Abstract
The rapid adoption of electronic health records (EHR) provides a comprehensive source for exploratory and predictive analytic to support clinical decision-making. In this paper, we investigate how to utilize EHR to tailor treatments to individual patients based on their likelihood to respond to a therapy. We construct a heterogeneous graph which includes two domains (patients and drugs) and encodes three relationships (patient similarity, drug similarity, and patient-drug prior associations). We describe a novel approach for performing a label propagation procedure to spread the label information representing the effectiveness of different drugs for different patients over this heterogeneous graph. The proposed method has been applied on a real-world EHR dataset to help identify personalized treatments for hypercholesterolemia. The experimental results demonstrate the effectiveness of the approach and suggest that the combination of appropriate patient similarity and drug similarity analytics could lead to actionable insights for personalized medicine. Particularly, by leveraging drug similarity in combination with patient similarity, our method could perform well even on new or rarely used drugs for which there are few records of known past performance.
Introduction
In contrast to the one-size-fits-all medicine, personalized medicine aims to tailor treatment to the individual characteristics of each patient. This requires the ability to classify patients into subgroups with predictable response to a specific treatment. The field of pharmacogenetics/pharmacogenomics has made important contributions to this problem for more than 50 years1. Ideally, personalized medicine will enable targeted prescription of any given treatment to only the likely responders, to avoid adverse reactions and expensive treatments in non-responders. Although there are already many examples of personalized medicine by leveraging genetics/genomics information in current practice2, such information is not yet widely available in everyday clinical practice, and is insufficient since it only addresses one of many factors affecting response to medication.
With the tremendous growth of the adoption of EHR, various sources of clinical information (e.g., demographics, diagnostic history, medications, laboratory test results, vital signs) are becoming available about patients. Recently, some treatment comparison studies3, 4 were conducted based on data from EHR of a cohort of clinically similar patients who received the treatments previously and whose outcomes were recorded. There are also some studies5, 6 of combining clinical and genetics/genomics information in selecting optimal clinical treatments. Existing approaches using clinical information for personalized medicine rely on large amounts of real-world data regarding the target treatment itself, which may not be available for new drugs or rarely-used treatments.
Drug similarity analytics aims to find drugs which display similar pharmacological characteristics to the drug of interest. The similarity analytics is usually conducted based on one or more types of drug characteristics (e.g., chemical structures, biological targets, indications, side-effects, and gene expression profiles). Drug similarity analytics has been widely used in drug repositioning7–9, drug side-effects prediction10, drug-target interactions prediction11, and drug-drug interactions prediction12, 13 applications. This approach has been shown to deliver competitive or even better accuracy to more complex, feature-vector-based methods9, 11 (e.g., support vector machines, random forests). In this study, we used drug similarity analytics to transmit EHR clinical information from well-studied drugs (i.e., drugs with many EHR records) to rarely-studied drugs (i.e., drugs with no or few EHR records).
Patient similarity analytics aims to find patients who display similar clinical characteristics to the patient of interest. The goal is to derive clinically meaningful distance metrics to measure the similarity between patients represented by their key clinical indicators. The resulting individualized insight of patient similarity analytics includes suggestions on how to manage care delivery to the patient (especially for patients has multiple diseases), and predictions of health issues that could arise in the future (because patients with similar characteristics had experienced such health issues). With the right patient similarity in place, patient similarity analytics have been used in the target patient retrieval14, medical prognosis15, 16, risk stratification17, 18, and clinical pathway analysis19 tasks.
In this study, we used patient similarity analytics to transmit EHR treatment information from training patients (i.e., patients with known effective treatments) to target patients (i.e., patients with no known effective treatment information).
In this paper, we construct a heterogeneous graph which includes two domains (i.e., patients and drugs) and encodes three relationships (i.e., patient similarity, drug similarity and patient-drug prior associations), and propose a heterogeneous label propagation algorithm which can be used to generate personalized drug recommendations by leveraging patient similarity and drug similarity analytics. To our best knowledge, the heterogeneous graph formulation of the EHR data has not been proposed in any previous literature. The label propagation model over heterogeneous graph by leveraging both patient similarity and drug similarity analytics is also significantly different from existing label propagation models.
