Abstract
Statin medications are often prescribed to ameliorate a patient’s risk of cardiovascular events due in part to cholesterol reduction. We developed and evaluated an algorithm that can accurately identify subjects with major adverse cardiac events (MACE) while on statins using electronic medical record (EMR) data. The algorithm also identifies subjects experiencing their first MACE while on statins for primary prevention. The algorithm achieved 90% to 97% PPVs in identification of MACE cases as compared against physician review. By applying the algorithm to EMR data in BioVU, cases and controls were identified and used subsequently to replicate known associations with eight genetic variants. We replicated 6/8 previously reported genetic associations with cardiovascular diseases or lipid metabolism disorders. Our results demonstrated that the algorithm can be used to accurately identify subjects with MACE and MACE while on statins. Consequently, future e studies can be conducted to investigate and validate the relationship between statins and MACE using real-world clinical data.
Introduction
Cardiovascular disease (CVD) is the leading cause of death worldwide. Recent mortality data show that CVD accounted for 32.8% of all deaths in the U.S.1 Many randomized clinical trials (RCTs) have shown that HMG-CoA reductase inhibitors (“statins”) significantly reduce the frequency of major adverse cardiac events (MACE) in patients at risk.2–7 Statins are one of the most commonly prescribed medications, and are generally well-tolerated.8 Given their clinical importance, they have been a frequent focus of investigation in electronic medical records (EMRs). We sought to develop a highly accurate algorithm to enable study of statin efficacy, measured as MACE while on statins, in EMRs. This algorithm can be used for later clinical and genomic studies.
Since 2000, EMRs have been widely implemented through the U.S.9 The deployment of EMRs not only improves patient care but also generates huge clinical practice-based datasets ideal for evaluating previous findings from randomized controlled trials (RCTs).10–13 Although useful for research, EMR data often requires carefully constructed algorithms to accurately identify phenotypes for clinical and genomic study10,14–16; this is especially true for pharmacogenomic studies in the EMR, since they require knowledge of the temporal relationship between exposures and outcomes. Once accurate algorithms are identified, studies can be conducted to investigate relevant relationships, e.g., between statins and MACE, using real-world clinical data.
Background
MACE can be defined as cardiac death, nonfatal acute myocardial infarction (AMI), or target lesion revascularization. Previously, several investigators have explored the possibility of identifying MACE subjects using EMR data. In 1996, Pladevall et al., reported that the accuracy of using the International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) code 410 to identify definite MI was 92%.17 Similarly, Petersen et al. in 1999 found that the positive predictive value (PPV) of AMI codes in the primary position was 96%. In addition, they also reported that the sensitivity and specificity of Current Procedural Terminology (CPT) coding were, respectively, 96% and 99% for coronary catheterization, 95% and 100% for coronary artery bypass graft surgery, and 90% and 99% for percutaneous transluminal coronary angioplasty.18 In 2002, Austin et al., examined the use of a discharge diagnosis of AMI and the PPV was 88%.19 In 2004, Kiyota et al., additionally required hospitalization lasting at least 3 days. Their results reflected a slightly improved PPV of 94%.20 Two recent studies, by Varas-Lorenzo et al. in 200821 and by Preciosa et al. in 201322, reported that ICD9-CM codes had a PPV of 95% and 96%, respectively. Generally, these results suggest that ICD-9-CM codes have been widely used for MACE subject identification and yield PPVs in the mid to high 90% range.23 However, all these studies were performed on primary/secondary discharge codes only (thus representing inpatient-generated codes, which typically result from professional coders). Such information is not available for many deidentified EMR datasets, i.e. it may not be clear if a code is for the principal or discharge diagnosis. Thus, the approach of simply using ICD9-CM codes may not generalize to a broad clinical research setting. Another important issue is identifying first MACE events. The recognition of such events empowers researchers to evaluate the effectiveness of a treatment for either primary or secondary prevention of MACE, therefore, has a foreseeable and significant impact on clinical practice.
