Abstract
Clinical research data generated by a federation of collection mechanisms and systems often produces highly dissimilar data with varying quality. Poor data quality can result in the inefficient use of research data or can even require the repetition of the performed studies, a costly process. This work presents two tools for improving data quality of clinical research data relying on the National Cancer Institute’s Common Data Elements as a standard representation of possible questions and data elements to A: automatically suggest CDE annotations for already collected data based on semantic and syntactic analysis utilizing the Unified Medical Language System (UMLS) Terminology Services’ Metathesaurus and B: annotate and constrain new clinical research questions though a simple-to-use “CDE Browser.” In this work, these tools are built and tested on the open-source LimeSurvey software and research data analyzed and identified to contain various data quality issues captured by the Comprehensive Research Informatics Suite (CRIS) at the University of Arkansas for Medical Sciences.
Introduction
With emerging healthcare technologies and systems becoming increasingly reliant on the efficient and expedient transfer of data between disparate systems, the assessment and maintenance of data quality of healthcare and clinical data has become prominent areas of research and effort in the electronic healthcare frontier1. Though many standards, technologies, and vocabularies exist to aid in supplying a maintainable level of data quality in healthcare systems, such as the HL7 messaging standard2, caCORE3, or the Unified Medical Language System4, there still exists significant hurdles and inadequacies in current methods for ensuring high data quality5, 6, 7, 8. As a facet of the entire quality problem presented by healthcare and clinical data, this work focuses on the data quality of clinical research data, describing and implementing two tools that utilize the National Cancer Institute’s (NCI) Common Data Elements (CDE)9 as a syntactic standard and the vocabulary accessed through the UMLS Terminology Service’s (UTS) Metathesaurus as a semantic standard for automated syntactic/semantic annotation of past clinical research data and as a library for computer-aided syntactic/semantic annotation and constraint of new clinical research questions for future studies. Through the supplied annotations, it is the hope of this work that data quality can be improved between disparate sources of clinical research through means of a standard semantic and syntactic representation of any and all produced research data as well as ensured data quality through enforced syntactic/semantic constraints. To explore the effectiveness of the proposed approach in achieving the aforementioned goals, two tools were developed, noted respectively as the Automated Annotation Tool (which annotates questions with CDEs based on minimizing semantic and syntactic distance between survey questions and potential CDEs) and the CDE Importer (a plugin for Limesurvey forcing users to annotate questions with CDE codes). These tools were implemented using the LimeSurvey software as a basis for clinical research data collection and run against clinical research data generated by the University of Arkansas for Medical Sciences (UAMS)’s Comprehensive Research Informatics Suite (CRIS). Here we describe our implementation focusing on relevant information for LimeSurvey as a clinical research tool, information concerning CDEs as defined by NCI, background information concerning UMLS, UTS, and the Metathesaurus, the methodology employed by the two tools (Automated Annotation Tool and CDE Importer) implemented in this work. We provide sample results of using the Automated Annotation Tool on live clinical research survey data captured with LimeSurvey, and final remarks detailing planned improvements on the Automated Annotation Tool and CDE Importer.
Background
Data Quality Processes
The Ten Step Process10 is used to assess, improve, and create high-quality information with long-lasting results. It should be considered an evolving and continuous process to improve the quality of the data with the following phases;
The Assessment Phase – this phase includes identifying business needs, analyzing information environment and conducting data quality assessment.
The Awareness Phase – studying the root causes of data quality problems identified in assessment phase.
The Action Phase –Implementing plans which are developed at the awareness phase.
In the context of data quality improvement cycle, the following phases were examined: “Assessment of actual environment”; “Awareness to understand true state of data and their impact on business”; and “Action to prevent future information quality problems and correction of existing problems.” These phases are essentially the main components of any improvement life cycle that can be used by individuals or teams alike. The improvement life cycle is the basis for the Ten-Step Process that provides explicit and detailed instructions for planning and executing quality improvement projects by combining data quality dimensions and business impact techniques.
