- MOODLEY S, BOBAT R. Expression of HLA-G1 at the placental interface of HIV-1 infected pregnant women and vertical transmission of HIV. Placenta. 2011;32:778–782. doi: 10.1016/j.placenta.2011.07.012. [DOI] [PubMed] [Google Scholar]
- NIU T, QIN ZS, XU X, LIU JS. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am J Hum Genet. 2002;70:157–69. doi: 10.1086/338446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAXIMADIS M, MINEVICH G, WINCHESTER R, SCHRAMM DB, GRAY GE, SHERMAN GG, COOVADIA AH, KUHN L, TIEMESSEN CT. KIR-HLA and maternal-infant HIV-1 transmission in sub-Saharan Africa. PLoS One. 2011;6:e16541. doi: 10.1371/journal.pone.0016541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEZZULLO JC. Logistic Regression, Version 05.07.20. 2005 http://statpages.org/logistic.html
- POLYCARPOU A, NTAIS C, KORBER BT, ELRICH HA, WINCHESTER R, KROGSTAD P, WOLINSKY S, ROSTRON T, ROWLAND-JONES SL, AMMANN AJ, IOANNIDIS JP, ARIEL P. Association between maternal and infant class I and II HLA alleles and of their concordance with the risk of perinatal HIV type 1 transmission. AIDS Res Hum Retroviruses. 2002;18:741–6. doi: 10.1089/08892220260139477. [DOI] [PubMed] [Google Scholar]
- REBMANN V, VAN DER VEN K, PASSLER M, PFEIFFER K, KREBS D, GROSSE-WILDE H. Association of soluble HLA-G plasma levels with HLA-G alleles. Tissue Antigens. 2001;57:15–21. doi: 10.1034/j.1399-0039.2001.057001015.x. [DOI] [PubMed] [Google Scholar]
- ROBINSON J, HALLIWELL JA, MCWILLIAM H, LOPEZ R, PARHAM P, MARSH SG. The IMGT/HLA database. Nucleic Acids Res. 2013;41:D1222–7. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROUSSEAU P, LE DISCORDE M, MOUILLOT G, MARCOU C, CAROSELLA ED, MOREAU P. The 14 bp deletion-insertion polymorphism in the 3' UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol. 2003;64:1005–10. doi: 10.1016/j.humimm.2003.08.347. [DOI] [PubMed] [Google Scholar]
- SABBAGH A, LUISI P, CASTELLI EC, GINEAU L, COURTIN D, MILET J, MASSARO JD, LAAYOUNI H, MOREAU P, DONADI EA, GARCIA A. Worldwide genetic variation at the 3' untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. Genes Immun. 2014;15:95–106. doi: 10.1038/gene.2013.67. [DOI] [PubMed] [Google Scholar]
- SANCHES RS, MASSARO JD, DA CRUZ CASTELLI E, MENDES-JUNIOR CT, DA PENNA MATOS FM, MARTINEZ J, HADDAD SK, QUINTANA SM, CERVI MC, TSUDA LC, DONADI EA, FERNANDES APM. Human Leukocyte Antigen-G 14 Base Pairs Polymorphism in the Human Immunodeficiency Virus −1 Perinatal Transmission. Open Access Scientific Reports. 2013:2. [Google Scholar]
- SEGAT L, CATAMO E, FABRIS A, PADOVAN L, MORGUTTI M, CROVELLA S. HLA-G 3' UTR haplotypes and HIV vertical transmission. AIDS. 2009;23:1916–8. doi: 10.1097/QAD.0b013e32832f8104. [DOI] [PubMed] [Google Scholar]
- SEGAT L, CROVELLA S. HLA-G 14bp del/ins genetic variation: association with susceptibility to human immunodeficiency virus-1 vertical transmission but not with human immunodeficiency virus-1 infection through horizontal transmission. Tissue Antigens. 2012;80:12–3. doi: 10.1111/j.1399-0039.2012.01874.x. [DOI] [PubMed] [Google Scholar]
- SEGAT L, ZUPIN L, KIM HY, CATAMO E, THEA DM, KANKASA C, ALDROVANDI GM, KUHN L, CROVELLA S. HLA-G 14 bp deletion/insertion polymorphism and mother-to-child transmission of HIV. Tissue Antigens. 2014;83:161–7. doi: 10.1111/tan.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIZZANO F, TESTI M, ZITO L, CROCCHIOLO R, TROIANO M, MAZZI B, TURCHIANO G, TORCHIO M, PULTRONE C, GREGORI S, CHIESA R, GAZIEV J, SODANI P, MARKTEL S, AMOROSO A, RONCAROLO MG, LUCARELLI G, CICERI F, ANDREANI M, FLEISCHHAUER K. Genotypes and haplotypes in the 3' untranslated region of the HLA-G gene and their association with clinical outcome of hematopoietic stem cell transplantation for beta-thalassemia. Tissue Antigens. 2012;79:326–32. doi: 10.1111/j.1399-0039.2012.01862.x. [DOI] [PubMed] [Google Scholar]
- SOLIER C, AGUERRE-GIRR M, LENFANT F, CAMPAN A, BERREBI A, REBMANN V, GROSSE-WILDE H, LE BOUTEILLER P. Secretion of pro-apoptotic intron 4-retaining soluble HLA-G1 by human villous trophoblast. Eur J Immunol. 2002;32:3576–86. doi: 10.1002/1521-4141(200212)32:12<3576::AID-IMMU3576>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- TAHA TE. Mother-to-child transmission of HIV-1 in sub-Saharan Africa: past, present and future challenges. Life Sci. 2011;88:917–21. doi: 10.1016/j.lfs.2010.09.031. [DOI] [PubMed] [Google Scholar]
- TAN Z, RANDALL G, FAN J, CAMORETTI-MERCADO B, BROCKMAN-SCHNEIDER R, PAN L, SOLWAY J, GERN JE, LEMANSKE RF, NICOLAE D, OBER C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–34. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIBODEAU V, LAJOIE J, LABBE AC, ZANNOU MD, FOWKE KR, ALARY M, POUDRIER J, ROGER M. High level of soluble HLA-G in the female genital tract of Beninese commercial sex workers is associated with HIV-1 infection. PLoS One. 2011;6:e25185. doi: 10.1371/journal.pone.0025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURK WJ, KIMANI J, BIELAWNY T, WACHIHI C, BALL TB, PLUMMER FA, LUO M. Associations of human leukocyte antigen-G with resistance and susceptibility to HIV-1 infection in the Pumwani sex worker cohort. AIDS. 2013;27:7–15. doi: 10.1097/QAD.0b013e32835ab1f2. [DOI] [PubMed] [Google Scholar]
- VEIT TD, CHIES JA. Tolerance versus immune response -- microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl Immunol. 2009;20:229–31. doi: 10.1016/j.trim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- YIE SM, LI LH, XIAO R, LIBRACH CL. A single base-pair mutation in the 3'-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol Hum Reprod. 2008;14:649–53. doi: 10.1093/molehr/gan059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


