Abstract
Social media postings are rich in information that often remain hidden and inaccessible for automatic extraction due to inherent limitations of the site’s APIs, which mostly limit access via specific keyword-based searches (and limit both the number of keywords and the number of postings that are returned). When mining social media for drug mentions, one of the first problems to solve is how to derive a list of variants of the drug name (common misspellings) that can capture a sufficient number of postings. We present here an approach that filters the potential variants based on the intuition that, faced with the task of writing an unfamiliar, complex word (the drug name), users will tend to revert to phonetic spelling, and we thus give preference to variants that reflect the phonemes of the correct spelling. The algorithm allowed us to capture 50.4 – 56.0 % of the user comments using only about 18% of the variants.
Keywords: Information Retrieval, Natural Language Processing and Free Text Data Mining, Spelling-Error
Introduction
The question to ask in information extraction from social media postings is not whether valuable information is present in the user data, but how to find it among the millions of daily postings and how to work around the limitations that these sites necessarily impose on automatic requests.
Social networking postings can indeed be a treasure trove of data. Twitter alone observes around 58 million tweets(3) per day. Obtaining the right ones might be tricky, however, even when using the site-provided APIs (Application Programming Interface) for data collection. For example, Twitter provides Streaming and Search APIs to collect tweets, but in order to collect tweets for a particular topic, appropriate keywords should be first selected and given to the API. Twitter allows to track up to 400 keywords per application key, returning all matching Tweets up to a volume equal to the streaming cap (which is about 1% of the totality of all public streamed tweets). The GooglePlus API also restricts calls up to 1000 requests per day.
Our ongoing work(1)(2) to extract mentions of adverse reactions of drugs directly from patient comments posted on social networks has exposed the significance and nuances of a common problem: medical terms, and specifically, drug names are particularly difficult for users to spell correctly, and frankly, they usually make no obvious effort to do so when posting a message online. Thus, given that for automatic collection of postings related to the drugs the drug name is the keyword used to obtain the postings, including misspelled versions of the drug name as keywords is important. Consider the following examples of Tweets obtained for various spellings of Seroquel:
@PsychoIogicaI HA! Not if you’re on # Seroquil. EXTREMELY vivid dreams that stay in conscious memory. Very # Freaky ! Any idea why?
@BipolarBlogger did you ever try the Seriquel XR??? It has a less sedative effect and has a longer lasting effect
Gone from 50mg to 150mg of Serequel last night. Could barely wake up this morning and I feel like my body is made of lead
@AndrewH_Smith Is the Inderal helpful? And yeah, they are short lasting but non addictive. You could try Seraquel too but it's pretty strong
However, algorithms to generate word variants (using 1 or 2 edit distance and typographical –keyboarding- errors, for example) produce in excess of 300 or more variants per drug name of average length. If we consider that the total number of drugs currently listed in DrugBank(4) website is around 6800, about 2 million keywords to track postings related to all drugs would be necessary, exceeding the limit imposed for an instance of the Twitter Streaming API crawler with only 2 or 3 drugs. Even if this limit could be bypassed via multiple application instances, handling and deploying such a large number of query terms might be impractical and unnecessary, as many misspellings are not common enough to warrant monitoring. On the other hand, without including the common misspellings, many postings would be missed. In fact, the number of postings that use the most common misspellings of drug names often exceed those that use correct spelling. Thus, we are faced with the problem of generating misspelled variants for a drug name and then filtering them to select the most common ones in order to remain within the crawling API limitations when mining drug-related postings in social media.
