Abstract
Recent large-scale genome-wide association studies (GWAS) have identified a number of new genetic variants associated with breast cancer. However, the degree to which these genetic variants improve breast cancer diagnosis in concert with mammography remains unknown. We conducted a case-control study and collected mammography features and 77 genetic variants which reflect the state of the art GWAS findings on breast cancer. A naïve Bayes model was developed on the mammography features and these genetic variants. We observed that the incorporation of the genetic variants significantly improved breast cancer diagnosis based on mammographic findings.
Introduction
High hopes for using genetic profiling for personalized medicine have been, in part, driven by the rapid progress of genome-wide association studies, which continue identifying more common genetic variants associated with diseases with high population prevalence. In particular, the recent Collaborative Oncological Gene-environment Study (COGS) [1], which pooled large quantities of genetic data via a massive international collaboration, more than doubled the number of known susceptibility loci that are associated to common cancers (breast, ovarian and prostate cancers). For breast cancer, over 130 institutions have collaborated and identified 41 new breast cancer associated variants [2]. One way these genetic variants could be used in clinical breast cancer care is in individualized screening recommendations and personalized diagnosis. Early attempts to incorporate genetic variants into breast cancer risk models revealed modest improvements in risk prediction accuracy. For example, adding seven SNPs to the Gail model only increased the area under the ROC curve (AUROC) from 0.607 to 0.632 [3, 4]. When ten SNPs were added to the Gail model, the AUROC increased from 0.580 to 0.618 on another dataset [5]. Incorporating these genetic variants with the mammographic findings to assess individualized risk will be highly relevant to clinical breast cancer diagnosis. In our prior study, we showed that when 22 SNPs were added to the 49 mammography features—the standard descriptors collected by radiologists on mammograms—the AUROC of the model increased from 0.693 to 0.731 [6]. This increase is statistically significant (P=0.021) [6], but the 22 SNPs only reflect the discoveries from the breast cancer GWAS up to 2010.
In this paper, we incorporated the new genetic variants and consolidated a list of 77 SNPs which reflect the state of the art of breast cancer GWAS. A great proportion of the new SNPs were contributed by COGS [2]. 41 SNPs were identified through a meta-analysis of 9 GWAS on 10,052 cases and 12,575 controls, and further showed significant association (P<5×10−8) on 45,290 cases and 41,880 controls. The list also includes the 22 SNPs used in Liu et al. [6] as well as another 14 SNPs identified by several other recent studies [7–13]. We incorporated these genetic polymorphisms with the descriptors that radiologists observe on mammograms using the standardized lexicon in breast imaging, the Breast Imaging Reporting and Data System (BI-RADS). These mammography features included the shape and the margin of masses, the shape and the distribution of microcalcifications, background breast density and other associated findings. We built naïve Bayes models, using the 49 mammography features together with the 77 genetic variants. We observed that the inclusion of the genetic variants significantly improved the breast cancer diagnostic model. We discovered that the mammographic findings were more predictive for high-risk women, whereas the genetic variants were more predictive for low-risk women, which demonstrated the potential benefit of combining genetic variants and mammographic findings for personalized breast cancer diagnosis.
Data
[Subjects]
The Personalized Medicine Research Project [14] at the Marshfield Clinic was used as the sampling frame to identify cases and controls. The project was reviewed and approved by the Marshfield Clinic IRB. The subjects were from a retrospective case-control design, and used in our previous study [6]. Women with a plasma sample available, a diagnostic mammogram, and a breast biopsy within 12 months after the mammogram were included in the study. Cases were defined as women having a confirmed diagnosis of breast cancer obtained from the institutional cancer registry. Controls were confirmed through the Marshfield Clinic electronic medical records as never having had a breast cancer diagnosis by ICD-9 diagnosis code (and absence from cancer registry). Cases included both invasive breast cancer and ductal carcinoma in situ. We employed an age matching strategy to construct case and control groups that were similar in age distribution. We selected a control whose age was within five years of the age of each case. We decided to focus on high-frequency/low-penetrance SNPs that affect breast cancer risk as opposed to low frequency SNPs with high penetrance or intermediate penetrance. We excluded individuals who had a known high-penetrance genetic mutation.
