Abstract
Although influenza (flu) and respiratory syncytial virus (RSV) infections are extremely common in children under two years and resolve naturally, a subset develop severe disease resulting in hospitalization despite having no identifiable clinical risk factors. However, little is known about inherent host-specific genetic and immune mechanisms in this at-risk subpopulation. We therefore conducted a secondary analysis of statistically significant, differentially-expressed genes from a whole genome-wide case-control study of children less than two years of age hospitalized with flu or RSV, through the use of bipartite networks. The analysis revealed three clusters of cases common to both types of infection: core cases with high expression of genes in the network core implicated in hyperimmune responsiveness; periphery cases with lower expression of the same set of genes indicating medium-responsiveness; and control-like cases with a gene signature resembling that of the controls, indicating normal-responsiveness. These results provide testable hypotheses for at-risk subphenotypes and their respective pathophysiologies in both types of infections. We conclude by discussing alternate hypotheses for the results, and insights about how bipartite networks of gene expression across multiple phenotypes can help to identify complex patterns related to subphenotypes, with the translational goal of identifying targeted therapeutics.
Introduction
Most children by the age of two have been infected by RSV or influenza, both of which are associated with acute morbidity and mortality1. While many recover through a normal immune response, a subset develops severe disease requiring hospitalization. In fact, RSV is the leading cause of hospitalization for infants and young children worldwide, and is associated with long-term morbidity and risk for developing future chronic and recurrent wheezing2. Studies have identified epidemiological (e.g., second hand smoke, atopy3), and underlying medical conditions (e.g., congenital heart disease, prematurity or chronic lung disease4) as risk factors for flu and RSV infections associated with severe disease. Because the majority of children hospitalized with respiratory illnesses are previously healthy, researchers have hypothesized that the different clinical responses to such infections could be the result of host-specific genomic and immune responses that predispose patients to more severe disease5.
Unfortunately, much remains to be discovered about these differentiating host-specific genomic and immune mechanisms because most of the research has either been conducted (1) in vitro using infection assays of human cells; (2) mouse models; or (3) used single markers and univariate methods to analyze a limited repertoire of analytes as potential biomarkers in humans. For example, the latter approach has found that certain cytokines (IL-4, IL-13, IL-10, IL-8), cytokine receptors (IL-4 receptor α, IL-8 receptor), innate receptors (TLR4), and even non-immune proteins like surfactant could be used as potential biomarkers to predict severe RSV infection6. While such studies have provided key insights into biomarkers activated in mice and certain patient populations, to the best of our knowledge no studies have used multivariate methods to analyze the heterogeneity of responses using whole-genome human data during naturally acquired flu or RSV infections. Such multivariate analysis could help to identify molecular biomarkers for at-risk subsets of patient, and in the case of RSV, pathways that predispose them to later chronic recurrent wheezing and asthma.
We therefore used bipartite networks to conduct a multivariate analysis of patients with either flu or RSV, with the goal of identifying subphenotypes and molecular pathways that were common to both types of infections. Such an approach could result in general approaches to identify and treat at-risk patients with either type of infection.
Methods
Our research began with the question: How do differentially expressed genes that are common to flu and RSV infections co-express across patients with either type of infection? To address our research question, we made critical decisions related to data selection, and data analysis as discussed below:
Data Selection
Our study was based on a secondary analysis of a publicly available dataset downloaded from the Gene Expression Omnibus (ID: 200034205). The data consisted of 28 flu and 51 RSV previously-healthy infected children less than 2 years of age, with confirmed microbiologic diagnosis of infection, and who were hospitalized due to severe illness. The data also included 22 age, gender, and race matched healthy controls. As reported in the primary study7, peripheral blood mononuclear cells (PBMCs) of naturally-infected subjects were collected between 42 to 72 hours after hospital admission, and their disease severity score (aggregated measure of percutaneous O2 saturation, respiratory rate, subcostal retractions, general appearance, and auscultation) was recorded on a scale of 1–15. The cellular RNA was extracted, their expression measured using the whole-genome Affymetrix HG-U133 plus 2.0 chip array, and the results adjusted using a single standard curve. Furthermore, the primary study identified statistically-significant (FDR corrected) genes in each infection, and selected 18 highly-significant, differentially-expressed genes that were common to both infections for univariate analysis, with the goal of identifying pathways associated with the top-ranked genes in both illnesses.