This paper describes a method to generate most probable misspelled drug name variants for querying social media postings. The method is based on the intuitive notion that people will tend to spell drug names phonetically, the “default” used by young children when spelling an unfamiliar word. Including these most probable misspelled variants allows us to collect valuable posts from social networking sites that we would have missed otherwise. There has been some prior work to this effect, but in general, most focus on correcting or detecting misspellings, not generating them. For example, Senger, et al.,(6) proposed an auto-correction algorithm to prevent errors in drug spelling. The web site Drugs.com allows a user to type a drug name phonetically (These approaches assume that all the text is available and then apply algorithms for spelling correction, on the contrary, in our task we have to generate keywords first to crawl the data from social media. An example of the later includes an approach to generate spelling variants for proper nouns, proposed by Bhagat and Hovy(5), in order to detect names of foreign places and people published after transliteration. Spelling mistakes of drug names can occur because of pronunciation error and typing errors. Hence it is important to consider both of these error types while generating probable misspelled versions of a drug name… Thus, our task is to generate a balanced list of keywords that can give maximum coverage (extracting a good portion of the useful comments) from social media sites. The rest of this paper covers the methods, evaluation, and results used for this task. For ease of reference, we will refer to the Twitter Streaming API, GooglePlus API and Facebook API collectively as the “crawling API”. Small variations in the APIs themselves are not relevant to the task.
Methods
Considering the possible ways the misspellings may occur in drug names, we sought to develop an algorithm to generate a list of all the likely misspelled variants of a drug name based on a simple 1-edit distance algorithm, and then filtering it using phonetic spelling. We evaluated the approach as to its ability to generate a list with maximum coverage of social networking postings for a minimum list size. For evaluation purposes, we thoroughly examined social network postings for 4 drugs – Paxil, Prozac, Seroquel and Olanzapine. The number of tweets collected directly from the user interface for Twitter for Paxil were 334 using 18 variants, for Prozac were 186 using 18 variants, for Seroquel 146 using 17 variants and for Olanzapine were 89 using 15 variants.
Tools and Dataset
We utilize three different social media resources in our system: Facebook, Twitter and GooglePlus. There are many phonetic spelling algorithms available. We choose to use the LOGIOS Lexicon Tool (7) and Metaphone library (8,9) as they are one of the most common APIs. We used these libraries to get variants with similar pronunciation for a drug name. The LOGIOS Lexicon Tool generates a list of different pronunciations by expanding the original word into machine readable pronunciation which is encoded using the modified form of Arpabet system(10).
The Metaphone phonetic algorithm is an improvement over the Soundex phonetic algorithm, where the words are encoded to the same representation so that they can be grouped despite minor differences. For example, Table 1 shows the encodings of words using the CMU pronunciation (11) and the Metaphone library. The words with similar pronunciations of “Prozac” obtained by the CMU library are “Prozak” and “Proxac”, whereas the similar pronunciation words obtained from Metaphone encoding are “Przac” and “Prozak”.
Table 1.
CMU and Metaphone encoding
| Word Variants | CMU Expanded Pronunciation | Metaphone Encoding |
|---|---|---|
| PROZAC* | P R OW Z AE K | PROZACPRSK |
| POZAC | P AA Z AH K | POZACPSK |
| PRZAC | P R Z AE K | PRZACPRSK |
| PROAC | P R OW AE K | PROACPRK |
| PROZAK | P R OW Z AE K | PROZAKPRSK |
| PROXAC | P R OW Z AE K | PROXACPRKS |
correct spelling of the word
Figure 1 shows the flowchart for the method used in this paper. The first step is to generate all variants of a word within 1-edit distance (Levenshtein distance, words that vary from the original by a single-character insertion, deletion, or substitution). The number of variants at even 1-edit distance to a drug name are very large and the count shoots up for 2 or more edit distances. Consider the drug “Paxil”: with only 5 characters in length, the number of variants obtained with 1-edit distance are 238. For the drug “Olanzapine” with length of 10 characters, there are 503 words within 1-edit distance. Table 2 shows the number of variants of drug names obtained by just 1-edit distance.
Figure 1.
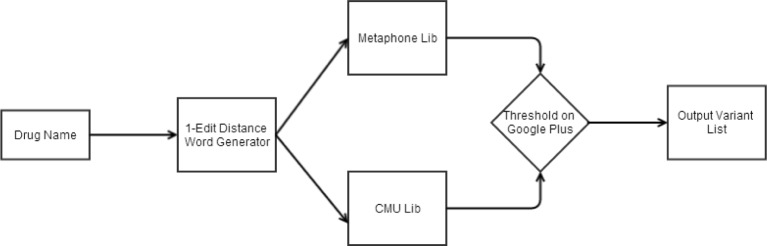
Control Flow Chart
Table 2.
