Summary
The plant cell wall is an important factor for determining cell shape, function and response to the environment. Secondary cell walls, such as those found in xylem, are composed of cellulose, hemicelluloses and lignin and account for the bulk of plant biomass. The coordination between transcriptional regulation of synthesis for each polymer is complex and vital to cell function. A regulatory hierarchy of developmental switches has been proposed, although the full complement of regulators remains unknown. Here, we present a protein-DNA network between Arabidopsis transcription factors and secondary cell wall metabolic genes with gene expression regulated by a series of feed-forward loops. This model allowed us to develop and validate new hypotheses about secondary wall gene regulation under abiotic stress. Distinct stresses are able to perturb targeted genes to potentially promote functional adaptation. These interactions will serve as a foundation for understanding the regulation of a complex, integral plant component.
Plant cell shape and function are in large part determined by the cell wall. Almost all cells have a primary wall surrounding the plasma membrane. Specialized cell types differentiate by depositing a secondary cell wall upon cessation of cell elongation. In addition to providing mechanical support for water transport and a barrier against invading pathogens, the polymers contained within the wall are an important renewable resource for humans as dietary fiber, as raw material for paper and pulp manufacturing, and as a potential feedstock for biofuel production. Secondary cell walls account for the bulk of renewable plant biomass available globally.
The secondary cell wall consists of three types of polymers - cellulose, hemicelluloses and lignin and is found in xylem, fibers and anther cells. Cellulose microfibrils form a main load-bearing network. Hemicelluloses include xylans, glucans, and mannans. Lignin is a complex phenylpropanoid polymer that imparts “water-proofing” capacity as well as mechanical strength, rigidity, and environmental protection. Despite the importance of the plant secondary cell wall, our knowledge of the precise regulatory mechanisms that give rise to these metabolites is limited. The expression of cell wall associated genes is tightly spatiotemporally co-regulated1,2. However, the pervasive functional redundancy within transcription factor families, the combinatorial complexity of regulation, and activity in a small number of cell types render functional characterization from single gene experiments difficult. A model of master regulators has been proposed with NAC domain and Homeobox HD-ZIP Class III (HD-ZIPIII) transcription factors initiating cell specification and secondary cell wall synthesis. In this model, VASCULAR-RELATED NAC DOMAIN6 (VND6) and VND7 are sufficient but not necessary to regulate xylem vessel formation; additionally, the HD-ZIPIII transcription factor PHABULOSA (PHB) also regulates vessel formation, and acts in a highly redundant manner with four other HD-ZIPIII factors3. In anthers, two NAC domain transcription factors, NAC SECONDARY WALL THICKENING1 (NST1) and NST2, are sufficient to drive the secondary cell wall biosynthetic program, but act redundantly4. Thus, regulation of this process is highly redundant and combinatorial. However, no comprehensive map of interactions has been developed at cell type-resolution over time, nor have upstream regulators been identified. We therefore chose to pursue a network-based approach to comprehensively characterize the transcriptional regulation of secondary cell wall biosynthesis.
To systematically map this regulatory network at cell type-resolution, we used a combination of high spatial resolution gene expression data5 and the literature1,6 to identify fifty genes implicated in xylem cell specification. These included transcription factors and enzymes implicated in cellulose, hemicellulose and lignin biosynthesis that are expressed in root xylem cells (Supplementary Table 1; Methods). Selection of both developmental regulators and downstream functional genes allowed us to interrogate upstream regulatory events that determine xylem specification and differentiation associated with secondary cell wall synthesis. Promoter sequences were screened using an enhanced yeast one hybrid (Y1H) assay against 467 (89%) of root xylem-expressed transcription factors7. Protein interactions were identified for 45 of the promoters (Supplementary Table 2). The final network comprises 242 genes and 617 protein-DNA interactions (Fig. 1A; http://gturco.github.io/trenzalore/stress_network). Thirteen of the transcription factors have been previously identified as having a role in xylem development or secondary cell wall biosynthesis. Six of the transcription factors were previously shown to bind to these promoters and a further nine of the protein-DNA interactions were implied in gene expression studies, i.e. without demonstrating direct binding6,8–11. These interactions represent independent validation of our approach (Supplementary Table 2, Extended Data Fig. 1). All together, the network contains 601 novel interactions, although false negatives and false positives are a component of all network approaches12.
