Abstract
The tranSMART knowledge management and high-content analysis platform is a flexible software framework featuring novel research capabilities. It enables analysis of integrated data for the purposes of hypothesis generation, hypothesis validation, and cohort discovery in translational research. tranSMART bridges the prolific world of basic science and clinical practice data at the point of care by merging multiple types of data from disparate sources into a common environment. The application supports data harmonization and integration with analytical pipelines. The application code was released into the open source community in January 2012, with 32 instances in operation. tranSMART’s extensible data model and corresponding data integration processes, rapid data analysis features, and open source nature make it an indispensable tool in translational or clinical research.
Background
Translation of biomedical research to patient care has been a difficult path, fraught with barriers and a paucity of information technology solutions. Funding structures have led to a landscape of siloed knowledge where collaboration and sharing were not facilitated.1 Software tools and data handling processes have been lacking, with little success in standardizing data integration practices.2 The pace of applying research findings into the clinical practice was also woefully slow.3,4,5 These issues were rapidly gaining notice in the early 2000’s, as identified in seminal publications.6 In 2003, the National Institutes of Health defined the Roadmap to Medical Research, and identified translation as a “vital component of research and health-care improvement.”6 However, the journey to bring new findings to the point of care has been stymied at the clinical level.3,4,6 Much of the research activity has resided in basic science and clinical trials (vs the clinical environment), where significant incentives exist to encourage development of new treatment options and modalities.
The demand for translating basic research into clinical practice has resonated in the pharmaceutical domain.7 Johnson & Johnson (J&J) identified a “significant challenge in the lack of translatability of preclinical models into meaningful biological knowledge.”8 J&J partnered with Recombinant Data Corporation in 2008 to develop “a knowledge management platform that would provide access to all R&D data as well as advanced analytics.”8 The first iteration of tranSMART was deployed in 2010; it utilized components of the i2b2 (Informatics for Integrating Biology and the Bedside; http://www.i2b2.org) schema,9 and incorporated search capabilities. A parallel goal was to release the software as open source to encourage precompetitive data sharing among pharmaceutical entities and academic institutions, including the Cancer Institute of New Jersey.8 Since its open source release in January 2012, tranSMART has continued to mature into a robust translational research knowledge management and analysis platform.
Materials and Methods
Application Infrastructure
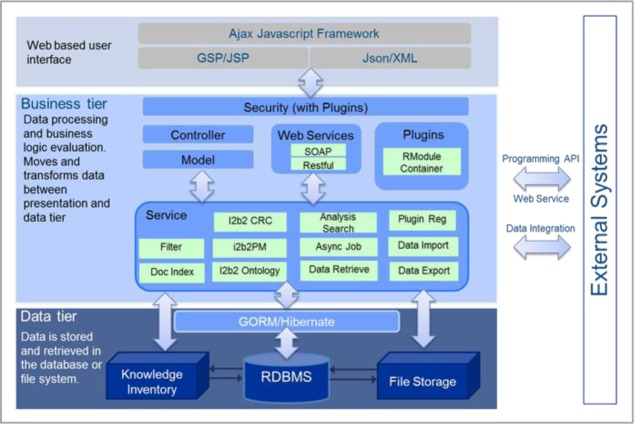
tranSMART version 1.1 employs an N-tier architecture (see Figure 1) and is built on Grails – a rapid web application development framework based on a Java platform (http://www.grails.org). The N-tier architecture typically separates application needs into a data-handling tier, a business logic tier, and the presentation tier. Here the data-handling tier is supported by a relational database with a series of purpose-built schemas to house various data types. The business logic tier code base is also written in Grails. The presentation layer is a web-based application. As a framework, Grails was selected for tranSMART because of its value in facilitating the development of robust web applications. By its nature, the framework lends itself to extensibility, making it a good fit for open source development. Functionality can be added to Grails applications through the plugin infrastructure, with plugins that are custom built or sourced through the Grails community.
Figure 1.
