Abstract
In this paper, we are using “structurally congruent concepts” in pairs of terminologies to suggest methods for harmonizing the terminologies. Two concepts are structurally congruent if they are children of the same more general concept and parents of the same more specific concept in two different terminologies. We show that structurally congruent concepts can be interpreted in six useful ways, e.g., as new synonyms. All structurally congruent concepts were found for six terminologies from the UMLS, each paired with SNOMED CT. In total, 1384 concept pairs were discovered. Concepts from a sample of 241 pairs were analyzed by a human expert. It was found that 59.3% indicated alternative classifications of the same general concept. This discovery allows an ontology designer to make existing, implicit knowledge explicit. Another 14.5% were newly discovered synonyms, 23.6% suggested the import of a concept into a terminology and 2.5% indicated errors in a terminology.
Introduction
Semantic interoperability is one of the big challenges in biomedical informatics. In order to enrich the semantics and coverage of a terminology and facilitate translational biomedical informatics to be utilized in clinical and research applications, semantic harmonization efforts have recently been extended for various terminologies, e.g. SNOMED CT [1]. However, structural methodologies for semantic harmonization of terminologies have not been studied sufficiently. Weng et al. [2] discussed a conceptual design of a collaborative system for semantic harmonization. Three key design principles were defined: (1) reuse, (2) collaboration, (3) harmonization as modeling. The BRIDG model was presented as a user-centric semantic harmonization framework [3]. The harmonization in the BRIDG model is based on the concept definitions, attributes, and concept relationships. Due to the fact that BRIDG participants are distributed across organizations and no implementation-specific information is provided, it may be hard to use this approach directly by application-oriented users. Tao et al. have discussed the importance of ontology harmonization before using ontologies to annotate clinical data [4]. In this paper, we are approaching semantic harmonization by analyzing the relationships between structurally congruent concepts from pairs of terminologies in the UMLS. An outline of the implementation details for finding such structurally congruent pairs is provided.
Auditing of terminologies may uncover problems such as omissions [5]. Previously, we have developed algorithmic and mixed human-computer auditing methods for the UMLS and some of its source terminologies [6, 7]. Auditing may also discover concepts that are synonymous in real life but are coded as different in the UMLS. Occasionally two terminologies in overlapping domains “cut the world at different joints,” which makes ontology alignment [8] and ontology integration difficult. In such a situation, the same conceptual knowledge may be classified in (often orthogonal) different ways. We call these “alternative classifications.” In this paper, we are describing the use of structural congruency in pairs of terminologies to alert a human auditor to possible cases of harmonization and correction. Due to the importance of SNOMED CT (abbreviated as “SNOMED”), we focus on its concepts.
Background
SNOMED CT (Systematized Nomenclature of Medicine – Clinical Terms) [9–11] is considered to be of increasing importance in Medical Informatics. One reason for this status is related to government mandates of using Electronic Health Record systems, meaningful use and incentive payments to physicians. By 2015, SNOMED will become the standard terminology for EHR encoding of diagnoses and problem lists [12]. SNOMED is to be used to “enable a user to electronically record, modify, and retrieve a patient’s problem list for longitudinal care (i.e., over multiple office visits).” Thus, in this paper, we are focusing on categorizing the relationships between structurally congruent concepts, one from SNOMED, the other from six reference terminologies. The Unified Medical Language System’s (UMLS) [13–16] Metathesaurus [17, 18] is an excellent source of pairs of terminologies with matched concepts. The 2012AB Metathesaurus contains more than 2.8 million concepts and 8.6 million unique concept names from about 160 source vocabularies [19]. SNOMED is also included in the UMLS.
Previously, Bodenreider performed a study of redundant relations and similarity across families of terminologies and discussed the relationship between redundancy and semantic consistency [20]. Bodenreider observed ([21]) that it is the policy in the UMLS that ‘PAR’ represents an explicit parent-child relationship in a source, and ‘RB’ indicates an implied one (as interpreted by the UMLS editorial team). In this paper, we are focusing on explicit hierarchical relationships, thus only terminologies in the UMLS with ‘PAR’ links annotated with ‘IS_A’ relationship attributes were chosen. This current work is also marginally related to research on density and granularity of terminologies. Kumar et al. [22] lay out a comprehensive theory of granularity in the context of medical terminologies. Schulz et al. identify granularity-related problems with “cross-granularity integration” in the biomedical domain [23]. Rector et al.’s analysis provides logical formulations of important distinctions between density and related properties [24].