Statistics of misspelled variants.
| Paxil | Prozac | Seroquel | Olanzapine | |
|---|---|---|---|---|
| Levenshtein (1-edit) distance words | 238 | 291 | 397 | 503 |
| CMU lib words generated | 21 | 18 | 27 | 31 |
| Metaphone words generated | 79 | 103 | 121 | 338 |
| Combining the two lists | 85 | 104 | 119 | 327 |
| Keywords selected by proposed algorithm | 18 | 18 | 17 | 15 |
Moving further, in order to compare the misspelled variants to the original word in pronunciation, we applied the CMU pronunciation and Metaphone algorithm to find those having the same pronunciation originating from the different spellings. For example, “Prozac” and “Prozak” have the same pronunciation. The results obtained from both libraries were useful, as they resulted in a significant number of social network postings, so we couldn’t right out eliminate any of them. For example, for the words “Paxil” and “Paxcil”, CMU lists the same pronunciation, whereas Metaphone does not, while Metaphone considers “Paxil” and “Paxial” as having similar pronunciation, and CMU does not. However, just combining the variants obtained from the two algorithms still results in a very large number of words, considering the limitations of the crawling API. For example, for the word “Paxil”, the number of variants in the combined list is 85 words (those with the same pronunciation as the original word). Table 2 shows the number of words obtained from the CMU and the Metaphone libraries, as well as the combination of both.
Thus, phonetic pronunciation alone, although it reduces the list by a third, might not be sufficient if one wishes to monitor more than a handful of drugs. To find out which of the given variants are common misspellings, we used the Google custom search API(12), issuing a query per variant composed of each of the selected misspelled variants that have the same pronunciation as the original drug name plus the word ‘drug’ (to reduce the “noise” generated by pages referring to other topics). Google hits were used as an estimate of how prevalent the misspelled word would be on the social networks. We then ranked the words according to the number of hits, and choose the top k, setting k to the rank where the rate of change of Google hits drops significantly. Using this threshold, the list of highly probable misspelled variants was set to the top k (18 for Paxil, as shown in Table 2).
The lists generated by the algorithm were used to obtain comments by users of the three social networking sites mentioned before (Twitter, Facebook, and Google Plus).
Evaluation Method 1
We used four drugs for the purpose of experiments. In order to evaluate our proposed approach, we want to compute the fraction of useful posts that the method was able to capture from the crawling API using the variants generated by the algorithm. Since it is not possible to retrieve the exact number of posts corresponding to a variant from the API due to its limitations, we retrieved the comments using screen scraping in order to get a true measure of misspelled variants. For the evaluation of this algorithm we used coverage of the sampled list as an evaluation metric. The coverage of sampled list can be defined as,
where the sampled list is the list selected from our algorithm and complete list is combined output of the CMU and Metaphone libraries. Comments Coverage give us fraction of tweets the method was able to capture using the sampled list. The metric “keywords coverage” evaluates the fraction of keywords used as tracking words.
Evaluation Method 2
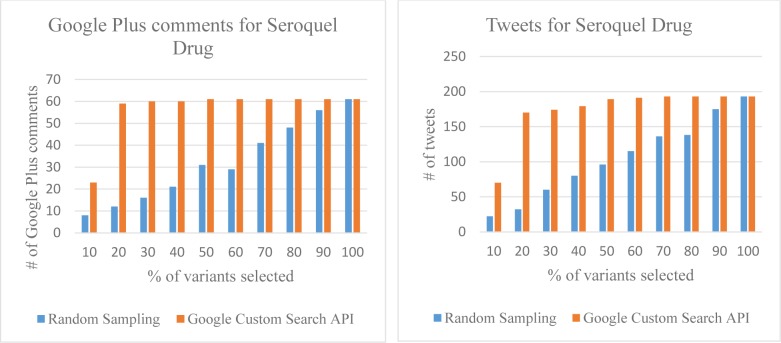
In this evaluation strategy we compared the results of our algorithm to a random keyword selector which is our baseline. We show that our algorithm produces a significant improvement over random keyword selector. The essence of our method is to capture a large amount of relevant data from a small variant list. Figure 3 show number of Google Plus comments and number of tweets collected respectively for random sample of variants and the variants sorted by Google Custom Search API.