N-tier Architecture of tranSMART
The Data Tier (see Figure 1) houses the data used in the tranSMART in relational databases, including raw data from files and databases as well as data normalized to common formats and ontologies (discussed in the Data section below). Supported file types include Microsoft Word documents, text files, PDFs, spreadsheets, and ‘omic data files. Data can be added to the tranSMART database through a suite of tools that support data extraction, transformation and loading (ETL). These tools load raw data that has been mapped via standard templates by a data curator into tranSMART schemas. Additional data files of any format can be associated on the patient level and linked via the file system. Grails makes use of a custom Object Relational Mapping system called GORM. GORM provides a connection between programmatic objects in Grails and underlying database objects. One significant advantage of the GORM layer is that it allows for the application to be database agnostic, thus enabling tranSMART to run on virtually any Relational Database Management System (e.g., Oracle, PostgreSQL). The Data Tier is made accessible through data services, which are predefined methods of communication between various components of the overall application. Data services reside in the business logic tier.
The Business Logic Tier (see Figure 1) encapsulates tranSMART’s application logic and provides services that implement tranSMART’s core functionality such as security, i2b2 features, data export and other plugins. Security in tranSMART is handled by the Spring Security Core open source Grails plug in. Spring Security is the de facto authorization plugin adopted by the Grails community; it allows for the rapid development of form-based or single sign-on authentication. Built into Spring Security is a role-based permissions management system. Another service is the i2b2 interface module that provides connectivity to the various individual i2b2 applications (known as cells) through web services and direct database calls. There is an export functionality that allows the user to download datasets in commonly available formats such as tab delimited text files, GSEA, and PLINK. The exported data can be used for further analysis in external statistical packages (e.g., Stata) Additionally, an Rmodules Grails plugin allows a researcher to specify a cohort of subjects with associated data of interest and run pre-defined analytics via an R statistical package. To do so, a non-programmer user chooses one of the available analyses and specifies the relevant input parameters. The data is subsequently pulled, formatted, and sent via a tranSMART API to R, where the relevant scripts are executed. The results are then presented in the tranSMART interface. Together, the Business Tier services are responsible for the core functionality of tranSMART.
tranSMART’s Presentation Tier (see Figure 1) relies on the data services to decouple the presentation layer from the data. JavaScript, along with popular libraries such as jQuery and ExtJS, is used to define the UI elements and the interactions among them. Asynchronous JavaScript and XML (AJAX) are used extensively to provide a rich interactive experience. JavaScript Object Notation (JSON) and XML provide a standard communication language between the presentation and business layers. Groovy Server Pages (GSP) are used by the Grails framework to display HTML content to the end user. Tag libraries can be built or downloaded to allow for more rapid development of common form and display elements.
Data, Data Store and Data Integration
The tranSMART knowledge management platform allows for the integration of data from a variety of data sources, across multiple data types. The data types that can be loaded into tranSMART include patient- or study-level clinical data (e.g., demographics, diagnosis, medications, lab results, etc.), subject-level high dimensional data (e.g., genotyping calls, gene expression arrays, protein expression arrays, etc.), study-level results and findings, as well as study descriptors in the form of metadata. The data can come from public sources (e.g., TCGA, GEO), or from internal sources (e.g., institutional clinical trials, etc.). Reference content brought into tranSMART can be proprietary (e.g., GeneGo, Ingenuity) or openly available (e.g., Entrez, MeSH). Additionally, the original data sources can be stored as files and accessed for export via the user interface for future research workflows.
Data integration is a vital step in ensuring downstream success of any data analysis undertaking. There are multiple issues in the data management workflow in a typical research environment. Raw data frequently have formatting problems and form discrepancies that are undetected or unintended at the original capture level, but can cause problems in subsequent activities, such as search or data analysis. Data often require some level of normalization or harmonization in order to be analyzed and utilized properly. In the typical research workflow, data is frequently cleaned to a very specific analytical purpose. If another type of analysis is desired, the data requires another round of time consuming data cleaning activities. If data is instead conformed to a common structure and representation of meaning, thus rendering it agnostic to the analysis, users can focus on studying the data rather than spending valuable time on data preparation.
We employ an internally developed best practices approach to data integration from various sources into a common environment (see Figure 2).10 This robust process systematizes the data integration effort into a series of well-defined steps and delegates specific tasks to staff members with appropriate skillsets. Data is mapped to a standard ontology that can be institution-specific or conform to an industry standard (e.g., SNOMED CT, SDTM).
Figure 2.