Getting to know the data
The core of the data utilized in this work originates with several research databases collected over the years by the Cancer Control Program of the Winthrop P. Rockefeller Cancer Institute. As initial assessment phase of the Ten Step Process, a data profiling process was implemented on the provided data and a number of data quality issues were identified15. Some of the main issues identified were:
Information obtained via an existing process generated difficult-to-classify values due to lack of standardization, consistency, and commonly accepted data elements.
Lack of clarity or absence of acceptable forms of responses.
Lack of data collection mechanisms to enforce constraints and quality controls.
Accuracy problems (i.e. incorrect values).
Completeness problems.
Data pattern problems.
Duplication problems.
The quality issues that were identified using this Ten Step Process were found to be addressable in either a manual or automated manner. Manual processes have been utilized in past works to address issues related to duplicate records (e.g. duplicate participant IDs). Conversely, this work provides automation for defining standardized annotations via CDEs, given that manual CDE annotation can be costly both in terms of time, money, and the required training to properly utilize CDEs.
UAMS CRIS
The Comprehensive Research Informatics Suite, or CRIS (formerly known as the Clinical Trials Informatics Suite) is a software suite developed and packaged by UAMS for distributed electronic maintenance and deployment of clinical trials and all related data. The suite provides functionality for subject registration, research study calendars and patient activities management, automated coding using standard medical vocabularies for supplied free text, electronic participant recruitment for clinical trials, date tracking, data reporting, and electronic data capture using tools such as OpenClinica and LimeSurvey. The tools developed in this work are built to utilize and annotate the data captured through CRIS, and all test data used in this work was captured using the CRIS system.
LimeSurvey
LimeSurvey is one of the primary applications for capturing clinical research data using UAMS’s CRIS. This open source, free survey software provides an extremely flexible platform and wide host of tools for developing surveys and survey questions through an intuitive interface. More notably, LimeSurvey offers excellent tools for constraining the syntax of possible answers that can be provided by survey users, such as the base question type (e.g. string, numeric, multiple choice, date, etc.) or through constrains such as minimum/maximum characters allowed, minimum/maximum values allowed, as well as user-defined regular expressions.
NCI Common Data Elements
Common Data Elements, or CDEs, are standardized metadata constructs that can describe both the syntactic and semantic constraints of an entity, such as a patient name or a street address. The National Cancer Institute (NCI) developed CDEs specifically for cancer research (though it now includes a wide variety of contexts) to address the data control issue present with the creation of new, dissimilar data elements per individual researcher. The main resource for accessing the all NCI CDEs is through the Cancer Data Standards Repository, or caDSR, which offers available web services such as the REST API for programmatically querying caDSR and retrieving CDEs. Manual queries can be performed with NCI‟s CDE Browser (not to be confused with the CDE Importer implemented and explained in this work).
Each NCI CDE has various attributes that make it particularly useful for the goals of this work. At the basis of each CDE is a “data element” that describes top level attributes such as a unique identifier, a preferred name, preferred question text (when utilized in clinical trials research), a workflow status (of being integrated into caDSR), all relevant contexts, any previous versions of the CDE, as well as other related information. Semantic information is captured in terms of the “data element concept,” providing unique codes that link to concepts defined in NCI’s Thesaurus, specifically the CDE’s “Object” and “Property” codes describing, in turn, the real world entity and attributes described by a given CDE. Each CDE also contains a “value domain” describing the syntactic constraints defined by the CDE, such as the data type, minimum/maximum character length, minimum/maximum values allowed, any permissible values for enumerated value domains, etc. With both the “data element concept” and “value domain,” each CDE contains sufficient metadata to describe a concept both syntactically and semantically. For this reason, CDEs were chosen in this work to annotate clinical research data and apply constraints on new clinical studies. Furthermore, despite shortcomings in the CDE database and design, current studies suggest that CDEs are an effective tool for providing data quality assurance11, 12, 13.