[Genetic Variants]
Our study included the 77 genetic variants (in Table 1) which were identified by the recent large-scale genome-wide association studies. 22 of these SNPs were evaluated in the previous study of Liu et al. (2013) [6]. Among the 55 new SNPs, 41 were identified by COGS [2], and 14 SNPs were included based on several other recent studies [7–13]. It is estimated that the current list of SNPs explains 14% of familial breast cancer risk [2].
Table 1.
The 77 SNPs identified to be associated to breast cancer.
| SNP | Chr | Ref | Notes1 | SNP | Chr | Ref | Notes |
|---|---|---|---|---|---|---|---|
| rs11249433 | 1 | [20] | WL | rs2380205 | 10 | [12] | |
| rs616488 | 1 | [2] | rs10995190 | 10 | [12] | ||
| rs1045485 | 2 | [21] | GWL | rs704010 | 10 | [12] | |
| rs17468277 | 2 | [22] | L | rs2981579 | 10 | [12] | |
| rs4666451 | 2 | [23] | L | rs7072776 | 10 | [2] | |
| rs13387042 | 2 | [20, 24] | GWL | rs7904519 | 10 | [2] | |
| rs4849887 | 2 | [2] | rs11199914 | 10 | [2] | ||
| rs2016394 | 2 | [2] | rs11814448 | 10 | [2] | ||
| rs1550623 | 2 | [2] | rs2107425 | 11 | [23] | L | |
| rs16857609 | 2 | [2] | rs3817198 | 11 | [20, 23] | GWL | |
| rs4973768 | 3 | [25] | L | rs614367 | 11 | [12] | |
| rs6762644 | 3 | [2] | rs3903072 | 11 | [2] | ||
| rs12493607 | 3 | [2] | rs11820646 | 11 | [2] | ||
| rs9790517 | 4 | [2] | rs6220 | 12 | [26, 27] | L | |
| rs6828523 | 4 | [2] | rs1292011 | 12 | [10, 13] | ||
| rs10941679 | 5 | [20, 28] | WL | rs17356907 | 12 | [2] | |
| rs30099 | 5 | [23] | L | rs10771399 | 12 | [2] | |
| rs889312 | 5 | [23] | GWL | rs12422552 | 12 | [2] | |
| rs981782 | 5 | [23] | L | rs11571833 | 13 | [2] | |
| rs1353747 | 5 | [2] | rs999737 | 14 | [20] | WL | |
| rs1432679 | 5 | [2] | rs2236007 | 14 | [2] | ||
| rs10069690 | 5 | [2] | rs2588809 | 14 | [2] | ||
| rs10472076 | 5 | [2] | rs9417 64 | 14 | [2] | ||
| rs2046210 | 6 | [29] | L | rs3803662 | 16 | [20, 23, 24] | GWL |
| rs2180341 | 6 | [30] | L | rs8051542 | 16 | [23] | L |
| rs17530068 | 6 | [9] | rs12443621 | 16 | [23] | L | |
| rs3757318 | 6 | [12] | rs13329835 | 16 | [2] | ||
| rs11242675 | 6 | [2] | rs17817449 | 16 | [2] | ||
| rs204247 | 6 | [2] | rs6504950 | 17 | [25] | L | |
| r s720475 | 7 | [2] | rs1436904 | 18 | [2] | ||
| rs13281615 | 8 | [20, 23] | GWL | rs527616 | 18 | [2] | |
| rs969344 4 | 8 | [2] | rs8170 | 19 | [8] | ||
| rs11780156 | 8 | [2] | rs4808801 | 19 | [2] | ||
| rs6472903 | 8 | [2] | rs3760982 | 19 | [2] | ||
| rs2943559 | 8 | [2] | rs2284378 | 20 | [9] | ||
| rs1011970 | 9 | [12] | rs2823093 | 21 | [10] | ||
| rs865686 | 9 | [7, 11, 13] | rs132390 | 22 | [2] | ||
| rs10759243 | 9 | [2] | rs6001930 | 22 | [2] | ||
| rs2981582 | 10 | [20, 23, 28, 30, 31] | GWL |
[Mammography Features]
Mammography is the most common breast cancer screening test, and the only one supported by multiple randomized trials demonstrating reduction in mortality rate [15]. There is a long history of development and codification of features observed by radiologists on mammograms. The American College of Radiology developed the BI-RADS lexicon to standardize mammographic findings and recommendations. The BI-RADS lexicon consists of 49 descriptors, including the characteristics of masses and microcalcifications, background breast density and other associated findings. Mammography data was historically recorded as free text reports in the electronic health record, and thus it was difficult to directly access the information contained therein. We used a parser to extract these mammography features from the text reports; the parser was shown to outperform manual extraction [16, 17]. After extraction, each mammography feature took the value “present” or “not present” except that the variable mass size was discretized into three values, “not present”, “small” and “large”, depending on whether there was a reported mass size and whether any dimension was larger than 30mm.