Recent studies have begun using EMR data for pharmacological studies. Drug response phenotypes can be challenging to identify accurately, as they require presence of a medication during the timing of an event. 24 In a recent paper, we described our methods for extracting information and constructing full dose-response curves for simvastatin and atorvastatin using EMR data. 25 Advanced techniques, e.g. natural language processing (NLP) and ontology, were used to retrieve medication and laboratory data from structured and unstructured EMRs. Other examples of pharmacological studies include pharmacogenetic studies of clopidogrel and CYP2C19 variants, in which manual review was ultimately required to achieve PPV26, and the affect of common variants with warfarin stable-dose international normalized ratios (INRs), which was able to be performed entirely using informatics techniques.27 Other clinical studies have used NLP, sometimes with laboratory data, to replicate known drug adverse events and suggest some others, though formal assessments of the PPV of each drug-event pair were not provided.28,29
In this manuscript, we introduce an algorithm to identify subjects with MACE while on statins from EMRs. We report its performance compared to manual chart review and a genetic validation study. Compared to other efforts, our algorithm involves all diagnosis codes as well as laboratory data and simple NLP, instead of just primary discharge codes; it also assesses concurrent statin use, and includes a determination of first MACE.
Methods
MACE Algorithm development
We used commonly captured EMR data, including ICD9-CM codes, CPT codes, and laboratory test results to develop an approach to identify MACE. We used all diagnosis codes rather than primary discharge codes alone so that our approach would be widely generalizable.
We categorized a MACE event as either AMI or revascularization. Qualifying cases of AMI while on statins were required to have ≥2 AMI relevant ICD9-CM Codes (410.* or 411.*) within a 5-day window and an abnormal laboratory test (Table 1). An abnormal laboratory test was defined as either troponin ≥0.10 ng/ml or both creatinine kinase (CK) MB fraction to CK ratio≥3.0 and CK-MB ≥10.0 ng/mL. In addition, a statin must have been prescribed prior to the AMI event ≥180 days (Figure 1). We chose slightly higher thresholds than usual to ensure the accuracy of the algorithm. The duration of 180 days was chosen empirically to represent a time course for which a patient would have significant statin exposure before their event and to make it easier to ascertain whether the patient had remained on the medicine. Statins were either simvastatin (Zocor), fluvastatin(Lescol, Canef, Vastin), atorvastatin (Lipitor), pravastatin(Pravachol, Selektine), lovastatin(Mevacor), cerivastatin (Baycol, Lipobay), or rosuvastatin(Crestor). Medications were identified using records from electronic prescribing tools and processing of free text notes using MedEx30.
Table 1.
Algorithm for identifying subjects with MACE while on statins.
| AMI on statin | • ≥ 2 AMI Codes (410.* or 411.*) within a 5-Day Window • Abnormal lab within the same time window defined by ○ Troponin-I ≥ 0.10 ng/ml) ○ or Troponin-T ≥ 0.10 ng/ml, ○ or CK-MB/CK ratio ≥ 3.0 and CK-MB ≥ 10.0 ng/mL • Statin prescribed prior to the AMI event ≥180 days |
| 1st AMI on statin | • AMI on statin • No AMI codes (410 – 412) assigned before the AMI event • No MACE history defined by NLP |
| Revascularization while on statin | • Any CPT code for angioplasty, stent, or CABG • statin prescribed prior to the procedure ≥180 days |
| 1st Revascularization while on statin | • Revascularization while on statin • No revascularization codes assigned before the AMI event • No MACE history defined by NLP |
Figure 1.