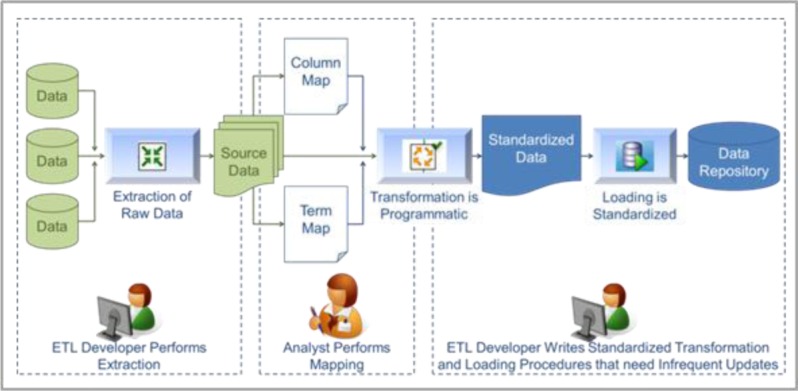
Best Practice Approach to Data Integration
The data integration process requires several discrete steps. First, under the direction of the principle scientist or system product owner, the data source is identified and selected for migration into the tranSMART environment. Next, an ETL developer performs the source data extraction and the data curation process is initiated. Both syntactic and semantic mappings are created in this step (see Figure 2). A knowledgeable analyst who follows standard practices performs the mapping of the data into the templates, ensuring high quality output. Once the mapping templates are filled out, an ETL developer can run the programmatic transformation of the source data into the standard format and load the output into tranSMART. In the last stage of the process, the data undergoes a quality assurance review to ensure integrity, and any defects in the data are noted and communicated.
The data model supporting tranSMART segregates the content into multiple stores and optimizes data structure to represent each type of content. For example, patient-centric clinical data is stored in the i2b2 star schema. High-dimensional data is housed in a different data store where each of the data types (e.g., SNP) retains its specific structure. There are many categories of data that are loaded into tranSMART (see Table 1). For levels 1–4, we employ definitions summarized by the Cancer Genome Atlas (http://www.cancergenome.nih.gov/), which indicate the degree of processing that the data has undergone. Master data are slowly changing elements such as patient identifiers, and Metadata are associated data of a structural or administrative nature. Another feature of the data model is that all the core data elements, such as genes, pathways, diseases, compounds, and concept codes, have a unique ID (UID). The crosswalks among different data stores are enabled using these UIDs. As data is reloaded or appended into the system, the relationships established with the resident data are maintained via the UIDs.
Table 1.
Data Categories Supported by tranSMART platform
| Category | Type | Description | Example | Usage | Storage |
|---|---|---|---|---|---|
| Level 1 | Raw | • Raw data from source platform • Not normalized |
• Raw binary machine reads • Data on the Case Report Form |
• Processing pipeline • Dataset Explorer export |
File system |
| Level 2 | Processed | • Normalized data through curation or data processing pipelines | • Clinical trial data • RMA or MASS normalized gene expression data • SNP data with calls and CNV |
• Dataset Explorer | Database: DeApp, i2b2DemoData |
| Level 3 | Interpreted | • Interpreted or aggregated data from processed data | • Z-scores for gene expression data • Survival times calculated at the end of a study |
• Dataset Explorer • Search |
Database: DeApp, BioMart |
| Level 4 | Summary and Findings | • Quantified association and analysis across multiple samples. • Published results |
• Fold changes • GWAS • Results from publications |
• Search | Database: BioMart |
| Master Data | Slow changing data | • Data about key business entities in the system. • Data might be from internal or external data source. |
• Study design • Platform specifications • User defined gene lists |
• Dataset Explorer • Search |
Database: i2b2Mctadata, i2b2DemoData, BioMart, SearchApp |
| Reference Data | Slow changing data used as reference | • Data from other system that’s used as identifier data or as a reference to other systems | • Affymetrix annotation files • Gene ID’S from Entrez • Disease lists from MeSH |
• Dataset Explorer • Search |
Database: DeApp, BioMart |
| MetaData - Structural | Metadata | • Data that describes data structure | • Data dictionary • Schema guide |
• Documentation | File system |
| MetaData – Administrative (Operational) | Metadata | • Data associated with application/data access and operation | • ETL auditing and QC results • Application access results |
• Search | Database: searchApp, rdc_cz |
Search
The Search performs fast and comprehensive queries against the tranSMART repository of research and reference data. This feature relies on the Apache Lucene Solr (http://lucene.apache.org/solr/) search server – an open source, robust search engine that supports near-real time indexing capabilities. Solr indexes files and table records when the data is initially loaded into the tranSMART databases. tranSMART allows click-through to externally available resources, such as PubMed and Entrez. Users benefit from a richer experience since searches for diseases or genes yield the Entrez ‘omic details or PubMed articles in addition to the data stored locally in tranSMART.