Figure 3.
Google Plus comments and Tweets for Custom Search API sorted variants and Random variants.
Results
Evaluation Method 1
Figure 2 show the plots for number of comments obtained from Twitter vs the number of drug variants. The X-axis represents the combined list of drug variants obtained from the CMU pronunciation and the Metaphone library and the Y-axis represents the Google hits for the variant. The drug variants were ranked according to the Google hits obtained from the custom search API. It is evident from Figure 2 that total number of comments does not increase by much after a particular point. The algorithm allowed us to capture 50.42 – 55.97% of comments using about 18.29% of the variants.
Figure 2.
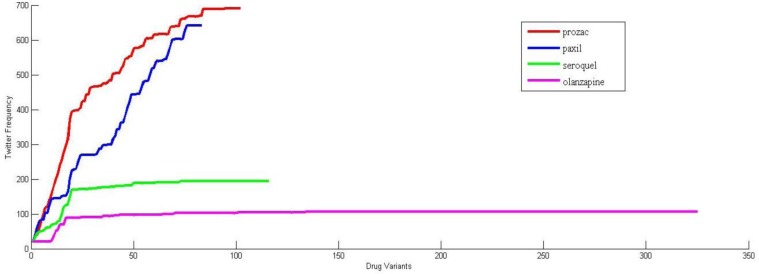
Number of Tweets vs Drug Variants.
Evaluation Method 2
Figure 3 shows that our method has an advantage when the keyword coverage is less, and that the method can capture useful data with minimal keyword coverage. For instance, for 20% keyword coverage the random selector captured 32 tweets while our approach captured 170 tweets for Seroquel.
Discussion
Our work focuses on finding a balance between the restrictions imposed by the crawling APIs and the many variants of a drug name needed to capture the large amount of data that hides behind these restrictions. Other approaches seek to correct misspelling by mapping the drug names from free text to standard nomenclature(13), but given the context of this work, these methods will fail in extracting data from social media given that all the data cannot be captured beforehand and then filtered out.
The main limitation of this algorithm is that some common misspellings that are due to typographical errors could be missed and might be commonly used. The false negative rate cannot be adequately computed since the universal set is not known. Moreover, the data obtained from social networking sites is complicated. For example, for the word Prozac, there are tweets that are not related to the drug “Local woodstock continues After the wild cats and a live connection with Ibiza now are the prozec mckenzie on stage … Tonight only”. The keyword coverage can be manipulated to achieve a higher comment coverage, adjustin for the number of drugs to track and API limits. Moreover, it is important to appreciate that crawling API is a resource which can be used in a better way if we have tracking words that are relevant.
Table 4.