In contrast to the above primary analysis, our secondary analysis of the data consisted of a multivariate analysis of all 101 subjects, and the above 18 genes that were common to both types of infection.
Data Analysis
Our analysis consisted of two steps: (1) exploratory visual analysis to identify emergent bipartite relationships between patients and genes; and (2) quantitative analysis suggested by the emergent visual patterns. This two-step method was motivated by our earlier studies8–10, which have demonstrated that bipartite networks can reveal complex patterns each prompting the use of quantitative methods that make the appropriate assumptions about the underlying data.
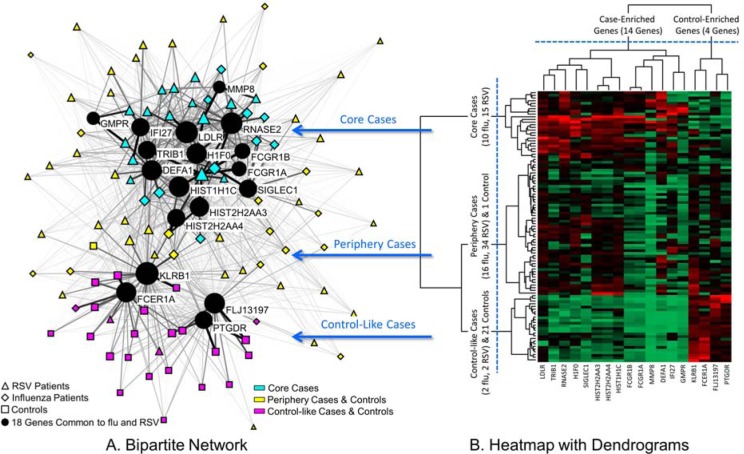
1. Exploratory Visual Analysis was conducted using network visualization and analysis11. Networks are increasingly being used to analyze a wide range of molecular phenomena such as gene and protein-protein interactions12–13, and to assess their relationships to diseases, symptoms, and syndromes. A network consists of nodes and edges; nodes represent one or more types of entities (e.g., patients or genes), and edges between the nodes represent a specific relationship between the entities. Figure 1 shows a bipartite network11 where edges exist only between patients and genes.
Figure 1.
A. A bipartite network (automatically laid out by the Kamada-Kawai algorithm14) shows how 18 genes (circular nodes) co-occur across 101 subjects (triangle, diamond and square nodes). The size of the nodes is proportional to the sum of the edge weights (representing normalized expression) that connect to them, and the thickness of edges is proportional to gene expression values. Therefore subjects with high total expression have large nodes, and higher expression is represented by thicker edges. The network has an overall distinct cluster topology that separates most cases from the controls, in addition to a core-periphery topology of cases. B. A heatmap with dendrogram generated through hierarchical clustering helped to identify the boundaries of three subject clusters, which were superimposed onto the network using colored nodes to denote cluster membership.
Edge weights in the network were used to represent the strength of the genes expression values for each patient-gene pair. Because the genes had different expression ranges, we used the min-max normalization method (which does a linear mapping of each genes expression to range from 0–1, and therefore preserves the relative distances between values to enable comparison). As shown in Figure 1, the edge thicknesses were drawn to be proportional to these normalized expression values. Node diameter was used to represent the sum of the edge weights connected to it (also referred to as the weighted degree centrality). This enabled a rapid visual inspection to determine for example, which patients have overall high aggregate expression values, and how such patients relate to the rest of the network. Finally, the node shape was used to represent phenotype or genes (triangles=RSV, diamonds=flu, squares=controls, circles=genes), and node color was used to represent members of a cluster based on hierarchical cluster analysis.