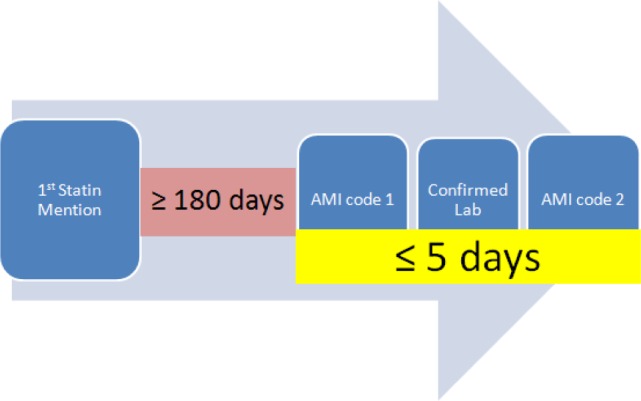
Overview of algorithm for determining AMI on statins
For qualified subjects with an AMI while on statins, we identified individuals with 1st AMI events, as those with no AMI codes (410 – 412) prior to the qualifying statin exposure-AMI event and with no other MACE history defined by applying NLP on previous notes. We used the KnowledgeMap Concept Indexer (KMCI)31,32, a general-purpose NLP engine, to parse a patient’s notes. Any non-negated keywords found, including AMI, MI, acute myocardial infarction, myocardial infarction, CABG, coronary artery bypass, cypher, taxus, BMS, DES, and stent, was considered as an indication of positive MACE history and thereby excluded as a subject.
Revascularization includes percutaneous coronary intervention (PCI) and coronary artery bypass grafting (CABG). To be a qualified subject with revascularization while on statins, one must have a revascularization CPT code and a statin must be prescribed prior to the procedure ≥180 days (Table 1). The CPT codes that we used included coronary artery bypass (33533-33536, 33510-33523), angioplasty (92980-92982, 92984, 92995, 92996), and stent (C1874-C1877). Individuals with 1st revascularizations while on statins were those whom met the above criteria and had no revascularization CPT codes and no revascularization history found by NLP prior to the MACE on statin event.
We similarly developed an algorithm to identify control subjects without MACE while on statin. We excluded patients with any AMI diagnosis or revascularization CPT codes, patients with previous history of AMI or revascularization defined by NLP. We also required controls to have had similar statin exposure in their EMRs matched with cases.
Manual chart review
We applied the algorithm on BioVU individuals at Vanderbilt University Medical Center (VUMC) 33 to identify possible cases. In brief, BioVU links a de-identified image of the Vanderbilt EMR to DNA extracted from blood samples (obtained during routine clinical care and about to be discarded). Each record and associated DNA sample is linked by a unique identifier generated by a one-way hash function. The resource has been considered as containing data for nonhuman subjects in accordance with the provisions of Title 45 of the Code of Federal Regulations part 46, as have the individual research studies utilizing the resource.33 As of 09/2013, BioVU contains > 170,000 unique individuals, including their dense longitudinal clinical records and associated blood samples.
From each category (AMI on statin, 1st AMI on statin, revascularization on statin, and 1st revascularization on statin), a group of 30 randomly selected cases was manually reviewed by two physicians. AMI on statin and 1st AMI on statin cases were reviewed by JCD, an internist. Revascularization on statin and 1st revascularization on statin cases were reviewed by PW, a cardiologist.
Genetic validation
To further illustrate the application of our algorithm, we performed a genotype and phenotype association study, also by leveraging BioVU resources. The study population consisted of the first 7747 European–Americans accrued into BioVU. The only selection criteria were that they met the general conditions for eligibility for BioVU; no clinical inclusion or exclusion criteria were applied. These subjects have already been genotyped in previous studies.34 In the current analysis, we identified 533 MACE cases and 2,642 MACE-free controls and compared the frequency of eight selected SNPs with previously known associations with cardiovascular diseases or lipid metabolism among cases and controls (Table 3): rs1045642 [pharmacogenetic predictors of lipid-lowering response to atorvastatin]35, rs440446 [ApoE gene, Variations in ApoE affect cholesterol metabolism, which in turn alter risk of heart disease and in particular a heart attack or a stroke]36, rs2200733[atrial fibrillation (AF) and ischemic stroke]37,38, rs405509 [CAD]39, rs1333049 [CAD]40,41, rs1800795 [CAD]42, rs1800888 [MACE after PCI]43, and rs1048101[hypertension] 44. SNPs were genotyped in DNA samples from these subjects. Genotyping was conducted using commercial Taqman Allelic discrimination assays available through Applied Biosystems, Inc. (ABI, Foster City, CA, USA). The case-control analyses was performed using PLINK, a free, open-source genetic analysis toolset (http://pngu.mgh.harvard.edu/~purcell/plink/).45 This platform was selected based on its efficiency, flexibility and ease of application. The primary outcome of this validation was to replicate these associations using our MACE algorithm.