Unified Medical Language System
The Unified Medical Language System, or UMLS, provides access to a large number of cross-referenced vocabularies that describe concepts semantically through relational mappings and semantic metadata. Two prominent components of UMLS include the Metathesaurus and Semantic Network, each which can be accessed and queried either manually or programmatically through the UMLS Terminology Services (UTS) by authorized users. The Metathesaurus provides access to “concepts” that contain relational mappings to an enormous number of vocabularies, such as SNOMED CT, RxNorm, and, most prominent to this work, the NCI Thesaurus. Each concept is defined by a preferred name, “atoms” which represent mapped concepts present in other vocabularies with their respective relations, and one or more semantic types. The semantic types map each concept to other concepts that share the same semantic type or are defined by a related semantic type as specified by the aforementioned Semantic Network. The Semantic Network is organized in a tree hierarchy, with more general semantic types defined at the root. Each semantic type, just as with the concepts, contains a list of related semantic types and their respective relations, as well as other auxiliary information. As UMLS contains semantic mappings to concepts defined in the NCI Thesaurus, it is utilized in this work to convert Concept Unique Identifiers (CUIs) extracted from question text using the MetaMap16 utility into NCI Thesaurus codes.
Methods
Automated Annotation Tool
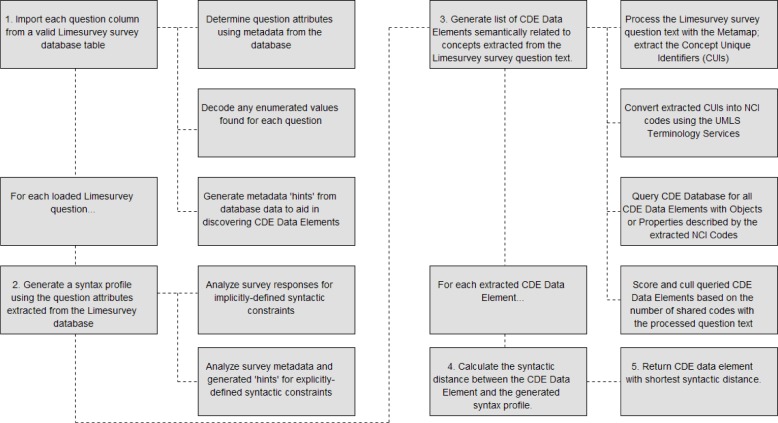
To provide syntactic/semantic annotation of past clinical survey data, the Automated Annotation Tool (AAT) was developed to automatically provide CDE annotations for clinical data with very little required human interaction. Given this, the implemented tool was designed to accept a valid LimeSurvey survey table and generate suggested CDE annotations based on minimizing semantic and syntactic distance between each survey question and their respective suggested CDEs. The process for generating the semantic and syntactic distance for that exists between a LimeSurvey question and a potential list of CDEs and thus determining the best CDE (in terms of minimum distance) is expressed in Figure 2 below:
Figure 2.
CDE-Question Distance Calculation Process
As shown in Figure 2, there are four primary steps taken to calculate the semantic and syntactic distance that exists between a given question and CDE data element. The first step encompasses extracting the questions and all of their relevant attributes found in the provided LimeSurvey table and database. This process includes determining the question text, question code, the LimeSurvey question type, and any user-specified constraints on the data (such as maximum character length). If the question contains enumerated values as answers, this step then also includes decoding the values and merging any shared enumeration lists. When available, certain metadata attributes can also be used to generate “hints” for the annotation process. One such hint can be generated with the LimeSurvey survey question type; given that many of the CDE data types can be mapped and directly compared to various LimeSurvey survey question types. While such hints allow the AAT to more accurately determine the syntactic distance that exists between a LimeSurvey survey question and a CDE, they are not necessary to perform the distance calculation.