Global patterns between subjects and genes in the network were visualized and analyzed using the Kamada-Kawai layout algorithm14 in Pajek (version 3.13). As shown in Figure 1, the algorithm pulls together nodes that are strongly connected, and pushes apart nodes that are not. This algorithm is fast but approximate and is well-suited for medium sized networks consisting of between 100-1000 nodes15. The result is that nodes with a similar pattern of connections (e.g., the gene nodes IFI27 and TRIB1 in the top of the network in Figure 1A) are placed close to each other.
A key advantage of a network representation is the simultaneous visualization of multiple raw values (patient-gene associations, expression values), aggregated values (sum of gene expression values), and emergent global patterns (clusters) in a uniform visual representation. Such a representation enables the rapid generation of hypotheses based on complex multivariate relationships, which can be verified through appropriate quantitative methods.
2. Quantitative Analysis was conducted using three measures to verify the insights derived from the exploratory visual analysis. These methods were selected based on their appropriateness to the emergent patterns in the network.
(a). Agglomerative Hierarchical Clustering
Because the network layout suggested a distinct clustering combined with a core-periphery topology (nodes with high overall edge weights in the core, and nodes with low overall edge weights in the periphery11), we used the agglomerative hierarchical clustering method. The clustering was done using the Euclidean dissimilarity measure with the Ward linkage function, and the number of clusters and their boundaries were determined based on natural breaks in the patient and gene dendrograms. The cluster boundaries were superimposed onto the network by using identical node color to denote cluster membership.
(b). Clusteredness
To test whether the clusters in the network could have occurred by chance, we compared the variance, skewness, and kurtosis of the dissimilarities in the data, to 1000 random permutations of this data. For each network permutation, we preserved the size of the network (number of nodes and edges), in addition to the edge weight distribution of each patient when analyzing the patient dendrogram, and the edge weight distribution for each gene when analyzing the gene dendrogram. Significant breaks in the patient, or gene dendrograms would result in a significantly larger variance, skewness, and kurtosis of the dissimilarity measures, compared to the same measures generated from the random networks.
(c). Relationship to Clinical Outcomes
Because the cases had a core-periphery topology, we used the Mann Whitney U test to analyze whether the patients in the case core had a higher overall gene expression (using weighted degree centrality calculated by adding each patient’s normalized gene expression across all genes) and higher disease severity (defined in the primary study as an aggregated measure of percutaneous O2 saturation, respiratory rate, subcostal retractions, general appearance, and auscultation) compared to the patients in the periphery. The same statistical test was used to compare the weighted degree centrality of the genes in each gene cluster.
Results
Patient Clusters
As shown in Figure 1A, the patients had a complex but understandable topology consisting of a majority of the cases on the top, and a majority of the controls at the bottom of the network. In addition, the cases on the top had a core-periphery topology, where there were patients with high overall gene expression in the center (henceforth referred to as the core cases), and many patients with low overall gene expression in the periphery (henceforth referred to as the periphery cases). Finally, there were four cases (pink diamonds and triangles) that were clustered with the controls at the bottom of the network (henceforth referred to as the control-like cases).
The boundaries of the above clusters were quantitatively verified through agglomerative hierarchical clustering. The vertical dendrogram in Figure 1B shows that there were three main subject clusters: the first consisting of 25 core cases (10 flu, 15 RSV), the second consisting of 50 periphery cases (16 flu, 34 RSV) with 1 control, and the third consisting of 4 control-like cases (2 flu, 2 RSV) clustered with 21 controls. These cluster boundaries were used to color nodes in the network to denote cluster membership as shown in Figure 1A.
To test whether the above clusters could have occurred by chance, we measured their clusteredness with respect to random permutations of the data. The subject clustering in the flu/RSV data was significant when compared to 1000 random networks based on variance of the dissimilarities (flu/RSV =2.75, Random-Mean=0.88, p<.001 two-tailed test), skewness of the distribution of dissimilarities (flu/RSV=5.55, Random-Mean=3.94, p<.001 two-tailed test), and kurtosis of the distribution of dissimilarities (flu/RSV=38.69, Random-Mean=25.03, p<.001 two-tailed test).