Table 3.
Association between eight SNPs previously reported to be associated with CV disease with MACE on statins in our population.
| Chr. | SNP | Gene/Association | Minor Allele Frequency | p-value |
|---|---|---|---|---|
| 7 | rs1045642 | ABCB1/predictors of lipid-lowering response to atorvastatin | 0.472 | 0.001 |
| 8 | rs1048101 | ADRA1A/hypertension | 0.457 | 0.006 |
| 19 | rs440446 | ApoE/cholesterol metabolism, heart disease, AMI, and stroke | 0.348 | 0.009 |
| 4 | rs2200733 | 4q25/atrial fibrillation(AF), ischemic stroke | 0.119 | 0.016 |
| 19 | rs405509 | ApoE/CAD | 0.474 | 0.032 |
| 5 | rs1800888 | ADRB2/MACE after undertaking PCI | 0.012 | 0.040 |
| 9 | rs1333049 | CDKN2B/CAD | 0.479 | 0.121 |
| 7 | rs1800795 | IL6/CAD | 0.418 | 0.940 |
Results
Table 2 summarizes manual chart review results that ranged from 90% to 97% positive predictive value (PPV) for MACE case identification. We observed some false positives that were caused by system coding errors, e.g. a “stent” code assigned for an esophageal stent placement. The algorithm performed well on identifying the 1st event (PPV ~90%). Some previous major events were missed because they happened long time ago (before 1990) and were not recorded in our current system.
Table 2.
Results of manual chart review
| Category | PPV |
|---|---|
| Any AMI event | 96.67% |
| 1st AMI event | 96.67% |
| Any AMI while on Statin | 90.00% |
| 1st AMI event while on Statin | 90.00% |
| Any revascularization event | 96.55% |
| 1st revascularization event | 89.66% |
| Any revascularization while on Statin | 96.55% |
| 1st revascularization event while on Statin | 89.66% |
A total of 533 MACE cases and 2,642 MACE-free controls were identified from 7747 subjects of the demonstration cohort. Eight pre-selected SNPs were genotyped for all 3175 subjects. Variants with call rate less than 99% were removed from final analyses. Case-control analysis successfully replicated six out of the eight previously reported associations with cardiovascular diseases or lipid metabolism disorders.
The validation results were shown in Table 3. The strongest association was observed from the variant located in ABCB1 gene (rs1045642). This SNP— rs1045642, has already been proven to influence the body response to atorvastatin35, therefore potentially affects our cardiovascular endpoint— MACE. Two SNPs (rs440446, rs405509) located in ApoE gene were replicated. Both of them play a critical role in cholesterol metabolism, which in turn affect the development of heart disease.36,39 Rs2200733 is another important cardiovascular relevant SNP that we replicated. Numerous studies have reported that it is strongly associated with CAD regardless of race.37,38,46–50 We also validated the associations between MACE and two adrenergic receptor SNPs rs1048101 and rs1800888. The former has been previously reported to be able to alter the alpha1-adrenergic receptor autoantibody production in hypertensive patients 44 while the latter is associated with a more aggressive CAD and adversely affects prognosis in a study of 330 patients undergoing PCI43.
Discussion
In this paper, we reported a novel algorithm for use in EMRs to accurately identify cases with MACE and 1st MACE while on statin. The algorithm achieved 90% to 97% PPVs for the identification of MACE cases as compared to clinician review. By applying the algorithm to EMR data of demonstration cohort in BioVU, cases and controls were identified and used subsequently to replicate six out of eight associations with known genetic variants. Our results demonstrated that the algorithm can be used to accurately identify cases with MACE while on statins.