Methodology
In this section we introduce the details of our method on how to combine patient and drug similarity analytics for personalized recommendations. There are three key components in our approach: drug similarity evaluation, patient similarity evaluation, and drug personalization.
Drug Similarity Evaluation
We used and compared chemical structure and drug target information to measure drug similarity. For chemical structure information, each drug was represented by an 881-dimensional binary profile whose elements encode for the presence or absence of each PubChem substructure by 1 or 0, respectively. Then we used the Tanimoto coefficient (TC), also known as the Jaccard index, to compute chemical structure similarities between all drug pairs. The TC between two vectors A and B is defined as the ratio between the number of features in the intersection to the union of both fingerprints: TC(A,B) = |A∩B|/|A∪B|. For drug target information, we collected all target proteins for each drug from DrugBank20. Then we calculated the pairwise drug target similarity between drugs dx and dy based on the average of sequence similarities of their target protein sets:
where given a drug d, we presented its target protein set as P(d); then |P(d)| is the size of the target protein set of drug d. The sequence similarity function of two proteins SW was calculated as a Smith-Waterman sequence alignment score21.
Patient Similarity Evaluation
We used co-occurring ICD9 diagnosis code information to measure patient similarity for simplicity and consistency purposes. In particular, we aggregated the longitudinal records of individual patients into a set of patient feature vectors, where each patient is a binary vector of ICD9 diagnosis categories. Then we used TC to compute similarities between all patient vectors.
Drug Personalization
As stated in the introduction, the basic question we want to answer for personalized medicine is “whether drug A is likely to be effective for specific patient B”. To take into consideration the specific condition of patient B as well as the characteristics of drug A, we propose to leverage the information of the patients who are clinically similar to patient B as well as the drugs which are similar to drug A. Moreover, we also considered the prior associations between patients and drugs, which were measured by the TC between ICD9 diagnosis of patients and ICD9-format drug indications from MEDI database22 (MEDI is an ensemble medication indication resource, which was created based on multiple commonly used medication resources by leveraging natural language processing techniques). In this way, we constructed a heterogeneous graph illustrated in Figure 1, which includes two domains (patients and drugs) and encodes three relationships (patient similarity, drug similarity and patient-drug prior associations). In the following we present a concrete heterogeneous label propagation algorithm to answer the question proposed at the beginning of this paragraph.
Figure 1.
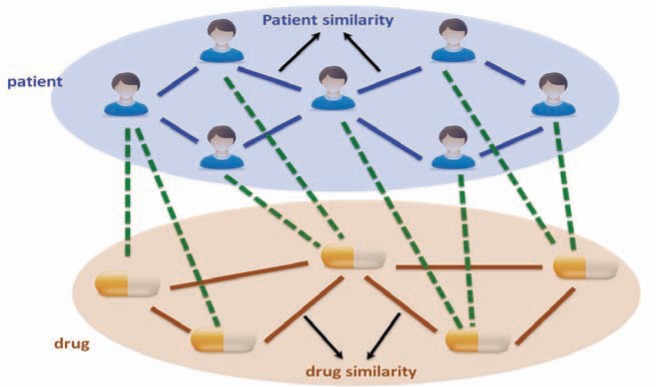
Illustration of the proposed heterogeneous label propagation method. The heterogeneous graph constructed with patients and drugs, where patient is one domain and drug is another domain. There are three types of relationships encoded in this graph: patient similarities, which are the blue edges; drug similarities, which are the yellow edges; patient-drug prior associations, which are the green dashed edges.