After each question is extracted, the AAT then proceeds to generate a syntax profile utilizing the extracted attributes from step 1 in Figure 2. All extracted explicitly-defined constraints (i.e. defined in the survey metadata) are loaded into the syntax profile; all other implicitly-defined constraints are determined through statistical analysis of the answers provided for each survey question. Constraints specified in a fully initialized syntax profile include the base data type (string, numerical, and enumerated) mapped from the LimeSurvey question type, minimum and maximum character lengths, minimum and maximum numerical precision, minimum and maximum numerical value, permissible answers for enumerated data types, and finally any available metadata hints. A single syntax profile is generated per survey question; any syntax profiles representing distributed LimeSurvey questions, such as arrays, are merged after the extraction process to increase annotation accuracy and reduce redundant question analysis.
Using the question text contained in the syntax profile for each LimeSurvey question, the third step in the AAT process attempts to implicitly minimize semantic distance by querying CDEs such that the returned set only contains data elements that share one or more semantic concepts contained within the question text. It is from this returned set that the final CDE, the chosen annotation, is determined. To create the set of semantically-related CDEs, a list of Concept Unique Identifiers (CUIs) is first generated from the question text by means of the MetaMap Parser16. Developed by the National Library of Medicine, the MetaMap parser maps free text to concepts found within the UMLS Metathesaurus, in which each concept is uniquely identified by a given CUI. Thus, each LimeSurvey question is parsed by the MetaMap, returning a list of CUIs describing semantic concepts contained within the text. In order to query for CDEs, the returned list of CUIs has to be converted into NCI Thesaurus codes via the UMLS Terminology Services. Once the list has been successfully converted, the entire CDE database is queried for those CDE data elements who’s “Object” or “Property” codes contain at least one of the extracted codes from the question text. The initial list of queried CDE data elements is then culled based on the number of total number of codes each discovered CDE shares with the question text, resulting in a final set of CDEs assured (by means of the MetaMap and NCI Thesaurus) to be the most semantically similar to the concepts in the question text.
Once all CDE data elements are discovered, and the syntax profile for the LimeSurvey question is initialized, the final step, the distance calculation, can be performed. In this step, the value domain of each discovered CDE data element is compared against the syntax profile of the LimeSurvey question being processed. The syntactic distance that exists between the LimeSurvey survey question being processed and a given CDE is defined by nine different distance calculations: minimum value distance, maximum value distance, precision distance, minimum character length distance, maximum character length distance, data type distance, enumerated value count distance, enumerated value text distance, and question text distance. Each respective distance calculation is shown below:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
In the above equations, all values pertaining to a CDE value domain are denoted with the ‘CDE’ subscript; all values pertaining to a LimeSurvey survey question syntax profile are denoted with the ‘LSQ’ subscript. Functions Min and Max refer to the functions that, respectively, return the minimum and maximum value from either two arguments or from a set of supplied numbers. Map refers to a function that accepts a LimeSurvey data type “hint” or a condensed data type (i.e. string, numeric, or enumerated) and returns the set of CDE data types mapped to the given LimeSurvey data type or condensed type. SmithWaterman refers to a generic implementation of the classical Smith-Waterman alignment algorithm14 returning the alignment score divided by the maximum character length of the two supplied strings. NormDis is defined by the equation below:
| (10) |
Granted the above, Equations 1–5 calculate the minimum value distance, maximum value distance, precision distance, minimum character distance, and maximum character distance by calculating the normalized distance (Equation 11) that exists between each value. If either the CDE value domain or the LimeSurvey question syntax profile does not contain values for one of aforementioned attributes, the maximum distance is assumed (normalized to 1.0) unless both the CDE value domain and the LimeSurvey question both lack a value for the same attribute. For instance, if both the syntax profile and the CDE value domain represent a string data type, both the syntax profile and the CDE value domain will be lacking all numerical constraints, such as maximum value. In this instance the minimum distance is assumed (normalized to 0.0).