A BI-RADS assessment category was assigned to each mammogram by the interpreting radiologist, which indicated the radiologist’s assessment of the absence or presence of breast carcinoma. In our study, the BI-RADS assessment category took values, with an order of increasing probability of malignancy, of 1, 2, 3, 0, 4a, 4, 4b, 4c and 5. We used the BI-RADS assessment category as the predictions from the radiologists. Our study only included diagnostic mammograms, and all the screening mammograms were excluded. For cases, we selected the mammograms within one year prior to diagnosis. For controls, we selected the mammograms within one year prior to biopsy. If there were multiple diagnostic mammograms during that one year time period, we selected the mammogram with a more suspicious BI-RADS category, with subsequent tiebreakers being recency and the number of extracted features.
Model
We built breast cancer diagnosis models using Naïve Bayes, which can be regarded as the weighted average of risk factors. Naive Bayes assumes that all features are conditionally independent of one another given the class [18]. Although this assumption seems strong, it generally works well in practical problems and provides easy interpretation of the risk contribution from different factors. In our experiments, we used the Naïve Bayes implementation in WEKA [19].
In total, we constructed three types of models on different sets of features. The first model was built purely on the 49 mammography features, namely the Breast Imaging model. The second type of model was based purely on genetic variants, namely the genetic models. Since we would like to align our study with previous work, we tested three sets of genetic variants. The first set consisted of the 10 SNPs in the study of Wacholder et al. (2010) [5]. The second included the 22 SNPs in the study of Liu et al. (2013) [6]. The last set was our full list of the 77 SNPs. We denote the three genetic models as Genetic-10, Genetic-22 and Genetic-77 models. The third type of model was built on the 49 mammography features and the genetic variants together, namely the combined models. Since we had three sets of genetic variants with different sizes, we had three combined models, namely Combined-10, Combined-22 and Combined-77 models. In both the genetic models and the combined models, we handled the genetic variants in the following way rather than using original genotypes of each SNP. We only introduced one additional variable, the total count of risky alleles the person carries in the DNA. This way of coding genetic variants was used in several models such as [5], and is helpful to build risk models when each SNP only has a small contribution to the risk.
We treated the BI-RADS category scores from the radiologists as the predictions from the radiologists, namely the baseline clinical assessment. We constructed ROC curves for each model, and used the area under the curve (AUC) as a measure of performance. We also provided the precision-recall (PR) curves for the models. We evaluated the models using 10-fold cross-validation.
Results
We identified 362 cases and 377 controls. Among the cases, there were 358 Caucasians, three non-Caucasians and one case whose race information was unknown. Among the controls, there were 373 Caucasians and four non-Caucasians. We do not disclose the race/ethnicity information of these non-Caucasians for privacy concerns. Subject characteristics including age distribution and family history of breast cancer are described in Table 2. There were more young people (age <50) in the case group than in the control group, and the proportion of elderly people (age ≥65) was roughly the same in the case group and in the control group. For the family history of breast cancer, we observed a considerable larger proportion of people with family history in the case group (45.3%) than in the control group (33.7%), which demonstrated the family aggregation of breast cancer.
Table 2.