Suppose we have a set of patients P={p1, p2,…, pn}, where n is the number of patients with pi representing the i-th patient, and a set of drugs D={d1 ,d2 ,…, dm}, where m is the number of drugs with dj representing the j-th drug. Let Sp be the patient similarity matrix of size n×n with its (i,j)-th entry representing the similarity between pi and pj; Sd be the drug similarity matrix of size m×m with its (i,j)-th entry representing the similarity between di and dj (in this study, the drug similarity comes from either chemical structure or drug target information source); and R be the patient-drug prior association matrix of size n×m with its (i,j)-th entry representing the association between pi and dj (in this study, the prior association comes from TC of patient diagnosis codes and drug indications). Then we can form a composite (n+m) × (n+m) patient-drug similarity matrix A by concatenating the three matrices as . For each drug d, we constructed a corresponding effectiveness vector y=[y1, y2,…, yn yn+1,…,yn+m]T where yk=1 (k=1,2,…,n) if d is an effective treatment for patient k, yk=1 (k=n+1,n+2,…,n+m) if d is the (k-n)-th drug, otherwise yk=0. In this way, the effectiveness vector for each drug is just like a “label” vector on the heterogeneous graph shown on Figure 1, where it has nonzero entries if the drug is effective for the corresponding nodes (for patients) or is the node itself (for drug nodes). The goal is to predict the values of those zero entries (for patient nodes, those are the entries indicating whether this drug will be effective or not for them; for drug nodes, those are the entries indicating whether this drug would be similar to them in real-world clinical usage). If we concatenate all effectiveness vectors for the m drugs, we can form a drug effectiveness matrix Y=[y1, y2,…, ym]. Then we adopted a label propagation procedure to spread the label information in Y for the whole graph. Over this heterogeneous graph, patients propagate their known effective treatments to other patients based on the patient similarity analytics, and drugs propagate their target effective patients to other drugs based on the drug similarity analytics simultaneously to derive the relevance between nodes until achieving a steady state. After label propagation, possibilistic label (i.e., the possibility when a drug is effective for a patient) matrix F can be obtained by a formula F=(1−μ)(I−μW)−1Y (for details please refer to Wang and Zhang23). In this formula, W is a normalized form of the similarity matrix A, and 0<μ<1 is a parameter that determine the influence of a node’s neighbors relative to its provided label.
Results
In this section we present experimental evaluation results of the proposed heterogeneous label propagation method on a treatment recommendation task for individual patients.
Data Description
Our real-world dataset contains 3-year longitudinal EHR of 110,157 patients. We selected hypercholesterolemia as our target disease for conducting experimental evaluations. There are 8 cholesterol-lowering drugs and 273,525 Low-Density Lipoprotein (LDL) lab-test records in the dataset. A patient, whose LDL level is below 130 mg/dL, is considered to be “well-controlled”. To define an effective drug for a patient, we selected the patients who take only one cholesterol-lowering drug within a 60-day treatment window and remain “well-controlled” for at least two consecutive lab assessments. We obtained 1219 distinct patients and 4 statin cholesterol-lowering drugs (i.e., Atorvastatin effectively treats 97 patients, Lovastatin effectively treats 221 patients, Pravastatin effectively treats 24 patients, and Simvastatin effectively treats 877 patients). The drug similarities from chemical structures and drug targets were calculated respectively. The patient similarities were calculated based on the ICD9 diagnosis codes within the 90-day patient assessment window prior to the first day a patient takes a drug within the 60-day treatment window. Then we constructed a heterogeneous graph based on our proposed method.
For illustration, Figure 2 depicts the definition of an effective drug for a given patient and assessment of patient diagnosis condition prior to treatments.
Figure 2.
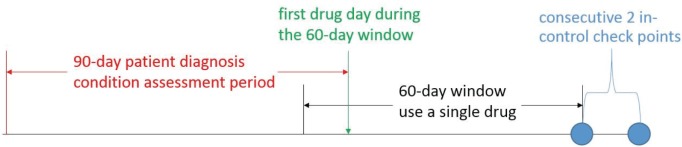
Assessments of patient diagnosis condition prior to treatments and definition of the effective drug for a single patient over time. Blue circles represent “well-controlled” LDL assessments (LDL < 130 mg/dL).