Equation 6 calculates the distance that exists between the CDE value domain’s data type and the data type defined by the LimeSurvey question’s syntax profile. Using the Map function described in the prior paragraphs, the syntax profile’s data type can be mapped to a defined set of CDE data types. With the generated set, the distance equation simply performs an intersection between the CDE value domain’s data type and the generated set to determine if the syntax profile’s data type is related to the CDE value domain’s data type. The size of the resulting set (which will either be 1 if the two data types are related or 0 if they are disjoint) is subtracted by 1 to generate the normalized data type score.
Equation 7 calculates the normalized distance between the count of enumerated values of the CDE value domain and the count of enumerated values of the question syntax profile. Equation 8 calculates the normalized accumulated minimum text difference between each enumerated value from both the CDE value domain and the question syntax profile. To do this, the equation determines the minimum text alignment between two components from both the CDE value domain enumerated values list and the question syntax profile enumerated values list. This minimum alignment is summated with all other minimum alignments, with the resulting summation divided by the minimum of the size of the CDE value domain enumerated values list and the size of the question syntax profile enumerated values list. Equations 7 and 8 only apply to instances in which both or either CDE value domain and question syntax profile represent enumerated data. Just as with equations 1–5, if neither the value domain nor the syntax profile represents enumerated data, the minimum distance is assumed; if only one of either the value domain or the syntax profile represents enumerated data, the maximum distance is assumed.
Finally, Equation 9 simply finds the minimum alignment distance that exists between the LimeSurvey survey question’s tokenized question text and the CDE value domain’s name, question text, or preferred definition.
Once all syntactic distance calculations are performed for a given CDE data element and a LimeSurvey survey question, all resulting values are weighted and added together, resulting in a final normalized score produced between 0.0 and 1.0, with 0.0 representing a CDE data element-LimeSurvey question pair that is assumed to be syntactically and semantically identical. This 4-step process of culling CDEs semantically and calculating the syntactic distance is repeated for every CDE data element discovered with a given set of search terms, for each set of search terms generated from a given LimeSurvey question, for each LimeSurvey question from a given LimeSurvey survey. The CDE data element with the minimum score for a given LimeSurvey question is suggested to be the proper annotation for that LimeSurvey question, with each annotation detailing the discovered syntax and semantic profiles of the question as well as the value domain and semantic profiles of the discovered CDE.
CDE Importer
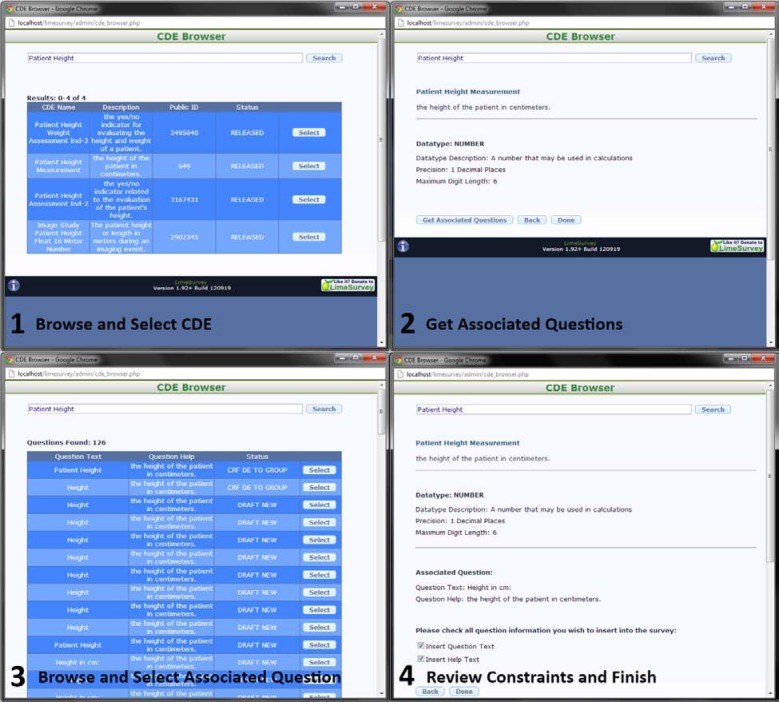
To apply syntactic constraints and annotate new clinical surveys with CDEs, the second tool, the CDE Importer, was developed as an plug in for the LimeSurvey to allow users to browse for and insert constraints defined by CDEs for each of their survey questions, explicitly annotating the survey questions with the CDE syntactic and, implicitly, semantic information in the same process. The four primary steps for annotating new survey questions with the CDE Browser include 1: browsing for and selecting a CDE using search terms, 2: discovering any associated questions defined for the selected CDE, 3: browsing for and selecting from the list of returned associated questions (if there are any), and 4: review constraints and finalize any insertion options. Screenshots of the application executing each of these four tasks is shown in Figure 3 below:
Figure 3.