The distribution of age and family history of breast cancer.
| Cases | Controls | All | Cases | Controls | All | ||
|---|---|---|---|---|---|---|---|
| Age Group | Family History | ||||||
| <50 | 81 (22.4%) | 58 (15.4%) | 139 (18.8%) | Yes | 164 (45.3%) | 127 (33.7%) | 291 (39.4%) |
| 50–65 | 123 (34.0%) | 168 (44.6%) | 291 (39.4%) | No | 188 (51.9%) | 236 (62.6%) | 424 (57.4%) |
| >65 | 158 (43.6%) | 151(40.0%) | 309 (41.8%) | N/A | 10 (2.8%) | 14 (3.7%) | 24 (3.2%) |
The Performance of the Three Combined Models
The ROC and the PR curves for the baseline clinical assessment, the Breast Imaging model and the three combined models are provided in Figure 1. For each model, we vertically average [32] the ROC curves from the ten replications of the 10-fold cross-validation to obtain the final curve; we do likewise for the PR curves. The area under the ROC curves for the Breast Imaging model, the Combined-10 model, the Combined-22 model and the Combined-77 model are 0.693, 0.712, 0.733 and 0.760. The ROC curve of the Combined-77 model almost completely dominates the ROC curve of the Breast Imaging model, which suggests that the 77 genetic variants can help to improve breast cancer diagnosis based on mammographic findings. We perform a two-sided paired t-test on the area under the ten ROC curves of the Breast Imaging model and the area under the ten ROC curves of the combined model from the 10-fold cross-validation, and the difference between them is significant with a P-value 0.00047. We further compare the AUROC of the Combined-77 model and the Combined-22 model with a two-sided paired t-test, and the difference between them is significant with a P-value 0.0046, which demonstrates the discriminative power of the 55 recently identified SNPs. From PR curves, we note that the combined models dominate the Breast Imaging model and the baseline clinical assessment in the high recall region (>0.8) in which clinicians operate, and therefore we want to optimize.
Figure 1.
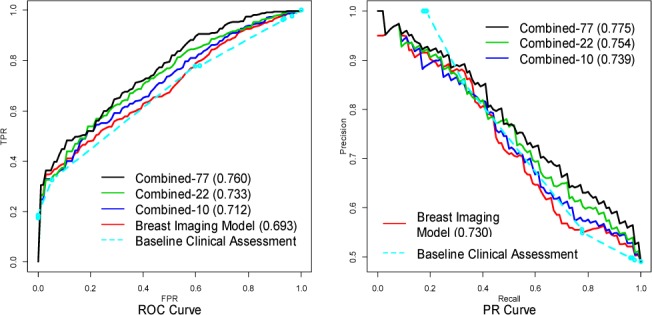
The ROC curves and PR curves for the baseline clinical assessment, the Breast Imaging model the three combined models.
The Performance of the Three Genetic Models
Furthermore, we compare the discriminative power of the three genetic models, namely the Genetic-10 model, the Genetic-22 model and the Genetic-77 model. The ROC curves and the PR curves for the three genetic models are provided in Figure 2, respectively. For each model, we vertically average the curves from the 10-fold cross-valuation to obtain the final curve. The area under the ROC curves for the Genetic-10 model, the Genetic-22 model and the Genetic-77 model are 0.591, 0.622 and 0.684, which demonstrates that the more associated SNPs the genetic model includes, the more discriminative the model becomes. We also use a two-sided paired t-test to compare the area under the ROC curves yielded by the three genetic models. The Genetic-77 model outperforms both the Genetic-22 model (P=0.028) and the Genetic-10 model (P=0.0068).
Figure 2.

The ROC and PR curves for the three genetic models.
Comparing Breast Imaging and Genetic-77 Model
We compare the performance of the Breast Imaging model, the Genetic-77 model and the Combined-77 model. The corresponding ROC curves and the PR curves for the three models are shown in Figure 3. We observe that the mammography features are more predictive for women with a high probability of cancer (low FPR region in ROC space) whereas genetic variants are more predictive for women with a low probability of cancer (mid/high FPR region in ROC space). Note that the Genetic-77 model describes the patient’s inherited breast cancer risk in DNA. However, after the patient starts developing malignant features on mammograms, mammographic findings (Breast Imaging model) provide superior discrimination. Still, knowing the genetic information can further improve the accuracy of breast cancer diagnosis even at higher baseline risk.