Method Comparison
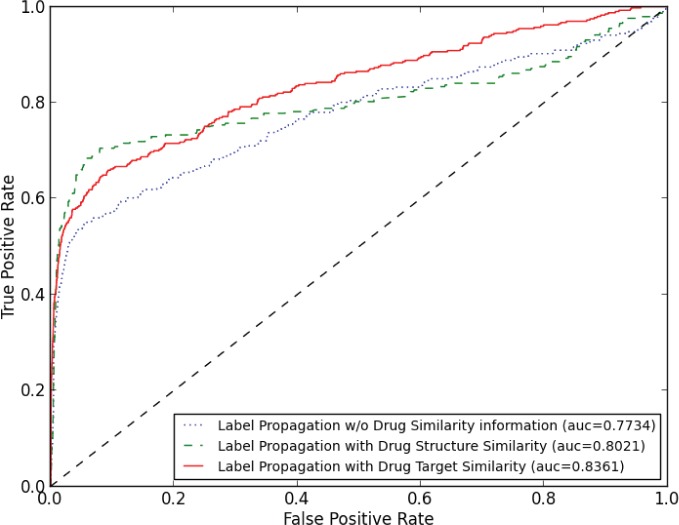
We used a 10-fold cross-validation scheme to evaluate treatment recommendation algorithms. To obtain robust results, we performed 50 independent cross-validation runs, in each of which a different random partition of the dataset to 10 parts was used. In our comparisons, we considered three treatment recommendation methods: (1) Label propagation using only patient information. The method propagates known effective treatments of training patients to testing patients based on the patient similarity analytics without considering drug information. (2) Heterogeneous label propagation using both patient and drug chemical structure information. The method propagates known effective treatments of training patients to the whole heterogeneous graph which is proposed in the methodology section. The drug similarity is calculated based on drugs’ chemical structures. (3) Heterogeneous label propagation using both patient and drug target information. The method propagates known effective treatments of training patients to the whole heterogeneous graph and the drug similarity is calculated based on drugs’ protein targets. Figure 3 shows the averaged ROC curves of 50 runs of the cross-validation for different methods based on the experiment.
Figure 3.
The averaged ROC comparison of three treatment recommendation strategies. Methods are sorted in legend of the figure according to their AUC score.
Figure 3 shows that label propagation algorithms are capable at treatment recommendation tasks. Without using any drug information, the label propagation algorithm obtains an averaged AUC score of 0.7734. When combining drug chemical structure or drug target information, heterogeneous label propagation algorithms obtain averaged AUC scores of 0.8021 or 0.8361 respectively. Analysis of the results revealed that rarely used treatments in the EHR data (e.g., Pravastatin only has 24 effective cases in the data, but it is very similar to Lovastatin from both structure and target perspectives) benefit from drug similarity analytics, thus the overall AUC scores were improved. Another observation is that heterogeneous label propagation using drug target similarity achieved a higher AUC score (0.8361) than the one using drug chemical structure similarity (0.8021). The results indicate that choosing an appropriate drug similarity measurement for the dataset will improve the performance of the heterogeneous label propagation. For example, Lovastatin is used to lower LDL by less than 30%, Simvastatin is used to lower LDL by 30% or more and treat the patients have heart disease and/or diabetes in the clinical settings24. Lovastatin and Simvastatin have very similar chemical structures, thus chemical structure similarity may not distinguish them well. Instead, Lovastatin and Simvastatin have different drug target sets (i.e., Lovastatin targets proteins 3-hydroxy-3-methylglutaryl-coenzyme A reductase, Integrin alpha-L, and Histone deacetylase 2; Simvastatin targets proteins 3-hydroxy-3-methylglutaryl-coenzyme A reductase, and Integrin beta-2), thus in this study drug target similarity may serve as a better similarity metric to recommend personalized treatments to patients.
Conclusion
We have proposed a heterogeneous label propagation method to support personalized medicine by leveraging patient similarity and drug similarity analytics. Experimental evaluation results on a real-world EHR dataset demonstrate the effectiveness of the proposed method and suggest that the combination of appropriate patient similarity and drug similarity analytics can help identify which drug is likely to be effective for a given patient. In future work we plan to apply the method to more drugs and more diseases, and explore more sophisticated drug and patient similarity measures.
