Abstract
It is increasingly recognized that redundant information in clinical notes within electronic health record (EHR) systems is ubiquitous, significant, and may negatively impact the secondary use of these notes for research and patient care. We investigated several automated methods to identify redundant versus relevant new information in clinical reports. These methods may provide a valuable approach to extract clinically pertinent information and further improve the accuracy of clinical information extraction systems. In this study, we used UMLS semantic types to extract several types of new information, including problems, medications, and laboratory information. Automatically identified new information highly correlated with manual reference standard annotations. Methods to identify different types of new information can potentially help to build up more robust information extraction systems for clinical researchers as well as aid clinicians and researchers in navigating clinical notes more effectively and quickly identify information pertaining to changes in health states.
Introduction
Electronic health record (EHR) systems provide significant opportunities to integrate and share health information and increase the efficiency of health care delivery, as well as re-use clinical data for research studies. Of the many functionalities of EHRs, clinical note documentation is an essential part of patient care. A large number of efforts in clinical research have focused on identification of patient cohorts that meet clinical eligibility criteria for studies, with some methods aiming to extract these criteria from both structured data and unstructured texts including phenotype extraction1, 2 and drug related information extraction3, 4. Structured information from billing and administrative codes (i.e., ICD and CPT codes), however, is often insufficient to accurately find patient cohorts for clinical research5. Unstructured text of clinical notes often contains the necessary more detailed information but information extraction (IE) systems based on clinical texts alone often have difficulty achieving adequate performance6, 7. For instance, when ICD-9 codes and IE from clinical texts are combined, a higher accuracy for detecting colorectal cancer can be achieved8.
Most EHR clinical documentation modules allow text from one note to be reused in subsequent notes (“copy-and-paste”). The practice of copying information from previous documents and pasting into the current clinical note being constructed is often used to shorten the time spent documenting. However, one of the unintended consequences of frequent copying and pasting of patient data especially with complicated care episodes or clinical courses is that copy-and-paste can create large amounts of replicated information resulting in longer and less readable notes than those seen previously with paper charts9–11.
Clinical texts with significant amounts of redundant information combined with large numbers of notes not only increase the cognitive burden and decision-making difficulties of clinicians11–16, but also may impact the accuracy and efficiency of IE systems17. Moreover, redundant information can also contain a mixture of outdated information or errors in the information copied making it difficult for clinicians to interpret information in these notes most effectively12. It has been suggested that considering the structure of texts and redundancy before implementing IE tasks may be valuable17, 18. Thus, effective classification of redundant and new information could potentially help to improve the performance of the IE systems for clinical research.
Large amounts of redundant information have been found in both inpatient and outpatient notes with automated methods12, 19, 20. Hammond et al. performed pair-wise comparisons to detect identical word sequence and found 12.5% of information in notes copied12. Wrenn et al. used global alignment techniques to quantify redundancy in inpatient clinical notes,19 finding large amounts of redundant information which increased over the course of a patient’s inpatient stay. Zhang et al. modified the Needleman-Wunsch algorithm, a classic global sequence alignment technique used in bioinformatics, to quantify redundancy in outpatient clinical notes with similar findings20.
Research has also focused upon methods to identify relevant new information and evaluation of the potential impact of redundant information on clinical practice. One of the recognized gaps in these approaches is that these methods do not intrinsically provide more details about the types of new information (e.g., medication, disorders, symptoms).
Categorization of new information may aid clinicians and researchers in finding specific types of new information more easily in a more purposeful manner within notes. The objective of this study was to extract specific types of relevant new information, specifically problem/disease (or comorbidities), medication, and laboratory.
Methods
The three-part methodological approach for this study included: 1) developing a reference standard of new information with information type; 2) identification of new information using an n-gram modeling technique modified for clinical texts; and 3) extraction of semantic types and key terms from identified new information.