CDE Importer for LimeSurvey
To initiate the CDE Browser application, the user must first create a new question using the LimeSurvey survey software. The CDE Browser application modifies the LimeSurvey software such that it disallows a user to create a question without an associated CDE, thus requiring that all questions have proper CDE annotations. Once the user has started to create a new question, they can choose to browse for CDEs by activating an added button next to the LimeSurvey question “Code” field (the CDE Browser inserts the selected CDE’s public ID as the code for a given question). This will generate a new window much like the one shown in frame 1 of Figure 3.
Once the user has opened the CDE Browser window, they can then supply any number of search terms and hit “search.” Utilizing the caDSR REST API provided by NCI, the CDE Browser will return the name, description, public ID, and status of each CDE, allowing the user to judge the appropriate CDE for their needs. If the user finds a CDE that is satisfactory, they can select it, bringing them to a window similar to frame 2 of Figure 3. The value domain, or syntactic description, of the selected CDE is shown, describing the data type, a description of the data type, and any constraints imposed by the selected CDE (such as maximum or minimum precision). If the selected CDE has an enumerated value domain, all permissible values for that CDE are shown with the option to select which permissible answers the user wants to insert into the survey (as many enumerated value domains contain overlapping values and/or redundant values).
The user can choose to stop at step 2, or they can complete the CDE annotation process by activating the “Get Associated Questions” button. Pressing this will search for any questions that are related to the selected CDE using the caDSR Rest API. If any associated questions are found, a screen similar to frame 3 in Figure 3 is shown, displaying each question’s preferred text, the question’s help text, and the question’s status. Just as with the CDE data element results, the user can use the displayed information to select the most appropriate question. Doing so will bring the user to a window much like frame 4, which again displays the CDE data element attributes and value domain, as well as the selected question text/help text with the options to insert these texts into the survey data. Selecting “Done” will automatically select the appropriate survey question type (based on the selected CDE’s value domain), insert any enumerated values the user has chosen to insert, insert all syntactic constraints defined by the CDE’s value domain, and insert any question text/help text the user has chosen to insert into the appropriate database table supplied by the LimeSurvey software. At this point, the survey question is considered to be annotated and thus the LimeSurvey software will allow the new question to be added to the current survey.
Results
To test the effectiveness of the Automated Annotation Tool, a random LimeSurvey survey extracted from data captured with UAMS’s CRIS was analyzed with Automated Annotation Tool. The survey contained 80 questions and was manually annotated with CDEs to provide a ground truth for what is determined to be a “correct” annotation. Each manual designation was either given a ‘weak’ or ‘strong’ flag, describing, respectively, if the annotation almost exactly described the LimeSurvey question or if the annotation is only weakly semantically related. The only tool used to manually discover these CDEs was the aforementioned NCI’s CDE Browser (utilizing simple text search). Of the 80 questions, 49 were determined to be weak annotations.
Once each Limesurvey survey question was given a “ground truth” annotation, the AAT was run on the survey, producing annotations that were determined to either be an exact match, semantically related, or a complete miss. An example of a random selection of results spanning these three types of annotations is shown in Table 1 below:
Table 1.