Furthermore, the weighted degree centrality (sum of edge weights) of the core cases (Median=4.55) was significantly different (U=49.00, p<.001, two-tailed test) compared to the periphery cases (Median=2.52) suggesting that the overall gene expression of the patients in the core was higher compared to those in the periphery. Finally, the disease severity of core cases (Median=7) was significantly higher (U=261.50, p<.001, two-tailed test) compared to periphery cases (Median=2). Finally, there was no significant difference (χ2(2, N=79)=0.86, p=0.652) in the proportion of flu vs. RSV patients across the three case clusters, suggesting that the gene-based clustering was common across both types of infection.
Gene Clusters
As shown in Figure 1, the genes fell into two clusters, whose boundaries were quantitatively verified through hierarchical clustering. As shown by the horizontal dendrogram in Figure 1B, there was a large cluster of 14 genes (LDLR, HIST2H2AA3, HIST1H1C, HIST2H2AA4, FCGR1A, TRIB1, SIGLEC1, FCGR1B, IFI27, DEFA1, MMP8, GMPR, RNASE2, H1F0) at the top of the network, and a smaller cluster of the 4 genes (FLJ13197, PTGDR, KLRB1, FCER1A) at the bottom. Based on the results previously published on the same data, the cluster of 14 genes consisted of all up-regulated genes, whereas the cluster of 4 genes consisted of all down-regulated genes. The bipartite network also revealed the inter-cluster relationships: the median gene expression of the 14 genes of the 25 core cases (Median=4.22) was significantly higher (U =16, p< .001, two-tailed test) compared to the 50 periphery cases (Median=1.95). This pattern can also be seen in the high expression values (shown in mostly red cells) in the upper left-hand corner of the heatmap in Figure 1B.
The clusteredness of the above gene clusters was significant when compared to 1000 random networks based on variance of the dissimilarities (flu/RSV=2.91, Random-Mean=0.24, p<.001 two-tailed test), skewness of the distribution of dissimilarities (flu/RSV=2.01, Random-Mean=0.80, p<.001 two-tailed test), and kurtosis of the distribution of dissimilarities (flu/RSV=7.81, Random-Mean=3.16, p<.001 two-tailed test).
Discussion
The bipartite visualization and quantitative verifications revealed not only a strong separation of the cases from the controls, but also a core-periphery topology for the cases. This complex but understandable topology helped to identify three possible subphenotypes and their potential pathways. (1) The core cases have significantly higher expression of 14 up-regulated genes, which included 4 histone genes, 4 genes with to date have unknown function in antiviral response, and 6 immune-related genes each of which has a well-known non-overlapping antiviral function. The latter set included RNASE2 which induces direct damage to viruses, IFI27 and DEFA1 which produce specific and general microbicidal protein responses respectively, FCGR1 which among a multitude of immune functions is primarily involved in activation of the monocyte, macrophages and dendritic cells for efficient antigen presentation, SIGLEC1 a type I interferon dependent sialoadhesin, and MMP8 a tissue remodeling enzyme (Collagenase 2). An Ingenuity Pathway Analysis (IPA) of the 14 genes suggested an indirect but strong interferon signature including TNFα and IL-6 cytokines involved in antiviral and innate inflammatory responses. Because the core cases also had a significantly higher disease severity score, we hypothesize that these patients represent a distinct at-risk subphenotype that are hyper responsive to pathways targeted to viral clearance, and possibly carry a risk for long-term epithelial cell damage. (2) The periphery cases have a medium expression of all 18 genes and therefore suggest a second subphenotype with a subdued anti-viral response relative to the above hyperresponders. (3) The control-like cases have a high expression of 4 down-regulated genes, and low expression of the 14 up-regulated genes, and therefore mirror the expression patterns in uninfected controls. The results therefore suggest that the down-regulation of these 4 genes indicates a “protective” phenotype making them similar to the uninfected controls. Existing literature on these genes provide some confirmatory evidence. While the exact role of the high-affinity receptor which binds to the constant portion of IgE (FcER1) is unknown in viral pathogenesis, SNPs included on this gene have been shown to be associated with severe RSV disease5. Additionally, KLRB1 which has been shown to have inhibitory functions on natural killer (NK) cells16 was downregulated, suggesting an enhanced antiviral response in patients resembling the immune response of controls. Finally, PTGDR a receptor important in mast cell function was downregulated, but the exact role of this receptor in viral infection is still unknown. Overall, control-like cases suggests a third subphenotype which have a “just enough” response to the virus, without overt stimulation of virally induced genes, and therefore potentially with reduced bystander damage.