Methods
Our method is based on comparing two medical terminologies from the UMLS. We formally define the targets of our investigation as follows.
Definition: The concepts X (from Terminology 1) and Y (of Terminology 2) are called “structurally congruent” if:
-
a)
Both concepts X and Y have the same parent A in Terminology 1 and in Terminology 2.
-
b)
Both concepts X and Y have the same child B in Terminology 1 and in Terminology 2.
-
c)
The concept X does not appear anywhere in Terminology 2.
-
d)
The concept Y does not appear anywhere in Terminology 1.
-
e)
There is no synonymy relationship and no hierarchical relationship between X and Y (in the UMLS).
Figure 1 shows an abstract layout of two structurally congruent concepts to elucidate the above definition.
Figure 1.
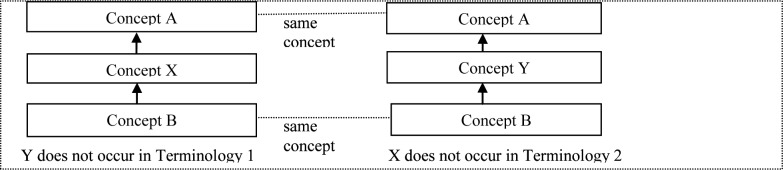
An abstract layout of structurally congruent concepts
It is hypothesized that there are six possible cases for how X and Y may relate to each other.
-
1)
The concepts X and Y are alternative classifications. That means that concept A may be validly assigned X and Y as its children. However, these two assignments are indicative of two different ways of clustering the grandchildren of A. Furthermore, concept B may be correctly classified as a child of X and as a child of Y. However, Terminology 1 omits the classification by Y and Terminology 2 omits the classification by X.
-
2)
It holds that B IS_A Y, Y IS_A X, and X IS_A A. In other words, Y may be inserted as a child of X into Terminology 1, thereby adding more detailed information to Terminology 1. Similarly, X may be inserted as a parent of Y into Terminology 2. Such insertions should only be done with approval of a subject matter expert.
-
3)
It holds that B IS_A, X IS_A Y, and Y IS_A A. This is the mirror case of Case 2) in that now X may be inserted as a child of Y into Terminology 2 and Y may be inserted as a parent of X into Terminology 1.
-
4)
Concept X is a real world synonym of concept Y, which was previously not recognized by the UMLS editors.
-
5)
There might be a structural error in Terminology 1, e.g., X is not really a child of A.
-
6)
There might be a structural error in Terminology 2.
Every one of these six cases may be utilized in a human review, possibly leading to an improvement and harmonization of both terminologies. To further probe the potential of this idea, we performed the following study. Six terminologies were selected from the 2012AB release of the UMLS to function as reference terminologies for SNOMED. (Note: It is a coincidence that there are six cases and six terminologies.) Only English-language terminologies using the “PAR” relationship annotated with “IS_A” relationship attributes were chosen. They are MEDCIN3_2012_07_16, National Cancer Institute Thesaurus (NCI2012_02D), Gene Ontology (GO20 12_04_03), Medical Entities Dictionary (CPM2003), UMDNS: product category thesaurus (UMD2012) and Foundational Model of Anatomy Ontology (FMA3_1). Due to the fact that the University of Washington Digital Anatomist (UWDA) consists of the Anatomy component and selected structural relationships of FMA, UWDA was excluded even though it also uses “PAR” relationships and “IS_A” relationship attributes. The algorithms were implemented in the Oracle Relational Database Management System (RDBMS) native programming language PL/SQL. The algorithms were used for finding all structurally congruent pairs of concepts, one taken from the list of six reference terminologies, the other one being the July 2012 version of SNOMED. The UMLS is well known to contain many cycles [21, 25], which were eliminated during processing.