Dataset Explorer
Dataset Explorer is the principle access point for primary study data. Within Dataset Explorer, the user is presented with data in an organized hierarchical fashion. Using drag and drop, the user can execute a variety of analytic workflows (or pre-defined statistical algorithms). The user is able to specify a cohort of patients, choose an available analysis modality, and set the relevant parameters. Once the selections are made, the scripted pipelines are executed. The results are returned to the user and presented in a graphical fashion that best matches the completed analysis. The user may continue to explore the data further by altering cohort details or analytic parameters and running other pre-scripted analyses. Thus the user is able to generate a hypothesis within tranSMART using one dataset and then test this hypothesis on a different dataset. Generated results can be saved or exported. No knowledge of statistical scripting languages is necessary to analyze the data because the user is presented with a predefined menu of powerful analysis options.
tranSMART performs its default analyses without allowing end-users direct access to raw data, thus preserving the integrity of the underlying sources. For researchers with deeper statistical programming knowledge, tranSMART allows direct manipulation of the data via export or direct connection to R. The latter uses a set of pipelines from the tranSMART database to the R statistical package and a custom command library to support direct interaction with the data in R.
Security
The tranSMART application uses a role-based security model to enable the use of the platform across large organizations. User authentication can be integrated into an organization’s existing infrastructure to streamline user management. The security model allows an organization to control data access in accordance with internal policies governing the use of research data. Security is based on the user’s role, where defined roles have different levels of data access. Many implementers have integrated security with proprietary single sign-on solutions, however there is also interest in utilizing open source options, such as Shibboleth (http://shibboleth.net/).
Results
The tranSMART application code became available as open source in January 2012 and is licensed through GPL 3. Currently, there are 32 implementations of tranSMART (see Table 2) in operation, with 11 implementations with pharmaceutical companies, 6 with research institutions, and 5 with non-profit organizations. Also, the tranSMART community has developed a strong web presence. The website, http://transmartproject.org/, is the hub of the open source community. The site acts as a portal for the central repository of code (both core framework and contributions), as well as the wiki with project information and active discussion groups. There is a significant social media presence with handles on LinkedIn, Twitter (@transmartapp) and Google Groups. The community is thriving with active engagement, collaboration and contributions.
Table 2.
tranSMART Implementations
| Organization Types | # Instances |
|---|---|
| AMC | 4 |
| Biopharma | 11 |
| Cancer Center | 2 |
| Commercial Software | 1 |
| Government | 4 |
| Non-profit | 5 |
| Research | G |
University of Michigan, Pistoia Alliance and Imperial College of London initiated the tranSMART Foundation (http://www.transmartfoundation.org) in 2013. The goal of the tranSMART Foundation is to coordinate development of the next generation of tranSMART as it evolves into a global platform solution for clinical and translational research.
There have already been significant contributions to the tranSMART application programming interface (API) since its release. Dataset Explorer has been improved with the addition of a data export feature, as well as a number of new statistical analyses in the Advanced Workflows. The R advanced analysis functions were contributed by the open source community and include: box plot with ANOVA, scatter plot with linear regression, table with Fisher test, survival analysis, heat map, hierarchical clustering, k-means clustering, marker selection, principle component analysis (PCA), correlation analysis, and line graph. The Search feature has been expanded with the incorporation of a faceted search capability as well as the extension of search to support Genome Wide Association Studies. The open source community is demonstrating excitement about the potential of tranSMART and is actively working to expand on the current platform with additional features in the development pipeline.