One could argue that the above result could also be the result of the progression of infection over time. For example, the core cases could be at the peak of infection, the periphery cases could be later in the infection, and the control-like cases could be recovering from the infection. However, an additional analysis revealed that the 3 case clusters were not significantly different (H(2, N=79)=2.56, p=0.278) in time of sample collection after hospitalization. There is of course the possibility that the children were infected at very different times before hospitalization, but controlling such a variable is practically impossible in the analysis of human infections. Therefore, we provide two explanations for why sample collection time is probably not an adequate explanation for the results: (1) Because all case samples were collected from patients that were hospitalized indicating severe illness, a resolution of such severity in the short time window of 42-72 hours is unlikely to occur. (2) The gene expression changes in the PBMCs of the patients suggest a specific induced innate immune response (e.g., Toll-like receptor) to viruses. Such signaling pathways (which induce interferon secretion and contribute to anti-viral immunity) last several days which exceeds the sample collection time window in this study. We therefore propose that the three case clusters are more likely the result of inherent host differences in anti-viral responses, and therefore represent distinct subphenotypes.
Conclusions and Future Research
Several epidemiological, clinical, and genetic risk factors have been examined to identify children at risk for severe acute RSV and influenza infection, and for long-term sequelae. However, to the best of our knowledge, no study has applied multivariate methods to analyze gene expression data from naturally-infected children with flu or RSV with the goal of identifying the heterogeneity of their host response and the respective pathways involved. Here we presented a multivariate analysis of gene expression in human data using bipartite networks.
We believe our study makes three biological and methodological contributions. (1) We have shown evidence for the existence of three subphenotypes that are common to flu and RSV. While there might be other subphenotypes and underlying pathways that are unique to each disease, we were specifically interested in subphenotypes and pathways that were common to both diseases, with the goal of providing insights into future therapeutic targets that address multiple types of respiratory infections. The study therefore has helped to identify data-driven hypotheses for subphenotypes that can be tested in future studies. (2) We have provided biological inferences for the genesis of the three subphenotypes, and argued why time of data collection alone is not an adequate explanation for the results. (3) We have demonstrated the utility of bipartite networks to reveal a complex but understandable combination topology consisting of distinct clustering, in addition to a core-periphery topology. Such an understanding of relationships in data is difficult using unipartite methods such as k-means (which can identify for example either patient or gene clusters but not both simultaneously), or even bipartite heatmaps with dendrograms (shown in Figure 2B), which are more useful for confirming a topology, rather than for discovering complex associations8. Furthermore, methods like modularity11 (used to identify disjoint clusters in networks) are also not designed to discover such combination topologies, therefore demonstrating the advantages of bipartite network visualizations to guide in the comprehension of complex multivariate associations, and in the selection of appropriate quantitative methods for verification. Given the importance of visualizations to detect such complex topologies, our current research is examining visualizations for “big data” containing hundreds of thousands of patients and variables.
A limitation of our study is that we examined only those genes that were common to both infections, and there might be subphenotypes which are unique to each infection type. Furthermore, we analyzed subphenotypes in only one dataset. Therefore our future research aims to examine genes that are specific to each infection type from the same dataset, and test them in a new dataset in collaboration with the authors (who collaborated in the current study) of the primary study. In that respect, this project demonstrates the promise of the Open Science17 movement, where publicly available data not only enables new hypotheses to be generated from existing data, but can also motivate new interdisciplinary collaborations among researchers who would not have ordinarily been motivated to work together. Such collaborations should help accelerate discoveries in complex phenomena such as disease subphenotypes and their pathways, with the goal of translating them into effective and well-targeted therapeutics.