Data collection
Outpatient EHR notes were retrieved from the University of Minnesota Medical Center affiliated Fairview Health Services. For this study, we randomly selected 100 geriatric patients with multiple co-morbidities, allowing for relatively large numbers of longitudinal notes in the outpatient clinic setting. To simplify the study, we limited the notes to office visit notes arranged chronologically. These notes were extracted in text format from the Epic™ EHR system between 06/2005 and 06/2011. Institutional review board approval was obtained and informed consent waived for this minimal risk study.
Automated methods to identify new information semantic types
We used a hybrid method with n-gram models and heuristic information previously developed to identify new information in clinical documents21. In brief, after text pre-processing, n-gram models with classic and TF-IDF stopword removal, lexical normalization, and heuristic rules were used to remove note formatting and adjustments by section. After obtaining new information within each note, this text was mapped to the UMLS22 using MetaMap23 with options to allow acronym/abbreviation variants (-a) and NegEx results (–negex). From this, we extracted semantic types using scores of 600 and over as the cutoff. To simplify the analysis, we restricted our detailed analysis to the specific types to identify information about problem/disease, medication, and lab results (Table 1)22.
Table 1.
Sections and semantic types for identifying category of new information.
| Category | Semantic Types |
|---|---|
| Problem/Disease | [Disease or Syndrome], [Finding], [Sign or Symptom] |
| Medication | [Clinical Drug], [Organic Chemical, Pharmacologic Substance], [Biomedical or Dental Material] |
| Laboratory | [Laboratory Procedure], [Therapeutic or Preventive Procedure], [Diagnostic Procedure], [Amino Acid, Peptide, or Protein], [Biologically Active Substance] |
Calculation of various types of new information proportion of patient notes
To calculate the NIP of each note, the new information algorithm was trained on previous n (e.g., 1, 2, …) notes to predict the new information of (n+1)th note for the whole corpus (100 patients). NIP was defined as the number of sentence (at least contain one piece of new information) divided by the total number of sentences of each note. We used the same method to further quantify NIP on the number (at a sentence or statement level) of various types of NIP for each note including disease (NDIP), medication (NMIP) and lab results (NLIP) based upon the identified semantic types of new information (Table 1). We then plotted NIP of various types for each patient over time. For the purposes of graphical display of notes temporally, we adjusted the dates of patient notes by a random offset of +/− 1 to 364 days.
Manually reviewed annotation as gold standard
Two medical intern physicians (aged 26 and 30) were asked to identify new and clinically relevant information based on all preceding documents chronologically for each patient using their clinical judgment. They were also asked to classify new information into the following types: problem, medication, laboratory, procedure or imaging, surgery history, family history, social history, and medical history. Longitudinal outpatient clinical notes from 15 patients with the last 3 notes for each patient annotated were selected for this study.
To maximize agreement, we first allowed the annotators to compare each other’s annotations on a small separate corpus to reach a consensus on the standards for judging relevant new information. To assess inter-rater reliability, each physician later manually annotated a separate set of 10 overlapping notes. Cohen’s Kappa statistic and percentage agreement were used to analyze agreement at a sentence or statement level.
In addition to the notes used to establish and measure inter-annotator agreement, 90 notes were annotated by medical interns. Forty notes were used for training and the remaining fifty notes for evaluation. Performance of automated methods was compared to the reference standard and then measured for precision and recall at a sentence or statement level.
A third resident (3rd year) manually reviewed chronologically ordered 100 office visit notes from five individual patients to note new information in the format of short key terms. We then compared this with the automatically computed new information proportion (NIP) measure for each note, and extracted biomedical terms of various categories. Precision and recall for three types of new information within these notes were calculated.
Results
Annotation evaluation and method performance
Two raters showed a good agreement (which was improved from previous evaluations21) with identifying new information with Cohen’s Kappa coefficient of 0.80 and percentage agreement of 97%. The precision and recall of the best method with n-grams are 0.81 and 0.84, respectively. The precisions values for extracting disease, medication, and laboratory new information are 0.67, 0.66, and 0.67; and the recall values are 0.72, 0.92, and 0.80, respectively.