Results
Table 1 shows the numbers of pairs of congruent concepts of six reference terminologies relative to SNOMED and the sizes of the samples we randomly chose for human review. The third column shows the number of pairs of congruent concepts found by the program. For reference terminologies with over 100 pairs of congruent concepts, random samples of 70 were chosen for human review; for the others, all of the congruent concepts were reviewed. In total, we reviewed 241 /1384 = 17.4% of all the congruent concept pairs discovered by the program.
Table 1.
Comparison of SNOMED CT with six reference terminologies
| Reference Terminology | Size of Terminology | # of Pairs of Congruent Concepts | Sample Size |
|---|---|---|---|
| MEDCIN3_2012_07_16 | 279529 | 655 | 70 |
| NCI2012_02D | 95523 | 582 | 70 |
| FMA3_1 | 82062 | 116 | 70 |
| UMD2012 | 15956 | 18 | 18 |
| GO2012_04_03 | 61925 | 6 | 6 |
| CPM2003 | 3078 | 7 | 7 |
| Total | – | 1384 | 241 |
The author GE, a medical informaticist and MD with many years of experience in auditing terminologies reviewed the sample. Table 2 shows the results according to the six cases defined in the Methods section. The results show that 59.3% are alternative classifications. Another 14.9% + 8.7% = 23.6% fall into the category where the congruent concept in the reference terminology could be imported into SNOMED, and vice versa.
Table 2.
Review results by reference terminology
| Reference Terminology | Sample Size | Alternative Classific. | Y IS_A X | X IS_A Y | Error in Trmgy 1 | Error in Trmgy 2 | Synonym |
|---|---|---|---|---|---|---|---|
| MEDCIN3_2012_07_16 | 70 | 44 | 10 | 7 | – | 1 | 8 |
| NCI2012_02D | 70 | 38 | 12 | 6 | – | 3 | 11 |
| GO2012_04_03 | 6 | 2 | – | 4 | – | – | – |
| CPM2003 | 7 | 5 | – | – | – | – | 2 |
| UMD2012 | 18 | 9 | 1 | – | – | – | 8 |
| FMA3_1 | 70 | 45 | 13 | 4 | 2 | – | 6 |
| Total | 241 | 143 | 36 | 21 | 2 | 4 | 35 |
| Percentage | 100% | 59.3% | 14.9% | 8.7% | 0.8% | 1.7% | 14.5% |
Figure 2 shows an example where congruent concepts were identified as alternative classifications. Thus, Eleventh posterior intercostal vein in the FMA is a classification by cardinality, while in SNOMED Lower right posterior intercostal vein is a classification by position.
Figure 2.
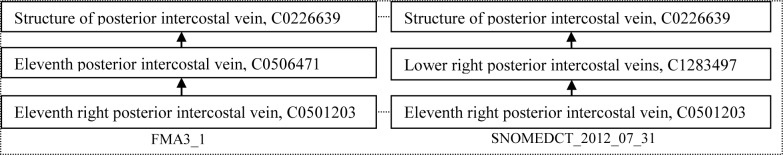
An example of alternative classification
The discovery of alternative classifications is useful, because it makes explicit the implicit assumptions of the ontology designers how they are viewing the world. This view could then be codified in the ontology. Figure 3 shows the utilization of the findings in Figure 2 by adding two new concepts (with labels shown in Italics.)
Figure 3.
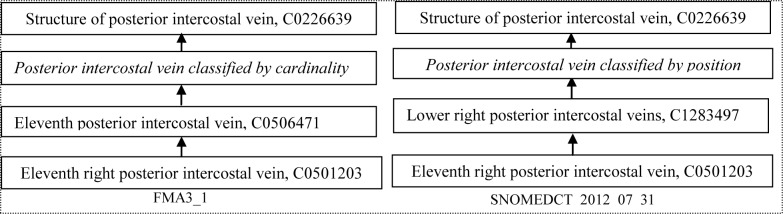
An example of making explicit an implicit assumption of the ontology designers
Figure 4 shows a case where one congruent concept was deemed a parent of the other by the auditor. In this example, the congruent concept Finding by Site or System can be a parent of Finding by site, thus the congruent concept Finding by Site or System from FMA may be added as a parent of Finding by site in SNOMED, and vice versa, if this is desirable in the judgment of the owners of the FMA and/or SNOMED.