Sample Automated Annotation Tool Results
| Question Text | Common Data Element | Assessment |
|---|---|---|
| Meeting Code | Coding Scheme Identifier | Exact Match |
| What is your annual household income from all sources? | Patient Household Annual Income Amount | Exact Match |
| Do you have any kind of health care coverage, including health insurance, prepaid plans such as HMOs, or government plans such as Medicare? | Registration Private Health Insurance Medicare Payment | Semantically Related |
| When was the last time you had an EKG? | Number of months to Last Clinical Assessment | Semantically Related |
| At least once a week, do you engage in regular activity such as brisk walking, jogging, bicycling, or another activity long enough to work up a sweat? | Blackout Week Day Week | Complete Miss |
| You are afraid of finding colon cancer if you were checked. Would you say you… | Malignant Neoplasm Biopsy Finding Indicator | Complete Miss |
In Table 1, one can see examples of the three types of annotations. The first and most desirable annotation type is naturally an exact match, in which the CDE annotation fully describes both the semantics and syntax of a given survey question. For example, “What is your annual household income from all sources,” is both semantically and syntactically described by the CDE ‘Patient Household Annual Income Amount.’ In many cases, however, the AAT cannot determine an exact match but instead returns a CDE that is weakly semantically related to the question text. For instance, the CDE “Number of Months to Last Clinical Assessment” is weakly semantically related to “When was the last time you had an EKG?” as both refer to an elapsed time frame concerning a medical assessment, but the CDE does not specifically refer to an EKG assessment. Finally, a complete miss describes an annotation in which the annotation supplied describes neither the syntax nor the semantics of the survey question, two examples of which can be seen in Table 1.
Table 2 below shows the results of running the AAT on the entire survey, giving the percentage of the 80 annotated questions that were said to be annotated with an exact match, a semantically related match, or a complete miss. The first row shows results that include survey questions that could not be manually discovered (i.e. “weak” annotations). The second row shows the annotation results on only those questions that could be strongly manually annotated.
Table 2.
Automated Annotation Tool Total Results
| Adjusted | Exact Match | Semantically Related | Complete Miss |
|---|---|---|---|
| No | 12.987% | 14.285% | 72.727% |
| Yes | 28.571% | 17.857% | 53.571% |
Conclusion
In this work, two tools, the Automated Annotation Tool and CDE Importer, are proposed and implemented to provide semantic and syntactic annotation for clinical research data to improve data quality of past clinical research data and constrain new clinical research to standard syntactic representations of survey questions and data elements. To test the effectiveness of the implemented Automated Annotation Tool, the tool was run against a randomly selected survey generated and maintained by UAMS’s CRIS. In general, as the samples in Table 1 and the results in Table 2 expressed, a small portion of suggested annotations are syntactically and semantically sound, however many of the results are complete misses. It is the thoughts of this team that this is possibly due to disjoint semantic information between what is extracted by the MetaMap, the NCI Thesaurus, and UMLS. Another possibility is that certain semantic codes could overpower other codes. For instance, in Table 1, the question text mentions ‘week,’ forcing the AAT to focus on ‘week’ as a concept code, resulting in an incorrect annotation ‘Blackout Day of the Week.” Granted this, the next aim of this work is to provide a more powerful mechanic for determining which of the extracted codes from a given question text is to be considered more relevant given the context of the question in order to determine a more semantically related pool of potential CDEs. Another aim is to remove reliance on syntactic analysis for determining which of the semantically culled CDEs are the most “correct,” as there is often very little correlation between the syntax profile of the survey question and the CDE syntax, even between correct annotations. Despite these shortcomings, the concepts exhibited by the tools implemented in this work display potential for future use and improvement for the goal of providing automated data quality assessment, improvement, and constraints for clinical research data.
Figure 1.