Evaluation for Twitter and GooglePlus
| Drug Name | Twitter Comments Coverage | GooglePlus Comments Coverage | Keyword Coverage |
|---|---|---|---|
| Prozac | 45.44138929 | 52.7607362 | 17.30769231 |
| Paxil | 25.85669782 | 54.54545455 | 21.17647059 |
| Seroquel | 65.28497 | 62.29508 | 14.40678 |
| Olanzapine | 65.09433962 | 54.28571429 | 4.573170732 |
| Average | 50.417 | 55.971 |
Appendix
Table Top k drug name variants and their corresponding Google Hits obtained from our algorithm
| Prozac | Paxil | Seroquel | Olanzapine | ||||
|---|---|---|---|---|---|---|---|
| Variant | Google Hits | Variant | Google Hits | Variant | Google Hits | Variant | Google Hits |
| prozact | 3960000 | paxl | 52300000 | seroquels | 1910000 | olanzapin | 1220000 |
| prozaac | 3160000 | pxil | 12200000 | seroqul | 1810000 | olanzapoine | 869000 |
| prozaqc | 1300000 | pexil | 10600000 | seroqual | 1810000 | olanzapines | 868000 |
| prozaxc | 1300000 | paxol | 2490000 | sroquel | 1800000 | olanzaoine | 864000 |
| prozax | 1270000 | paxial | 2340000 | seruquel | 1790000 | olanzaopine | 863000 |
| prozc | 1260000 | paxiol | 866000 | saroquel | 1760000 | olanzapne | 796000 |
| prozec | 1260000 | paxill | 856000 | seroqel | 1710000 | olanzaplne | 765000 |
| proazac | 1260000 | paxilk | 819000 | seroquell | 1230000 | olanzapuine | 734000 |
| prozzac | 1220000 | paxilo | 809000 | serocquel | 763000 | olanzapins | 567000 |
| prazac | 1210000 | paxils | 790000 | seroguel | 751000 | olanzpine | 565000 |
| proazc | 1180000 | paxilv | 750000 | seroquol | 742000 | olanzopine | 536000 |
| proxac | 1150000 | paxilj | 746000 | sereoquel | 676000 | olanzipine | 530000 |
| prozacs | 1120000 | paxiln | 738000 | seriquel | 615000 | olanazapine | 525000 |
| prizac | 1100000 | paxilq | 738000 | serroquel | 604000 | olanzepine | 386000 |
| przac | 1070000 | paxcil | 708000 | serequel | 111000 | olanzapinm | 6820 |
| porzac | 997000 | paxiul | 694000 | seraquel | 106000 | ||
| prozacc | 995000 | paxilz | 668000 | seroquela | 5580 | ||
| prozaq | 12500 | paxila | 5700 | ||||
References
- 1.Nikfarjam A, Gonzalez GH. Pattern mining for extraction of mentions of Adverse Drug Reactions from user comments. AMIA Annu Symp Proc. 2011 Jan;2011:1019–26. [PMC free article] [PubMed] [Google Scholar]
- 2.Leaman R, Wojtulewicz L, Sullivan R, Skariah A, Yang J, Gonzalez G. Towards Internet-Age Pharmacovigilance : Extracting Adverse Drug Reactions from User Posts to Health-Related Social Networks. 2010. Jul, pp. 117–25.
- 3.Twitter Statistics [Internet] Available from: http://www.statisticbrain.com/twitter-statistics/
- 4.DrugBank Statistics [Internet] Available from: http://www.drugbank.ca/stats.
- 5.Bhagat R, Hovy E, Way A, Rey M Del. Phonetic Models for Generating Spelling Variants.
- 6.Senger C, Kaltschmidt J, Schmitt SPW, Pruszydlo MG, Haefeli WE. Int J Med Inform. 12. Vol. 79. Elsevier; Ireland Ltd: 2010. Dec, Misspellings in drug information system queries: characteristics of drug name spelling errors and strategies for their prevention; pp. 832–9. [cited 2013 Oct 5] [DOI] [PubMed] [Google Scholar]
- 7.LOGIOS Lexicon Tool [Internet] Available from: http://www.speech.cs.cmu.edu/tools/lextool.html.
- 8.Metaphone Wiki [Internet] Available from: http://en.wikipedia.org/wiki/Metaphone.
- 9.Soundex Wiki [Internet] Available from: http://en.wikipedia.org/wiki/Soundex.
- 10.Arpabet System [Internet] Available from: http://en.wikipedia.org/wiki/Arpabet.
- 11.CMU Pronouncing Dictionary [Internet] Available from: http://en.wikipedia.org/wiki/CMU_Pronouncing_Dictionary.
- 12.Google Custom Search API. Available from: https://developers.google.com/custom-search/
- 13.Levin MA, Krol M, Ph D, Doshi AM, Reich DL. Extraction and Mapping of Drug Names from Free Text to a Standardized Nomenclature AMIA 2007 Symposium Proceedings Page – 438 AMIA 2007 Symposium Proceedings Page – 439. 2007. pp. 438–42. [PMC free article] [PubMed]