Identification of various types of relevant new information
After calculating new information using the reference standards at the sentence level, we obtained the following percentage of various categories (e.g., lab, problem, medication) of relevant new information annotated by medical experts. The top three categories were problem (34.1%), medication (31.7%) and laboratory results (17.3%). Other types include procedures of imaging (5.0%), family history (2.8%) social history (2.7%), medical history (2.4%), surgery history (0.4%), and others (3.6%).
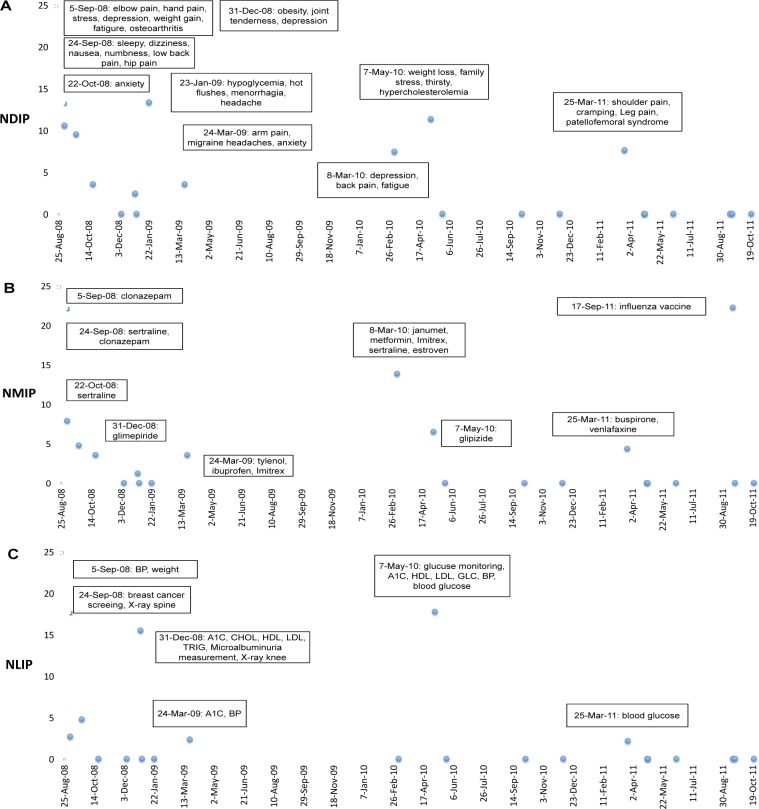
NIP, NDIP, NMIP, and NLIP were then calculated for each note where NIP = NDIP + NMIP + NLIP + NOIP. Note that NOIP represents other types of new information proportion (e.g., Mental Process). Individual patients were then selected and NIP, NDIP, NMIP, and NLIP plotted as illustrated in one patient in Figure 1 and Figure 2. Subjective new information for each note was also obtained for each note (Figure 1). Overall, notes with higher NIP correlated with more new information, and notes with lower NIP scores tended not to contain significant new information. Key biomedical concepts were extracted for each information category and were marked (using the automated new text extracted) for each note in Figure 2.
Figure 1.
New information proportion (NIP) of clinical notes an illustrative patient. Boxes contain summarized new information.
Figure 2.
Plot of (A) NDIP (disease), (B) NMIP (medication), and (C) NLIP (laboratory) over time for the same patient as Figure 1. Biomedical concepts for each note included in boxes. NDIP, new problem/disease information proportion; NMIP, new medication information proportion; NLIP, new laboratory information proportion.