Acknowledgments
This research was supported in part by a grant from IHII, UTMB, and NIH UL1TR000071 UTMB CTSA (ARB).
References
- 1.World Health Organization Acute Respiratory Infections. http://www.who.int/vaccine_research/documents/ARI07062010_2.pdf. Accessed August 28, 2013.
- 2.Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am J Respir Crit Care Med. 2000 May;161(5):1501–7. doi: 10.1164/ajrccm.161.5.9906076. [DOI] [PubMed] [Google Scholar]
- 3.Welliver RC. Review of epidemiology and clinical risk factors for severe respiratory syncytial virus (RSV) infection. J Pediatr. 2003 Nov;143:S112–7. doi: 10.1067/s0022-3476(03)00508-0. [DOI] [PubMed] [Google Scholar]
- 4.Bhat N, Wright JG, Broder KR. Influenza-associated deaths among children in the United States, 2003–2004. N Engl J Med. 2005 Dec 15;353(24):2559–67. doi: 10.1056/NEJMoa051721. [DOI] [PubMed] [Google Scholar]
- 5.Janssen R, Bont L, Siezen CL, Hodemaekers HM, Ermers MJ, Doornbos G, van ’t Slot R, Wijmenga C, Goeman JJ, Kimpen JL, van Houwelingen HC, Kimman TG, Hoebee B. Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J Infect Dis. 2007 Sep 15;196(6):826–34. doi: 10.1086/520886. [DOI] [PubMed] [Google Scholar]
- 6.Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev. 2011 Jan;24(1):210–29. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, Ramilo O, Mejias A, Flaño E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol. 2012 May;86(10):5422–36. doi: 10.1128/JVI.06757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhavnani SK, Bellala G, Victor S, Bassler KE, Visweswaran S. The Role of Complementary Bipartite Visual Analytical Representations in the Analysis of SNPs: A Case Study in Ancestral Informative Markers. JAMIA. 2012;19:e5–e12. doi: 10.1136/amiajnl-2011-000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhavnani SK, Victor S, Calhoun WJ, Busse WW, Bleecker E, Castro M, Ju H, Brasier AR. How Cytokines Co-occur across Asthma Patients: From Bipartite Network Analysis to a Molecular-Based Classification. Journal of Biomedical Informatics. 2011;44:S24–S30. doi: 10.1016/j.jbi.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhavnani SK, Drake J, Bellala G, Dang B, Visweswaran S, Olano JP. How Cytokines Co-occur across Rickettsioses Patients: From Bipartite Visual Analytics to Mechanistic Inferences of a Cytokine Storm. AMIA Summit on Translational Bioinformatics. 2013 [PMC free article] [PubMed] [Google Scholar]
- 11.Newman M. Networks: An Introduction. Oxford University Press; 2010. [Google Scholar]
- 12.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007 May 22;104(21):8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008 Apr;18(4):644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamada T, Kawai S. An algorithm for drawing general undirected graphs. Information Processing Letters. 1989;31(1):7–15. [Google Scholar]
- 15.Nooy W, Mrvar A, Batagelj V. Exploratory Social Network Analysis with Pajek. New York, NY: Cambridge University Press; 2005. [Google Scholar]
- 16.Pozo D, Valés-Gómez M, Mavaddat N, Williamson SC, Chisholm SE, Reyburn H. CD161 (human NKR-P1A) signaling in NK cells involves the activation of acid sphingomyelinase. J Immunol. 2006 Feb 15;176(4):2397–406. doi: 10.4049/jimmunol.176.4.2397. [DOI] [PubMed] [Google Scholar]
- 17.Molloy JC. The Open Knowledge Foundation: Open Data Means Better Science. PLoS Biology. 2011;9 doi: 10.1371/journal.pbio.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]