Figure 3.
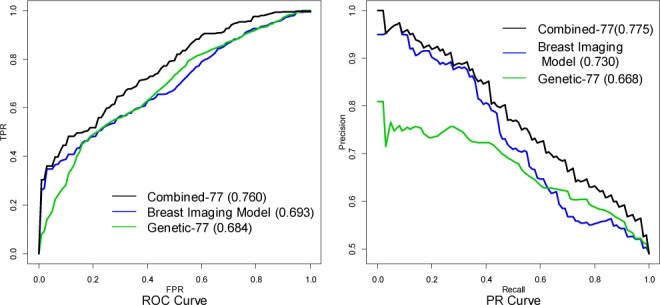
The ROC curves and PR curves for the Breast Imaging model, the Genetic-77 model and the Combined-77 model.
Discussion
The primary contribution of our study is to show that the genetic variants can significantly improve breast cancer diagnosis on mammographic findings, resulting in reduced false positives and alleviated risk of overdiagnosis. This result indicates promise for translating discoveries from massive collaborative GWAS into clinical breast cancer diagnosis. Our study includes the most up-to-date breast cancer associated SNPs, the majority identified and/or verified through the massive COGS (over 55k cases and over 54k controls), and therefore these new SNPs are credible and can explain a larger proportion of familial breast cancer risk. Indeed, we observe that the Combined-77 model significantly outperforms the Combined-22 model used in our previous study [6]. We also demonstrate that the Genetic-77 model significantly outperforms the Genetic-22 model. The increased discriminative power derived from the new 55 SNPs identified by recent published studies [2] highlights the rapid progress the breast cancer GWAS community has made since 2010. Furthermore, we make a novel discovery that mammography features are more predictive for high-risk women whereas genetic variants are more predictive for low-risk women, which explains the benefit of combining genetic variants and mammographic findings for personalized breast cancer diagnosis.
Our study, in a novel way, differs from the previous study of Wacholder et al. (2010) [5] which adds ten genetic variants to the Gail model, a risk model based on self-reported demographic and personal risk factors. The unique contribution in our study is that we include mammography features which represent richer phenotypic data directly relevant to breast cancer diagnosis and thus provide high signal. Therefore, our study contributes the potential clinical impact of translating exciting discoveries from GWAS to the patient experience at diagnosis. The additional discriminative power from these genetic variants can significantly rule out the false positives of mammogram screening, and therefore has the potential to decrease recommendations for unnecessary breast biopsies. Of course, it will be interesting to combine the epidemiology features in Gail model, the mammography features and the SNPs for more accurate personalized breast cancer diagnosis.
Limitations of our study include small sample size and the pitfalls of data extraction from text reports. We understand that parsing mammography features from text reports may introduce noise into the data. However, despite the challenges inherent in extracting accurate data, which may affect our results, we are encouraged that improvements in predictive accuracy remain, especially after observing the discriminative power of genetic factors alone in the genetic models. Furthermore, we recognize that methodological issues in our study may represent shortcomings but also signify opportunities for future investigation. First, we do not explicitly model how individual SNPs function to alter breast cancer risk, nor do we model potential SNP interactions [33]. Our current model only adds one extra feature which simply counts the totally number of risky alleles, assuming that the effect size of the genetic variants are the same and that the genetic effect of the genetic variants is non-mechanistic and additive. We do not model the individual SNPs for the curse of dimensionality concern; each individual SNP only confers a fairly mild relative risk and if we model them individually, the model will perform poorly on test data unless a larger cohort of training data is available. Modeling SNP-SNP interaction is even harder and requires more training data.
Second, we do not differentiate the different subtypes of breast cancers (for example, the estrogen-receptor status and progesterone-receptor status) in the current study. Breast cancer is a complex and heterogeneous disease with different subtypes, including two main subtypes of estrogen receptor (ER) negative tumors (basal-like and human epidermal growth factor receptor-2 positive/ER− subtype) and at least two types of ER positive tumors (luminal A and luminal B) [34, 35]. These molecular subtypes are important predictors of breast cancer mortality [36] and have different genetic susceptibility [37]. Therefore it is desirable to tease them apart in the pursuit of increasingly personalized breast cancer care.
