Abstract
The MURDOCK Study is longitudinal, large-scale epidemiological study for which participants’ medication use is collected as free text. In order to maximize utility of drug data, while minimizing cost due to manual expert intervention, we have developed a generalizable approach to automatically coding medication data using RxNorm and NDF-RT and their associated application program interfaces (APIs). Of 130,273 entries, we were able to accurately map 122,523 (94%) to RxNorm concepts, and 106,135 (85%) of those drug concepts to nodes under the Drug by VA Class branch of NDF-RT. This approach has enabled use of drug data in combination with other complementary information for cohort identification within an i2b2-based participant registry. The method may be generalized to other projects requiring coding of medication data from free-text.
Introduction and Background
Standardized drug terminologies are useful to facilitate data sharing, and to ensure semantic interoperability across organizations [1]. Even when medication data is not collected in a coded manner, RxNorm1 and the VA National Drug File Reference Terminology (NDF-RT)2 have, with certain caveats [1], proven useful for normalizing both structured and free text data from electronic health records (EHRs) [2, 3]. We have taken a similar approach to mapping participant-provided drug information to RxNorm and NDF-RT to enable cohort identification using the i2b2 platform [4].
Medication information is an important facet of a person’s medical history. Medication data from EHRs may be limited to prescriptions taken as an inpatient or prescribed by a clinician at the health care facility in question. In addition, over-the-counter medications, vitamins, and supplements may not be included. The work described here was done in the context of the MURDOCK Study, a long-term epidemiological study aimed at reclassifying human health and disease based on molecular mechanism rather than the macroscopic observations that have been used for hundreds of years [5]. Participants in the study provide blood and urine biospecimens, along with self-reported clinical, medication, demographic, lifestyle, and medical history data. They also provide consent to annual follow up and access to their EHRs. The MURDOCK Study has an advantage over EHR-only projects in that medication data is to be provided both from EHRs and as self-reported information. To date, approximately 9600 participants have been enrolled out of an ultimate goal of 50,000 participants.
Here we describe the successes, limitations, and caveats of coding free text medication data using RxNorm and NDF-RT in the context of patient-reported information and contrast these factors with those described previously using EHR-derived medication data [3].
Methods
A graphical overview of the method described in this manuscript is given in Error! Reference source not found.. Participant-reported medications were collected by study staff as free text (A) annually for all participants in a community-based registry [6]. These free text medication entries were mapped, where possible, to RxNorm CUIs (Concept Unique Identifiers) using the National Library of Medicine (NLM)’s RxNorm REST API (B). A hierarchical structure of drug classes was developed based on the NDF-RT’s “Drug Products by VA Class” subtree (C) by replacing drug concepts containing attributes such as route and dosage (e.g., ibuprofen 200mg oral) with ingredient concepts (e.g., ibuprofen) and brand name concepts (e.g., Advil) (D). Multiple or single ingredient RxNorm concepts in our dataset were mapped to this hierarchy, followed by brand name terms as leaf nodes to the ingredient or sets of ingredients.
Mapping free text medication data to RxNorm
Participants were instructed to list medications by generic or brand names, leaving out other attributes such as route, strength, and form. To facilitate the accurate collection of medication information, participants were requested to bring their medications to the enrollment visit. The NLM RxNorm API was run on free text data from all participants enrolled in the MURDOCK registry as of 7 June 2013. A total of 130,273 medication entries were present, representing 18,924 unique terms reported by 9432 participants. An additional 2,579 entries (16 unique) included non-medication terms, e.g. “no medications”, ”can’t afford drugs”, etc. and were excluded from the analysis.
An attempt was made to detect perfect string matches first (http://rxnav.nlm.nih.gov/REST/drugs?name=value). For those that did not return a perfect match the approximate match resource was used, e.g. http://rxnav.nlm.nih.gov/REST/approx?term=aspirin.3 This resource returns 0 or more RxNorm terms that match the input string, in this case aspirin. The output includes a set of potential matches along with their RxCUI, score (an integer between 1 and 100 that measures the similarity between the input string and the candidate RxNorm term), and rank. Details on the approximate match algorithm are available elsewhere [7]. In the case of multiple matches, a winner was chosen using the following rules:
Highest score, or in case of tie:
Non-proprietary > proprietary source terminology (RxNorm content is derived from 11 source terminologies, some of which are proprietary.)
○ Among non-proprietary RxNorm concepts > NDF-RT concepts
RxNorm concept type: ingredient name (IN) > brand name (BN)
Results were categorized as follows:
Category A: Perfect match (no score assigned)
Category B: Score == 100 for exactly 1 term, and that one is non-proprietary
Category C: Score == 100 for more than 1, and winner is non-proprietary
Category D: Score == 100 for proprietary only (whether 1 or more)
Category E: Match score < 100
Category F: No match found
Category E was further divided into E3: scores (s) < 50; E2: 50 ≤ s < 75; and E1: s ≥ 75.