Discussion
In the field of clinical research informatics, many researchers focused on improvement of clinical NLP and data mining methods applied to EHR clinical texts, but minimal research has focused on the impact of the redundant nature of clinical texts it being ubiquitous and an issue for clinicians in reviewing a patient’s notes. Cohen et al. recently reported that redundancy of clinical texts had impact on two text mining techniques: collocation extraction and topic modeling. The authors suggest examining the redundancy of a given corpus before implementing text mining techniques17. Thus, studies on automated methods to identify relevant new information represent a potential set of techniques to improve the information extraction process from clinical notes. Previous work has demonstrated that NIP measures may be useful in identifying notes with clinically relevant new information24. While notes with higher NIP scores usually correlate with new findings, some other pertinent questions include answering questions such as “Why are notes with high new information scores important?” and “What specific new information does this note contain?”. This study examines types of new information in several important categories by dividing original NIP scores into various types of new information. Such classification of new information can potentially help clinical researchers navigate to specific types of information in clinical more effectively.
In comparing annotations based on UMLS concepts by medical residents to automated methods there were several key findings. We found some types of problem/disease information where automated methods identified information that was not included in the physician-generated reference standard. In one example, symptoms of elbow pain, hand pain, and depression were identified in the reference standard but several other symptoms such as anhedonia and insomnia were not identified but were new. In contrast, with medications, automated methods incorrectly identified some medications. For example, (Figure 4b) new medications of clonazepam (5-Sep-08), sertraline (24-Sep-08), metformin (8-Mar-10), estroven (8-Mar-10), glipizide (7-May-10), and buspirone (25-Mar-11) were found via automated methods and by our expert annotators, but the method incorrectly found “janumet” from the sentence “…janumet was too expensive, so she did not take it.” (8-Mar-10). Although we used NegEx functionality in MetaMap to account for negation, our automated method did not effectively deal with the co-reference issue (it refers to janumet). Another example is “vernlafaxine” from the note (25-Mar-10) “…another future option may be to try venlafaxine”. Here, the physician only recommended the medicine instead of prescribing it accounting for another false negative example. Finally, with respect to laboratory information, there were examples where the physician annotator did not mark laboratory data. One reason for this is that glucose and hemoglobin A1C tests are routine monitoring tests, and clinicians will not focus on that unless there are significant changes of the results. We also faced mapping issues with respect to acronyms for laboratory procedures. For example, we had to translate “A1C” to its full name “Hemoglobin A1C” to be recognized by MetaMap. In follow-up studies, we may provide more detailed information (e.g., if the value excess the normal range) other than just listing laboratory name to aid clinicians to pay more attention to the specific lab results with unexpected values.
Our method has certain other limitations. Mapping techniques such as that provided with MetaMap do not give additional types of information such as changes in dosage for specific drugs. We also did not solve other semantic level issues as mentioned previously in the discussion, such as co-reference. Also, although we compared our results with annotated reference sample patient notes, this reference standard was not built at the concept level per sentence whereas our method used concepts at the biomedical term level, which was readily available. Currently, we have only looked at three types of new information; other types of information such as Mental Process could be another valuable set of semantic types to explore. In future research, we will also consider the use of specialized modules such as MedEx25 to extract more details associated with changes in medication use, other than just providing drug name.
Conclusion
We used the combination of language models and semantic types not only to identify new information, but also to extract key new information at the biomedical term level. We found that our ability to extract new key terms with our methods had good correlation with expert judgment. As these methods for new information detection are further developed, they can potentially help researchers avoid the biased clinical texts due to the redundant information and find the information type of interest. Moreover, it can also aid clinicians in finding notes and information within notes with more detailed types of new information, such as new diseases or medications.
Acknowledgments
This research was supported by the American Surgical Association Foundation Fellowship (GM), University of Minnesota (UMN) Institute for Health Informatics Seed Grant (GM & SP), Agency for Healthcare Research & Quality (#R01HS022085-01) (GM), National Library of Medicine (#R01LM009623-01) (SP) and the UMN Graduate School Doctoral Dissertation Fellowship (RZ). The authors thank Fairview Health Services for support of this research. The authors thank Dr. Janet T. Lee for annotation of the patient notes for this study.