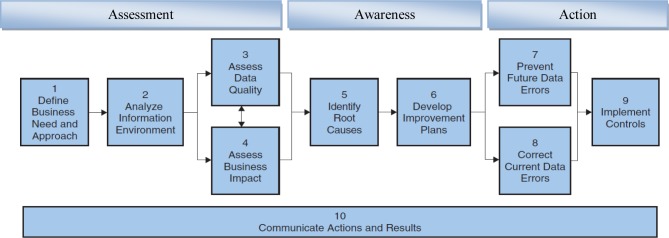
The Information and Data Quality Improvement Cycle and the Ten Steps Process10
Acknowledgments
This work was sponsored by the Winthrop P. Rockefeller Cancer Institute and by the award number UL1TR000039 from the National Center for Advancing Translational Sciences (NCATS). We also would like to thank University of Arkansas at Little Rock Information Quality Graduate Program for their support and guidance.
References
- 1.Gendron MS, D’Onofrio MJ. Data Quality in the Healthcare Industry. Data Quality [Internet] 2001 Sep;7(1) Available from: http://www.dataquality.com/901GD.htm. [Google Scholar]
- 2.Health Level Seven International [Internet]. [publisher unknown] [updated 2013; cited 2013 Mar 10]. Available from: http://www.hl7.org/
- 3.Covitx PA, et al. caCORE: A common infrastructure for cancer informatics. Bioinformatics. 2003;19(18):2404–12. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- 4.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32(1):267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodenreider O, Mitchell JA, McCray AT. Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. AMIA; 2002. pp. 61–65. [PMC free article] [PubMed] [Google Scholar]
- 6.Richesson RL, Krischer J. Data Standards in Clinical Research: Gaps, Overlaps, Challenges and Future Directions. J AM Med Inform Assoc. 2007;14:687–96. doi: 10.1197/jamia.M2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mead CN. Data interchange standards in healthcare IT—computable semantic interoperability: now possible but still difficult, do we really need a better mousetrap? J Health Inf Manag. 2006;20(10):71–8. [PubMed] [Google Scholar]
- 8.Tobias J, et al. The CAP cancer protocols – a case study of caCORE based data standards implementation to integrate with the Cancer Biomedical Informatics Grid. BMC Medical Informatics and Decision Making. 2006;6(25) doi: 10.1186/1472-6947-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiley A. CTEP Common Data Elements [Internet]. [publisher unknown] [updated 2012 Mar 16; cited 2013 Mar 10]. Available from: https://wiki.nci.nih.gov/display/caDSR/CTEP+Common+Data+Elements.
- 10.McGilvray D. Executing Data Quality Projects: ten steps to quality data and trusted information. Massachusetts: Morgan Kaufmann Publishers; 2008. pp. 19–23. [Google Scholar]
- 11.Patel AA, et al. The development of common data elements for a multi-institute prostate cancer tissue bank: The Cooperative Prostate Cancer Tissue Resource (CPCTR) experience. BMC Cancer. 2005;5(108) doi: 10.1186/1471-2407-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohanty SK, et al. The development and deployment of Common Data Elements for tissue banks for translational research in cancer – An emerging standard based approach for the Mesothelioma Virtual Tissue Bank. BMC Cancer [Internet] 2008;8(91) doi: 10.1186/1471-2407-8-91. Available from http://www.biomedcentral.com/1471-2407/8/91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadkarmi PM, Brandt CA. The Common Data Elements for Cancer Research: Remarks on Functions and Structure. Methods Inf Med. 2006;45(6):594–601. [PMC free article] [PubMed] [Google Scholar]
- 14.Smith TF, Waterman MS. Identification of Common Molecular Subsequences. Journal of Molecular Biology. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 15.Pushkarev V. Information Quality in Clinical Research Survey. UALR Information Quality Program Graduate Project. 2010 Apr; [Google Scholar]
- 16.Aronson A. Effective Mapping of Biomedical Text to the UMLS Metathesaurus: The MetaMap Program. AMIA; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]