Category A matches were inspected visually and determined to be 100% accurate. For each of the remaining categories and sub-categories (B, C, D, E1–E3), 100 term mappings were selected at random and reviewed by an analyst with clinical expertise who determined whether the mapping was correct. In addition, all Category B matches (n=17) were reviewed. Therefore, 517 approximate matches were manually reviewed and evaluated for accuracy.
Mapping to an NDF-RT-based hierarchy
In order to be able to query for drugs by class, as opposed to name or ingredients, it was necessary to map RxNorm terms to another terminology that included categorization. NDF-RT’s “Drug Products by VA Class” hierarchy has been used for this purpose. NDF-RT leaf nodes, however, include dose and route information, e.g. “ibuprofen, 20mg tablet, oral”. In contrast, the medication data collected (and thus the corresponding RxCUIs) generally included only drug names, not strength or route, e.g. “ibuprofen, 200mg tablet, oral.” It was therefore necessary to rebuild the NDF-RT hierarchy down to the drug name level, without route or dose information. This revised version was designed to include brand names relationships in the hierarchy so that, for example, a query for participants taking ibuprofen would also return participants who reported taking Advil, Midol, Motrin, etc. This was accomplished through the following steps:
For each of the original leaf nodes (e.g., carbamazepine 100mg tab, chewable and carbamazepine 100mg/5ml susp)
Identify the term type (TTY) of each RxCUI for each NDF-RT leaf node.
“Walk” the NDF-RT ontology back to ingredient[s]. This is done by taking advantage of the relationships between different term types within RxNorm, e.g. clinical drug form (SCDF) has a “has_ingredient” relationship to ingredient (IN).
If the drug comprised multiple ingredients, append a multiple-ingredient child node onto the initial hierarchy.
For those mapped only to a single ingredient, walk to ingredient and create a child node under NDF-RT.
If the RxNorm concept had no relationships:
Map NDF-RT product component[s] (converting from precise ingredient (PIN) to ingredient (IN) if applicable) to [multiple] ingredients and create a node if one does not already exist.
Parse drug name and use string match to identify ingredient or brand name and create a node if one does not already exist.
Create child nodes of the nodes created above for each brand associated with that ingredient (or set of ingredients).
In the modified NDF-RT VA Drug Class hierarchy, RxCUIs for MURDOCK drug entries corresponded directly to the Ingredient (or Multiple Ingredient) and Brand Name nodes.
Results
Mapping free text to RxNorm
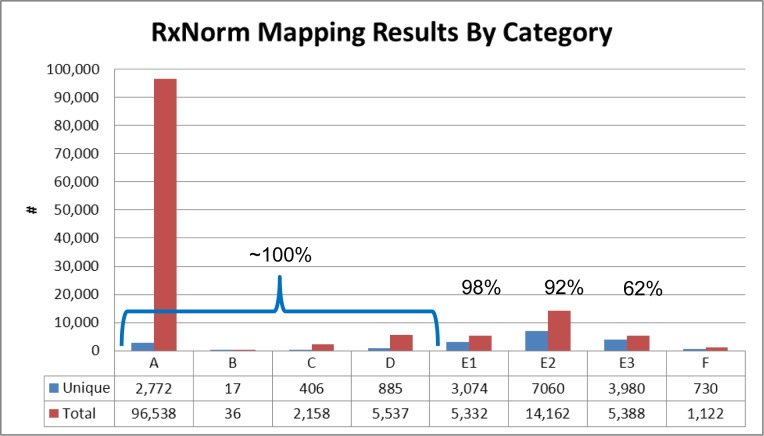
Out of 9432 participants, 8356 indicated taking one or more medications (including OTC medications, vitamins, and supplements) at one or more time points. The terms entered largely did not include dosages or delivery mechanism. This resulted in 130,273 total (18,924 unique) drug name entries. As illustrated in Figure 2, 99,538 entries (76%) of terms were perfect matches. On the other hand, the majority of the unique terms fell into category E (14,114 out of 18,924; 75%). This was to be expected as there are a number of different incorrect ways to spell a given drug name, and only one correct way.
Figure 2:
Distribution of input terms across scoring categories. A. Perfect matches; B: Score == 100 for exactly 1 term, and that one is non-proprietary; C: Score == 100 for more than 1, and winner is non-proprietary; D: Score == 100 for proprietary only (whether 1 or more); E1: 75 ≤ Match score < 100; E2: 50 ≤ Match score < 75; E3 Match score < 50; F: No match found.