References
- 1.Meyer UA. Pharmacogenetics - five decades of therapeutic lessons from genetic diversity. Nat Rev Genet. 2004;5:669–675. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- 2.Fernald GH, Capriotti E, Daneshjou R, Karczewski KJ, Altman RB. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27(13):1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuvirth H, Ozery-Flato M, Hu J, Laserson J, Kohn MS, Ebadollahi S, Rosen-Zvi M. Toward personalized care management of patients at risk: the diabetes case study; Proceedings of ACM international conference on knowledge discovery and data mining; 2011. pp. 395–403. [Google Scholar]
- 4.Liu L, Tang J, Cheng Y, Agrawal A, Liao WK, Choudhary A. Mining diabetes complication and treatment patterns for clinical decision support; Proceedings of ACM international conference on information and knowledge management; 2013. [Google Scholar]
- 5.Rosen-Zvi M, Altmann A, Prosperi M, Aharoni E, Neuvirth H, Sonnerborg A, Schülter E, Struck D, Peres Y, Incardona F, Kaiser R, Zazzi M, Lengauer T. Selecting anti-HIV therapies based on a variety of genomic and clinical factors. Bioinformatics. 2008;24(13):i399–i406. doi: 10.1093/bioinformatics/btn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett CC, Hauser K. Artificial intelligence framework for simulating clinical decision-making: a Markov decision process approach. Artificial Intelligence in Medicine. 2013;57(1):9–19. doi: 10.1016/j.artmed.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Mol Syst Biol. 2011;7:496. doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Lu Z. A new method for computational drug repositioning using drug pairwise similarity; Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, Agarwal P, Obradovic Z. Computational drug repositioning by ranking and integrating multiple data sources; Proceedings of European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases; 2013. pp. 579–594. [Google Scholar]
- 10.Lounkine E, Keiser M, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding H, Takigawa I, Mamitsuka H, Zhu S. Similarity-based machine learning methods for predicting drug-target interactions: a brief review. Brief Bioinform. 2013 doi: 10.1093/bib/bbt056. [DOI] [PubMed] [Google Scholar]
- 12.Gottlieb A, Stein GY, Oron Y, Ruppin E, Sharan R. INDI: a computational framework for inferring drug interactions and their associated recommendations. Mol Syst Biol. 2012;8:592. doi: 10.1038/msb.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilar S, Harpaz R, Uriarte E, Santana L, Rabadan R, Friedman C. Drug-drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc. 2012;19(6):1066–1074. doi: 10.1136/amiajnl-2012-000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Wang F, Hu J, Edabollahi S. Supervised patient similarity measure of heterogeneous patient records. SIGKDD Explorations. 2012;14(1):16–24. [Google Scholar]
- 15.Wang F, Hu J, Sun J. Medical prognosis based on patient similarity and expert feedback; Proceedings of International Conference on Pattern Recognition; 2012. pp. 1799–1802. [Google Scholar]
- 16.Chawla NV, Davis DA. Bringing big data to personalized healthcare: a patient-centered framework. J Gen Intern Med. 2013 doi: 10.1007/s11606-013-2455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Syed Z, Guttag JV. Unsupervised similarity-based risk stratification for cardiovascular events using long-term time-series data. Journal of Machine Learning Research. 2011:999–1024. [Google Scholar]
- 18.Roque FS, Jensen PB, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol. 2011;7(8):e1002141. doi: 10.1371/journal.pcbi.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Z, Dong W, Duan H, Li H. Similarity measure between patient traces for clinical pathway analysis: problem, method, and applications. IEEE Journal of Biomedical and Health Informatics. 2013 doi: 10.1109/JBHI.2013.2274281. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Knox C, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith TF, Waterman MS, Burks C. The statistical distribution of nucleic acid similarities. Nucleic Acids Res. 1985;13(2):645–656. doi: 10.1093/nar/13.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei WQ, Cronin RM, Xu H, Lasko TA, Bastarache L, Denny JC. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc. 2013;20(5):954–961. doi: 10.1136/amiajnl-2012-001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Zhang C. Label propagation through linear neighborhoods; Proceedings of International Conference on Machine Learning; 2006. pp. 985–992. [Google Scholar]
- 24.Best Buy Drugs (Consumer Reports) Evaluating statin drugs to treat high cholesterol and heart disease: comparing effectiveness, safety, and price; p. 9. [Google Scholar]