Discussion
tranSMART is an open source knowledge management and high content analytics platform that enables secondary use of clinical and translation research data. Typically, researchers do not have unfettered access to the necessary data because of poor availability. In addition, raw data requires significant curation to prepare it for analysis. tranSMART addresses data availability by acting as a catalog of research data and related knowledge such as reference data (e.g., probe to gene mapping, study metadata, links to publication repositories and 3rd party references, etc.). tranSMART enables rapid exploration of research trials data in an ad hoc fashion by allowing users to identify cohorts, and perform pre-defined univariate statistical analyses with the ease of a drag and drop interface and without prior statistical programming knowledge. tranSMART supports data sharing and promotes collaboration at the institution level by providing an environment where individual departments can access research data from the entire institution.
A major component to the success of tranSMART is the employment of a common data model. By defining a common structure with common meaning for data, the platform realizes upstream benefits by being able to scale data curation activities according to breadth and depth of source data available, as well as downstream benefits by reusing processing pipelines and analytic tools for optimal and efficient end user experience.
Success of an open source project depends largely on the community engagement. Necessary technological underpinnings for a healthy community have been put in place: robust web presence, development tools (code repository, issue tracker, developer forum) to support tranSMART in this purpose. A flourishing community, as exampled by the bevy of contributions to the source code, is the most telling feature of an open source project; tranSMART appears to be well on its way to achieving this goal.
The tranSMART application platform is enjoying a wave of rapid adoption across life sciences and health care provider sectors. About a year after release to open source, 32 institutions have implemented the platform for use in their research and development activities. Individual implementations range from proof of concept to full institutional implementations. The platform is in the early stages of adoption, but there are already publications demonstrating the use of tranSMART.11 The highest adoption is seen in the pharmaceutical industry, with noteworthy representation by government agencies, academic medical centers and research institutions. The tranSMART Foundation is collaborating internationally to support institutions outside the US in their use of tranSMART.
A combination of tranSMART’s architecture and associated data integration processes serve as the foundation for a robust translational research environment. tranSMART supports multiple data types of varying levels of complexity and relationships, and provides novel yet intuitive end-user components for accessing and analyzing large volumes of data quickly in a secure environment. These features bring together complex patient phenotypic and high dimensional ‘omics data, collected for both research and in EHRs, and enable the continuum of translational science from basic research to bedside in a single application platform.
References
- 1.Zerhouni EA. Translational and clinical science–time for a new vision. N Engl J Med. 2005;353(15):1621–1623. doi: 10.1056/NEJMsb053723. [DOI] [PubMed] [Google Scholar]
- 2.Sung NS, Crowley WF, Jr, Genel M, Salber P, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003;289(10):1278–1287. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 3.Contopoulos-Ioannidis DG, Alexiou GA, Gouvias TC, et al. Life cycle of translational research for medical interventions. Science. 2008;321:1298–1299. doi: 10.1126/science.1160622. [DOI] [PubMed] [Google Scholar]
- 4.Ioannidis JPA. Commentary: materializing research promises: opportunities, priorities and conflicts in translational medicine. J Transl Med. 2004;2(5):1–6. doi: 10.1186/1479-5876-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenfant C. Clinical research to clinical practice–lost in translation? N Engl J Med. 2003;349:868–874. doi: 10.1056/NEJMsa035507. [DOI] [PubMed] [Google Scholar]
- 6.Drolet BC, Lorenzi NM. Translational research: Understanding the Continuum from Bench to Bedside. Transl Res. 2011;157:1–5. doi: 10.1016/j.trsl.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Szalma S, Koka V, Khasanova T, et al. Effective knowledge management in translational medicine. J Transl Med. 2010;8(68):1–9. doi: 10.1186/1479-5876-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perakslis ED, Van Dam J, Szalma S. How informatics can potentiate precompetitive open-source collaboration to jump-start drug discovery and development. Clin Pharmacol and Ther. 2010:1–3. doi: 10.1038/clpt.2010.21. [DOI] [PubMed] [Google Scholar]
- 9.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aronzon D, Palchuk MB. Best practices in biomedical data extraction, transformation and load. AMIA TBI Summit. 2013 [Google Scholar]
- 11.Irgon J, Huang CC, Zhang Y, Talantov D, Bhanot G, Szalma S. Robust multi-tissue gene panel for cancer detection. BMC Cancer. 2010;10:319. doi: 10.1186/1471-2407-10-319. [DOI] [PMC free article] [PubMed] [Google Scholar]