Acknowledgments
The authors would like to acknowledge funding by the American Heart Association (13POST16470018), NIH/NLM (R01 LM 010685-01A1), T32 GM007569, UL1RR0 24975 (Vanderbilt CTSA, please double check), PGPoP (5 U19 HL065962-11), and PARC (U19 HL069757).
Reference
- 1.Roger VL, Go AS, Lloyd-Jones DM, et al. Executive summary: heart disease and stroke statistics–2012 update: a report from the American Heart Association. Circulation. 2012 Jan 3;125(1):188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 2.Baigent C, Keech A, Kearney PM, et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005 Oct 8;366(9493):1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 3.Delahoy PJ, Magliano DJ, Webb K, Grobler M, Liew D. The relationship between reduction in low-density lipoprotein cholesterol by statins and reduction in risk of cardiovascular outcomes: an updated meta-analysis. Clinical therapeutics. 2009 Feb;31(2):236–244. doi: 10.1016/j.clinthera.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Hao PP, Chen YG, Wang JL, et al. Meta-analysis of the role of high-dose statins administered prior to percutaneous coronary intervention in reducing major adverse cardiac events in patients with coronary artery disease. Clinical and experimental pharmacology & physiology. 2010 Apr;37(4):496–500. doi: 10.1111/j.1440-1681.2009.05339.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim MC, Ahn Y, Cho KH, et al. Early statin therapy within 48 hours decreased one-year major adverse cardiac events in patients with acute myocardial infarction. International heart journal. 2011;52(1):1–6. doi: 10.1536/ihj.52.1. [DOI] [PubMed] [Google Scholar]
- 6.Ridker PM, Pradhan A, MacFadyen JG, Libby P, Glynn RJ. Cardiovascular benefits and diabetes risks of statin therapy in primary prevention: an analysis from the JUPITER trial. Lancet. 2012 Aug 11;380(9841):565–571. doi: 10.1016/S0140-6736(12)61190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato H, Kinjo K, Ito H, et al. Effect of early use of low-dose pravastatin on major adverse cardiac events in patients with acute myocardial infarction: the OACIS-LIPID Study. Circulation journal: official journal of the Japanese Circulation Society. 2008 Jan;72(1):17–22. doi: 10.1253/circj.72.17. [DOI] [PubMed] [Google Scholar]
- 8.Maji D, Shaikh S, Solanki D, Gaurav K. Safety of statins. Indian journal of endocrinology and metabolism. 2013 Jul;17(4):636–646. doi: 10.4103/2230-8210.113754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shea S, Hripcsak G. Accelerating the use of electronic health records in physician practices. The New England journal of medicine. 2010 Jan 21;362(3):192–195. doi: 10.1056/NEJMp0910140. [DOI] [PubMed] [Google Scholar]
- 10.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther. 2011 Mar;89(3):379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarty CA, Wilke RA. Biobanking and pharmacogenomics. Pharmacogenomics. 2010 May;11(5):637–641. doi: 10.2217/pgs.10.13. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. American journal of human genetics. 2010 Apr 9;86(4):560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012 Jul;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei WQ, Leibson CL, Ransom JE, Kho AN, Chute CG. The absence of longitudinal data limits the accuracy of high-throughput clinical phenotyping for identifying type 2 diabetes mellitus subjects. International journal of medical informatics. 2013 Apr;82(4):239–247. doi: 10.1016/j.ijmedinf.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Science translational medicine. 2011 Apr 20;3(79):79re71. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei WQ, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association : JAMIA. 2012 Mar-Apr;19(2):219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pladevall M, Goff DC, Nichaman MZ, et al. An assessment of the validity of ICD Code 410 to identify hospital admissions for myocardial infarction: The Corpus Christi Heart Project. International journal of epidemiology. 1996 Oct;25(5):948–952. doi: 10.1093/ije/25.5.948. [DOI] [PubMed] [Google Scholar]