Figure 4.
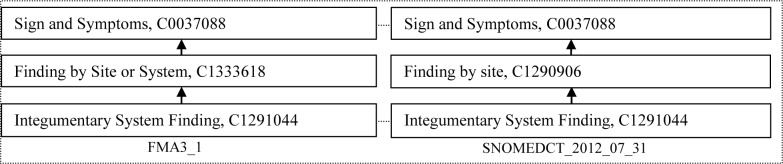
An example of one structurally congruent concept being a parent of the other
The congruent concepts Chemical Viewed Structurally from CPM and Chemical categorized structurally from SNOMED are deemed synonyms that were not recognized before by our auditor (Figure 5) and should be merged.
Figure 5.
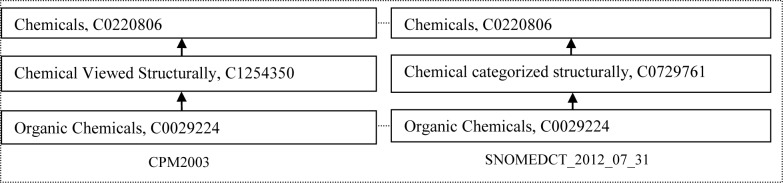
An example of one middle concept being synonymous of the other
During the review of the sample, a few errors within terminologies emerged. The concept from SNOMED Artificial Implant was deemed incorrect by the auditor because it should not be considered as “artificial,” in the structure with A = Prosthesis, C0175649, Y = Artificial Implants, C0021113, and B = Blood Vessel Prosthesis, C0005846.
Discussion
The UMLS provides many concept pairs from different terminologies, where algorithmically made structural observations raise the question how to harmonize those concepts. In this paper, we identified one such structural observation “structurally congruent concepts” and indicated the different ways how such a congruency can be resolved. However, the semantic harmonization cannot be done without the consent of terminology curators. Moreover, modeling differences between terminologies make semantic harmonization difficult. For UMD2012 (Table 2), eight pairs of congruent concepts were found to be synonyms. For GO, more cases where one congruent concept is a potential parent of the other were found than alternative classifications. For our cases 2) and 3), relevant work in MIREOT [26] defines a set of guidelines for importing classes from external ontologies and proposes an automated mechanism and a minimal information standard for selectively importing classes into an ontology. However, it only supports OBO foundry ontologies (OWL format). In this paper, all the terminologies are in UMLS RRF format. Thus, the import guidelines introduced in MIREOT cannot be used here directly.
A possible limitation of this work is that it uses SNOMED concepts and all reference terminology concepts in the formats that they were provided in by the UMLS. There may be differences between the original concept representation of SNOMED (or the reference terminologies) and the representation of SNOMED that is accessible through the UMLS.
Conclusions and Future Work
Six terminologies of the UMLS were compared with SNOMED with respect to structurally congruent concepts. In a sample study it was found that the great majority of cases corresponded to alternative analysis situations (143 out of 241, corresponding to 59.3%). The second most common situation indicated the possibility of adding more detail to SNOMED CT or the reference terminologies (57 out of 241, corresponding to 23.6%). In 35 cases new synonyms were discovered, and three pairs of concepts indicated errors. As future work, we plan to conduct a study to analyze structurally congruent concepts between pairs of any two META terminologies with explicitly defined hierarchical relationships, e.g., not limited to SNOMED CT being Terminology 2. We are also planning a more extensive evaluation of the results. The work in this paper was limited to pairs of structurally congruent concepts. However, we have noticed cases of congruency that involve three, four and even more concepts. An analysis of these cases is under way.