Nevertheless, we are encouraged by these promising results in our current study, especially after the disappointment [38] and caution [39] in the early years of translating GWAS discoveries to personalized risk prediction. We hope that the rapid progress being made through these massive collaborative studies together with our growing knowledge about breast cancer mechanisms and genotype-phenotype relationships will bring us even closer to the practical personalized breast cancer diagnosis and treatment.
Acknowledgments
The authors acknowledge the support of the Wisconsin Genomics Initiative, NCI grant R01CA127379-01 and its ARRA supplement 3R01CA127379-03S1, NIGMS grant R01GM097618-01, NLM grant R01LM011028-01, NIEHS grant 5R01ES017400-03, the UW Institute for Clinical and Translational Research (ICTR) and the UW Carbone Cancer Center.
Reference
- 1.Bahcall OG. iCOGS collection provides a collaborative model. Foreword. Nat Genet. 2013;45(4):343. doi: 10.1038/ng.2592. [DOI] [PubMed] [Google Scholar]
- 2.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–61. 361e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101(13):959–63. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–41. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wacholder S, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–93. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, et al. Genetic Variants Improve Breast Cancer Risk Prediction on Mammograms; American Medical Informatics Association Symposium; 2013. [PMC free article] [PubMed] [Google Scholar]
- 7.Warren H, et al. 9q31.2-rs865686 as a susceptibility locus for estrogen receptor-positive breast cancer: evidence from the Breast Cancer Association Consortium. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1783–91. doi: 10.1158/1055-9965.EPI-12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens KN, et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72(7):1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21(24):5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoussaini M, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44(3):312–8. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher O, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103(5):425–35. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 12.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42(10):885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty C, et al. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 15.Marmot M, et al. The benefits and harms of breast cancer screening: an independent review. British Journal of Cancer. 2013;108(11):2205–2240. doi: 10.1038/bjc.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houssam N, et al. Information Extraction for Clinical Data Mining: A Mammography Case Study; Proceedings of the 2009 IEEE International Conference on Data Mining Workshops; IEEE Computer Society; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Percha B, et al. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc. 2012;19(5):913–6. doi: 10.1136/amiajnl-2011-000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowd D, Domingos P. Naive Bayes models for probability estimation; Proceedings of the 22nd international conference on Machine learning; 2005. [Google Scholar]
- 19.Hall M, et al. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11(1):10–18. [Google Scholar]
- 20.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(17293864):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 22.Odefrey F, et al. Common genetic variants associated with breast cancer and mammographic density measures that predict disease. Cancer Res. 2010;70(20145138):1449–1458. doi: 10.1158/0008-5472.CAN-09-3495. [DOI] [PubMed] [Google Scholar]
- 23.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(17529967):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(19330027):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biong M, et al. Genotypes and haplotypes in the insulin-like growth factors, their receptors and binding proteins in relation to plasma metabolic levels and mammographic density. BMC Med Genomics. 2010;3(20302654):9–9. doi: 10.1186/1755-8794-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelemen LE, Sellers TA, Vachon CM. Can genes for mammographic density inform cancer aetiology? Nat Rev Cancer. 2008;8(18772892):812–823. doi: 10.1038/nrc2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(18438407):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 29.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(19219042):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gold B, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105(18326623):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(17529973):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T F. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27(8):861–874. [Google Scholar]
- 33.Turnbull C, et al. Gene-gene interactions in breast cancer susceptibility. Hum Mol Genet. 2012;21(4):958–62. doi: 10.1093/hmg/ddr525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carey LA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295(21):2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 35.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 36.Haque R, et al. Impact of breast cancer subtypes and treatment on survival: an analysis spanning two decades. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1848–55. doi: 10.1158/1055-9965.EPI-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Closas M, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45(4):392–8. 398e1–2. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–8. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 39.Kraft P, Hunter DJ. Genetic risk prediction–are we there yet? N Engl J Med. 2009;360(17):1701–3. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]