- 18.Petersen LA, Wright S, Normand SL, Daley J. Positive predictive value of the diagnosis of acute myocardial infarction in an administrative database. Journal of general internal medicine. 1999 Sep;14(9):555–558. doi: 10.1046/j.1525-1497.1999.10198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austin PC, Daly PA, Tu JV. A multicenter study of the coding accuracy of hospital discharge administrative data for patients admitted to cardiac care units in Ontario. American heart journal. 2002 Aug;144(2):290–296. doi: 10.1067/mhj.2002.123839. [DOI] [PubMed] [Google Scholar]
- 20.Kiyota Y, Schneeweiss S, Glynn RJ, Cannuscio CC, Avorn J, Solomon DH. Accuracy of Medicare claims-based diagnosis of acute myocardial infarction: estimating positive predictive value on the basis of review of hospital records. American heart journal. 2004 Jul;148(1):99–104. doi: 10.1016/j.ahj.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Varas-Lorenzo C, Castellsague J, Stang MR, Tomas L, Aguado J, Perez-Gutthann S. Positive predictive value of ICD-9 codes 410 and 411 in the identification of cases of acute coronary syndromes in the Saskatchewan Hospital automated database. Pharmacoepidemiology and drug safety. 2008 Aug;17(8):842–852. doi: 10.1002/pds.1619. [DOI] [PubMed] [Google Scholar]
- 22.Coloma PM, Valkhoff VE, Mazzaglia G, et al. Identification of acute myocardial infarction from electronic healthcare records using different disease coding systems: a validation study in three European countries. BMJ open. 2013;3(6) doi: 10.1136/bmjopen-2013-002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutrona SL, Toh S, Iyer A, et al. Design for validation of acute myocardial infarction cases in Mini-Sentinel. Pharmacoepidemiology and drug safety. 2012 Jan;21(Suppl 1):274–281. doi: 10.1002/pds.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden DM, Xu H, Denny JC, Wilke RA. Electronic medical records as a tool in clinical pharmacology: opportunities and challenges. Clin Pharmacol Ther. 2012 Jun;91(6):1083–1086. doi: 10.1038/clpt.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei WQ, Feng Q, Jiang L, et al. Characterization of Statin Dose-response within Electronic Medical Records. Clin Pharmacol Ther. 2013 doi: 10.1038/clpt.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaney JT, Ramirez AH, Bowton E, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther. 2012 Feb;91(2):257–263. doi: 10.1038/clpt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. Journal of the American Medical Informatics Association : JAMIA. 2011 Jul-Aug;18(4):387–391. doi: 10.1136/amiajnl-2011-000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther. 2011 Jul;90(1):133–142. doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LePendu P, Iyer SV, Bauer-Mehren A, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther. 2013 Jun;93(6):547–555. doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association : JAMIA. 2010 Jan-Feb;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denny JC, Peterson JF, Choma NN, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. Journal of the American Medical Informatics Association : JAMIA. 2010 Jul-Aug;17(4):383–388. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denny JC, Bastarache L, Sastre EA, Spickard A., 3rd Tracking medical students’ clinical experiences using natural language processing. Journal of biomedical informatics. 2009 Oct;42(5):781–789. doi: 10.1016/j.jbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010 May 1;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosales A, Alvear M, Cuevas A, Saavedra N, Zambrano T, Salazar LA. Identification of pharmacogenetic predictors of lipid-lowering response to atorvastatin in Chilean subjects with hypercholesterolemia. Clinica chimica acta; international journal of clinical chemistry. 2012 Feb 18;413(3–4):490–501. doi: 10.1016/j.cca.