Based on manual expert review of a random sample from each category, accuracy rates were determined to be 100% for categories A–D, and 98%, 92%, and 62% for E1, E2, and E3 respectively. Thus 104,269 (80%) of the total terms could be mapped to concepts in RxNorm with near 100% accuracy. Another 5332 (4%) could be mapped with approximately 98% accuracy, and 14,162 (11%) with accuracy of approximately 92%.
Choosing the appropriate threshold level below which to reject matches was subjective. While choosing a score of 100 or 75 would have maintained the highest level of accuracy, a significant number of entries (14,162 term entries; 7060 unique terms) fell into category E2. It was therefore decided to include through category E2 (scores ≥ 50) to maximize coverage, and keep the score to indicate confidence level.
Using a cutoff threshold of ≥ 50 (i.e. excluding only category E3), we were able to map 123,763 terms (Error! Reference source not found.). Extrapolating from our established rates of accuracy in each category, ~94% (122,523) of the total terms were mapped correctly, with 5% unmapped, and a “false positive rate” (i.e. mapped, but incorrectly) of approximately 1% (1,240 entries). Manual review suggests that missing and incorrect mappings were primarily over-the-counter medications, vitamins, minerals, and other supplements.
Mapping RxNorm to NDF-RT
104,653 (2158 unique) of the 123,763 (3137 unique) RxNorm terms (85% of the terms; 69% of the unique terms) obtained from mapping text to RxCUIs could be mapped directly to VA Class (Error! Reference source not found.). Many of the remaining 19,110 (969 unique) terms are not within scope for the NDF-RT, e.g. medical devices and brand names. Additional transformations, e.g. further tree walking, parsing and remapping, brought these numbers up to 106,135 (86%) of the total terms, representing 2790 (89%) unique RxCUIs.
Discussion and Conclusion
In this study, we designed and evaluated a method for automatically coding free-text medication data into standard terminologies. The method has some important strengths: 1) fully-automated; 2) accurate; 3) leverages publicly available tools and standard terminologies; and 4) can be applied to other uses cases that also involve free-text medication data.
In the planning phase of this project, one proposed option for drug coding was to use a hybrid model of automated and manual coding. However, cost estimates for professional coding at the level that is commonly performed for clinical trials were on the order of $1 million for full coding of 50,000 participants, including 25% manual coding. In evaluating coverage, accuracy, and cost of the mapping approach, it is important to consider the use case for which this system is intended. In the case of the MURDOCK Study, ascertainment of concomitant medications is a means to an end, namely identifying specific participant cohorts for follow-up studies. Importantly, this data itself is not to be used for, e.g. monitoring severe adverse events or FDA submission. Therefore, the automated coding approach we have described is a viable solution to standardization of free text drug data.
To evaluate the performance of the RxNorm API matching algorithms, Peters et al. mapped clinical drug terms (e.g., metoprolol succinate 200mg tab) from different drug formularies to RxNorm concepts [7]. The evaluation resulted in a recall of 61% to 76% of unique terms compared to 75% in our study, despite the fact that our data set had a large number of misspelled terms. This similar performance demonstrates the robustness of the RxNorm matching algorithm for different data sources, further indicating the generalizability of our method to other use cases and data sets.
Previous efforts to code existing medication data have been performed primarily on data collected through clinical care, whether structured or free text [3]. Our efforts to apply the RxNorm to participant-reported data adds additional complications in that fewer details were included (i.e. no dose or route, making mapping to NDF-RT more difficult) and participants are less familiar with drug names and spellings. On the other hand, the designation of medication data is more straightforward in this context than previously reported use of NLP to extract medication information from notes in EHRs. Our mapping success rate therefore shows immediate promise for easy application of RxNorm and its related APIs to participant-reported medications in research contexts.
Limitations
By relying on NDF-RT, we are constrained to using only the class assigned to each drug in the legacy VA Drug class system. However, many drugs fall into different categories and/or are used for multiple different indications. In order to use drug information to identify cohorts, particularly without knowledge of dosage, it was critical to also collect the reason for taking the drug. For example, the “antiparkinson agents” class of drugs in NDF-RT, is a reasonable start for identifying a cohort of Parkinson’s patients. However, because antiparkinson agents may be used for other conditions, e.g. restless leg syndrome, it is necessary to search both by drug type and reason taken in order to avoid false positives.
The accuracy threshold chosen in this study might not apply to all other systems or contexts. For example, investigators interested in the use of vitamins, minerals, and dietary supplements might have lowered the acceptable threshold, as those types of products tended to have lower matching scores. In contrast, a higher threshold might be justified for a use case in which false positives were particularly problematic.