References
- 1.Kullo IJ, Fan J, Pathak J, Sayoya GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010 Sep;17(5):568–74. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011 Apr 20;3(79):79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther. 2011 Jul;90(1):133–42. doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang XY, Hripcsak G, Markatou M, Friedman C. Active Computerized Pharmacovigilance Using Natural Language Processing, Statistics, and Electronic Health Records: A Feasibility Study. J Am Med Inform Assoc. 2009 May-Jun;16(3):328–37. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005 May;43(5):480–5. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- 6.Penz JF, Wilcox AB, Hurdle JF. Automated identification of adverse events related to central venous catheters. J Biomed Inform. 2007 Apr;40(2):174–82. doi: 10.1016/j.jbi.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Chase HS, Patel CO, Friedman C, Weng C. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc. 2008:404–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annu Symp Proc. 2011;2011:1564–72. [PMC free article] [PubMed] [Google Scholar]
- 9.Markel A. Copy and paste of electronic health records: a modern medical illness. Am J Med. 2010 May;123(5):e9. doi: 10.1016/j.amjmed.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Hirschtick RE. A piece of my mind. Copy-and-paste. JAMA. 2006 May 24;295(20):2335–6. doi: 10.1001/jama.295.20.2335. [DOI] [PubMed] [Google Scholar]
- 11.Yackel TR, Embi PJ. Copy-and-paste-and-paste. JAMA. 2006 Nov 15;296(19):2315. doi: 10.1001/jama.296.19.2315-a. author reply -6. [DOI] [PubMed] [Google Scholar]
- 12.Hammond KW, Helbig ST, Benson CC, Brathwaite-Sketoe BM. Are electronic medical records trustworthy? Observations on copying, pasting and duplication. AMIA Annu Symp Proc. 2003:269–73. [PMC free article] [PubMed] [Google Scholar]
- 13.Weir CR, Hurdle JF, Felgar MA, Hoffman JM, Roth B, Nebeker JR. Direct text entry in electronic progress notes. An evaluation of input errors. Methods Inf Med. 2003;42(1):61–7. [PubMed] [Google Scholar]
- 14.Hripcsak G, Vawdrey DK, Fred MR, Bostwick SB. Use of electronic clinical documentation: time spent and team interactions. J Am Med Inform Assn. 2011 Mar;18(2):112–7. doi: 10.1136/jamia.2010.008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel VL, Kaufman DR, Arocha JF. Emerging paradigms of cognition in medical decision-making. J Biomed Inform. 2002 Feb;35(1):52–75. doi: 10.1016/s1532-0464(02)00009-6. [DOI] [PubMed] [Google Scholar]
- 16.Reichert D, Kaufman D, Bloxham B, Chase H, Elhadad N. Cognitive analysis of the summarization of longitudinal patient records. AMIA Annu Symp Proc. 2010:667–71. [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen R, Elhadad M, Elhadad N. Redundancy in electronic health record corpora: analysis, impact on text mining performance and mitigation strategies. BMC Bioinformatics. 2013;14:10. doi: 10.1186/1471-2105-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downey D, Etzioni O, Soderland S. Analysis of a probabilistic model of redundancy in unsupervised information extraction. Artif Intell. 2010 Jul;174(11):726–48. [Google Scholar]
- 19.Wrenn JO, Stein DM, Bakken S, Stetson PD. Quantifying clinical narrative redundancy in an electronic health record. J Am Med Inform Assoc. 2010 Jan-Feb;17(1):49–53. doi: 10.1197/jamia.M3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang R, Pakhomov S, MaInnes BT, Melton GB. Evaluating Measures of Redundancy in Clinical Texts. AMIA Annu Symp Proc. 2011:1612–20. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R, Pakhomov S, Melton GB. Automated Identification of Relevant New Information in Clinical Narrative. IHI’12 ACM Interna Health Inform Sym Proc. 2012:837–41. [Google Scholar]
- 22.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010 May-Jun;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R, Pakhomov S, Lee JT, Melton GB. Navigating longitudinal clinical notes with an automated method for detecting new information. Stud Health Technol Inform. 2013;192:754–8. [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Jan-Feb;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]