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Andreotti G, Menashe I, Chen J, et al. Genetic determinants of serum lipid levels in Chinese subjects: a population-based study in Shanghai, China. European journal of epidemiology. 2009;24(12):763–774. doi: 10.1007/s10654-009-9402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gretarsdottir S, Thorleifsson G, Manolescu A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Annals of neurology. 2008 Oct;64(4):402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 38.Virani SS, Brautbar A, Lee VV, et al. Usefulness of single nucleotide polymorphism in chromosome 4q25 to predict in-hospital and long-term development of atrial fibrillation and survival in patients undergoing coronary artery bypass grafting. The American journal of cardiology. 2011 May 15;107(10):1504–1509. doi: 10.1016/j.amjcard.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fredriksson J, Anevski D, Almgren P, et al. Variation in GYS1 interacts with exercise and gender to predict cardiovascular mortality. PloS one. 2007;2(3):e285. doi: 10.1371/journal.pone.0000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karvanen J, Silander K, Kee F, et al. The impact of newly identified loci on coronary heart disease, stroke and total mortality in the MORGAM prospective cohorts. Genetic epidemiology. 2009 Apr;33(3):237–246. doi: 10.1002/gepi.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buysschaert I, Carruthers KF, Dunbar DR, et al. A variant at chromosome 9p21 is associated with recurrent myocardial infarction and cardiac death after acute coronary syndrome: the GRACE Genetics Study. European heart journal. 2010 May;31(9):1132–1141. doi: 10.1093/eurheartj/ehq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antonicelli R, Olivieri F, Bonafe M, et al. The interleukin-6 -174 G>C promoter polymorphism is associated with a higher risk of death after an acute coronary syndrome in male elderly patients. International journal of cardiology. 2005 Sep 1;103(3):266–271. doi: 10.1016/j.ijcard.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 43.Piscione F, Iaccarino G, Galasso G, et al. Effects of Ile164 polymorphism of beta2-adrenergic receptor gene on coronary artery disease. Journal of the American College of Cardiology. 2008 Oct 21;52(17):1381–1388. doi: 10.1016/j.jacc.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 44.Sun YX, Liao YH, Zhu F, et al. Association between ADRA1A gene polymorphism and autoantibodies against the alpha1-adrenergic receptor in hypertensive patients. Zhonghua xin xue guan bing za zhi. 2008 Oct;36(10):883–887. [PubMed] [Google Scholar]
- 45.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohanty S, Santangeli P, Bai R, et al. Variant rs2200733 on chromosome 4q25 confers increased risk of atrial fibrillation: evidence from a meta-analysis. Journal of cardiovascular electrophysiology. 2013 Feb;24(2):155–161. doi: 10.1111/jce.12017. [DOI] [PubMed] [Google Scholar]
- 47.Henningsen KM, Olesen MS, Haunsoe S, Svendsen JH. Association of rs2200733 at 4q25 with early onset of lone atrial fibrillation in young patients. Scandinavian cardiovascular journal : SCJ. 2011 Dec;45(6):324–326. doi: 10.3109/14017431.2011.594081. [DOI] [PubMed] [Google Scholar]
- 48.Wnuk M, Pera J, Jagiella J, et al. The rs2200733 variant on chromosome 4q25 is a risk factor for cardioembolic stroke related to atrial fibrillation in Polish patients. Neurologia i neurochirurgia polska. 2011 Mar-Apr;45(2):148–152. doi: 10.1016/s0028-3843(14)60026-8. [DOI] [PubMed] [Google Scholar]
- 49.Goodloe AH, Herron KJ, Olson TM. Uncovering an intermediate phenotype associated with rs2200733 at 4q25 in lone atrial fibrillation. The American journal of cardiology. 2011 Jun 15;107(12):1802–1805. doi: 10.1016/j.amjcard.2011.02.326. [DOI] [PubMed] [Google Scholar]
- 50.Lee KT, Yeh HY, Tung CP, et al. Association of RS2200733 but not RS10033464 on 4q25 with atrial fibrillation based on the recessive model in a Taiwanese population. Cardiology. 2010;116(3):151–156. doi: 10.1159/000318172. [DOI] [PubMed] [Google Scholar]