In addition to the evaluation above, which focuses on the proportion of total entries and unique entries that were accurately mapped to the drug terminologies, two additional questions one might ask are what percentage of people who tried to report taking a given drug do we know reported taking it (e.g. if “asprin” mapped correctly to aspirin, but “asparin” did not, we would only know about those who misspelled it one way and not the other), and what percentage of people do we think reported taking a drug who in actuality did not (e.g. askarin was mapped to aspirin, but the participant actually misspelled the brand name Akarin)? However, these questions are highly drug specific. This type of analysis may be done in the future if it becomes relevant for a specific use case.
Future work
The Anatomical Therapeutic Chemical (ATC) is a terminology maintained by the WHO Collaborating Centre for Drug Statistics Methodology. It offers categorization analogous to that provided by NDF-RT. During the course of the work described here, the Anatomical Therapeutic Chemical (ATC) classification system was mapped to RxNorm. Future work will include evaluation of ATC as an alternative to NDF-RT, as it appears to address some shortcomings of NDF-RT, for example providing more than one class for a given drug (NDF-RT currently does this only for a very few cases), or the presence of certain drug classes not present in NDF-RT (e.g., selective serotonin reuptake inhibitors). It has been shown that the overlap in drug-class pairs between NDF-RT and ATC is poor [8]. Time and user input will determine whether NDF-RT is sufficient for the task at hand in the MURDOCK registry, or whether an alternative terminology is needed.
Figure 1:
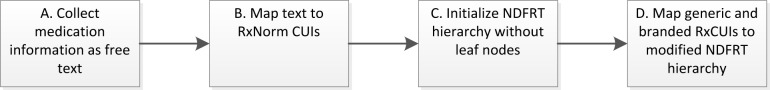
Methods overview.
Figure 3:
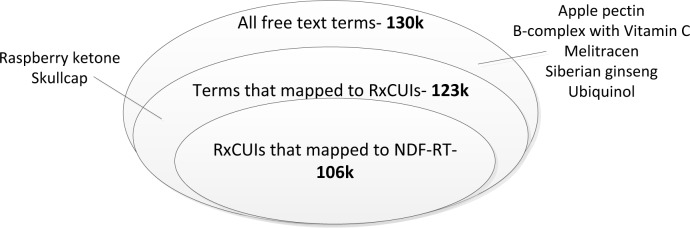
Successful mapping rates from terms to RxCUIs and from RxCUIs to NDF-RT classes. (Not to scale.)
Acknowledgments
This work was funded by Duke’s Clinical and Translational Science Award UL1RR024128 and a gift from David H. Murdock.
Footnotes
http://www.nlm.nih.gov/research/umls/rxnorm/ (Accessed 10/9/13)
http://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/NDFRT/ (Accessed 10/9/13)
The approxMatch function used here was released in September 2011. A similar function, approximateTerm, was released in May 2013 that gives similar results but provides additional output control, e.g. maximum number of entries returned.
References
- 1.Pathak J, Chute CG. Further revamping VA’s NDF-RT drug terminology for clinical research. J Am Med Inform Assoc. 2011;18(3):347–8. doi: 10.1136/amiajnl-2011-000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palchuk MB, et al. Enabling Hierarchical View of RxNorm with NDF-RT Drug Classes. AMIA Annu Symp Proc. 2010;2010:577–81. [PMC free article] [PubMed] [Google Scholar]
- 3.Pathak J, et al. Using RxNorm and NDF-RT to classify medication data extracted from electronic health records: experiences from the Rochester Epidemiology Project. AMIA Annu Symp Proc. 2011;2011:1089–98. [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy SN, et al. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc. 2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 5.Tenenbaum JD, et al. The MURDOCK Study: a long-term initiative for disease reclassification through advanced biomarker discovery and integration with electronic health records. Am J Transl Res. 2012;4(3):291–301. [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya S, et al. The Measurement to Understand Reclassification of Disease of Cabarrus/Kannapolis (MURDOCK) Study Community Registry and Biorepository. Am J Transl Res. 2012;4(4):458–70. [PMC free article] [PubMed] [Google Scholar]
- 7.Peters L, Kapusnik-Uner JE, Bodenreider O. Methods for managing variation in clinical drug names. AMIA Annu Symp Proc. 2010;2010:637–41. [PMC free article] [PubMed] [Google Scholar]
- 8.Mougin F, Burgun A, Bodenreider O. Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium. ACM; 2012. Comparing drug-class membership in ATC and NDF-RT. [Google Scholar]