References
- 1.IHTSDO SNOMED CT and LOINC to be linked by cooperative work. 2013. [cited September 29, 2013]; Available from: http://www.ihtsdo.org/about-ihtsdo/governance-and-advisory/harmonization/loinc/
- 2.Weng C, Fridsma DB. A call for collaborative semantics harmonization; AMIA Annu Symp Proc; Washington DC; 2006. [PMC free article] [PubMed] [Google Scholar]
- 3.Weng C, Gennari JH, Fridsma DB. User-centered semantic harmonization: a case study. J Biomed Inform. 2007 Jun;40(3):353–64. doi: 10.1016/j.jbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Tao C, Solbrig HR, Chute CG. CNTRO 2.0: A Harmonized Semantic Web Ontology for Temporal Relation Inferencing in Clinical Narratives. AMIA Summits Transl Sci Proc. 2011;2011:64–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Geller J, Perl Y, Halper M, Cornet R. Special issue on auditing of terminologies. J Biomed Inform. 2009 Jun;42(3):407–11. doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Gu H, Perl Y, Geller J, Halper M, Liu LM, Cimino JJ. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000 Jan-Feb;7(1):66–80. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Gu HH, Perl Y, Geller J. Structural group-based auditing of missing hierarchical relationships in UMLS. J Biomed Inform. 2009 Jun;42(3):452–67. doi: 10.1016/j.jbi.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shvaiko P, Euzenat J. Ontology Matching: State of the Art and Future Challenge. Knowledge and Data Engineering, IEEE Transactions. 2013;25(1):158–76. [Google Scholar]
- 9.Wilcke JR, Green JM, Spackman KA, et al. Concerning SNOMED-CT content for public health case reports. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):613. doi: 10.1136/jamia.2010.003756. author reply -4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stearns MQ, Price C, Spackman KA, Wang AY. SNOMED clinical terms: overview of the development process and project status. Proc AMIA Symp. 2001:662–6. [PMC free article] [PubMed] [Google Scholar]
- 11.SNOMED CT Homepage. [cited January 10, 2013]; Available from: http://www.ihtsdo.org.
- 12.US Department of Health and Human Services Health Information Technology: Initial Set of Standards, Implementation Specifications, and Certification Criteria for Electronic Health Records Technology. [cited May 21, 2013]; Available from: http://www.gpo.gov/fdsys/pkg/FR-2010-07-28/pdf/2010-17210.pdf.
- 13.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell KE, Oliver DE, Shortliffe EH. The Unified Medical Language System: toward a collaborative approach for solving terminologic problems. J Am Med Inform Assoc. 1998 Jan-Feb;5(1):12–6. doi: 10.1136/jamia.1998.0050012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998 Jan-Feb;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuyler PL, Hole WT, Tuttle MS, Sherertz DD. The UMLS Metathesaurus: representing different views of biomedical concepts. Bull Med Libr Assoc. 1993 Apr;81(2):217–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Tuttle M, Sherertz DD, E M, Olson N, Nelson S. Implementing Meta-1: The First Version of the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care. 1989:483–7. [Google Scholar]
- 19.Resource Description Framework (RDF) [cited March 3, 2013]; Available from: http://www.w3.org/RDF/
- 20.Bodenreider O. Strength in numbers: exploring redundancy in hierarchical relations across biomedical terminologies. AMIA Annu Symp Proc. 2003:101–5. [PMC free article] [PubMed] [Google Scholar]
- 21.Bodenreider O. Circular hierarchical relationships in the UMLS: etiology, diagnosis, treatment, complications and prevention. Proc AMIA Symp. 2001:57–61. [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Smith B, Novotny DD. Biomedical informatics and granularity. Comp Funct Genomics. 2004;5(6–7):501–8. doi: 10.1002/cfg.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz S, Boeker M, Stenzhorn H. How Granularity Issues Concern Biomedical Ontology Integration; Proceedings of the International Congress of the European Federation for Medical Informatics (MIE 2008); Gothenburg, Sweden. 2008. pp. 863–68. [PubMed] [Google Scholar]
- 24.Rector A, Rogers J, Bittner T. Granularity, scale and collectivity: when size does and does not matter. J Biomed Inform. 2006 Jun;39(3):333–49. doi: 10.1016/j.jbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Mougin F, Bodenreider O. Approaches to eliminating cycles in the UMLS Metathesaurus: naive vs. formal. AMIA Annu Symp Proc. 2005:550–4. [PMC free article] [PubMed] [Google Scholar]
- 26.Courtot M, Gibson F, Lister AL, Malone J. MIREOT: The Minimum Information to Reference an External Ontology Term. In: Smith B, editor. International Conference on Biomedical Ontology. Buffalo, New York, USA: 2009. pp. 87–90. [Google Scholar]