Figure 1. Regulators of xylem development and secondary cell wall biosynthesis.
(A) Gene regulatory network for secondary cell wall biosynthesis in Arabidopsis root xylem. Nodes-transcription factors or promoters, edges-protein-DNA interactions. Edges in feed-forward loops are red. (B) A sample feed-forward loop in red. (C) ‘Power edges’ between node sets. (D) The secondary wall network from sub-fragments of cell wall promoters.
Our Y1H approach revealed a highly interconnected regulatory network. On average, each cell wall gene promoter was bound by 5 transcription factors from 35 protein families with over-representation of AP2-EREBP, bHLH, C2H2, C2C2-GATA and GRAS gene families (Supplementary Table 3). Our network now adds an additional layer of gene regulation with novel factors upstream of VND6 and VND7 and supports feed forward loops9,11,13 as an overarching theme for regulation of this developmental process with a total of 96 such loops (Fig. 1A, B).
To organize the network, we employed a power graph compression approach to condense the network into overlapping node sets with similar connectivity. Protein-DNA interactions (edges) between proteins and promoters (nodes) in the original network were replaced by ‘power edges’ between overlapping ‘power nodes’14. A power edge exists between suites of transcription factors that bind to the same set of promoters. Using this approach, 24 power edges were observed (Supplementary Table 4; Fig. 1C). Some sets could be distinguished based on target gene function. For instance, one power edge connects 16 transcription factors with promoters of two lignin genes, 4CL1 and HCT, while another power edge connects three transcription factors with genes related to cellulose and hemicellulose biosynthesis such as CESA4, CESA7, IRX9, COBL4 and GUX2.
Using our network, we hypothesized that E2Fc is a key upstream regulator of VND6, VND7, and secondary cell wall biosynthesis genes. This hypothesis is based on our findings that E2Fc bound to 23 promoters including VND6, VND7, MYB46, cellulose, hemicellulose, and lignin associated genes (Fig. 2A). VND7 and MYB46 are also known to bind to the promoters of many of these genes as well9,13,15, creating a suite of feed forward loops. E2Fc is a known negative regulator of endoreduplication16,17. Before terminally differentiating, xylem cells elongate and likely undergo endoreduplication prior to secondary cell wall deposition. E2Fc can act as a transcriptional repressor16–18 as well as a transcriptional activator19–22 and here we report both. E2Fc activated VND7 expression in a dose-dependent manner (Fig. 2B and Extended Data Fig. 2A, B) in transient assays, but not in the presence of RETINOBLASTOMA-RELATED (RBR) protein, as is typical of E2F transcription factors (Extended Data Fig. 2C). In an E2Fc-overexpressor line with the N-terminus deleted to overcome post-translational degradation16,17, regulation of VND7 expression varied with extremely high or low E2Fc levels resulting in VND7 repression and moderate E2Fc levels resulting in VND7 activation (Extended Data Fig. 2B). The dynamic regulation was also observed in an E2Fc-knockdown line23, where transcript abundance of VND6 and VND7 were significantly increased (Fig. 2C). Based on our results, we propose that E2Fc acts in a complex, concentration-dependent manner to regulate gene expression either as an activator or a repressor. Coincident with the repression observed in E2Fc-knockdown lines, ectopic patches of lignin were observed near the root-shoot junction using phloroglucinol staining (Fig. 2D). Based on an Updegraff assay, a significant increase in crystalline cellulose in the knockdown line was observed (Fig. 2E).
Figure 2. E2Fc represses secondary cell wall gene biosynthesis.
(A) E2Fc-DNA interactions. Solid edges=Y1H, dashed edges=literature. (B) Bright field (top) and dark-field (bottom) of representative leaves (n=20) expressing VND7::LUC or together with 35S::E2Fc in 1:0.1, 1:1, 1:2, 1:5, and 1:10 ratios respectively. C) VND6 and VND7 expression relative to UBC10 control in an E2Fc RNAi line relative to wild-type. n= 2 biological replicates with 3 technical replicates. (D) Phloroglucinol staining of lignin (n=6xgenotype, representative images shown) and (E) crystalline cellulose in wild-type and E2Fc-knockdown roots (n=3×1000xgenotype). For all panels, *p<0.05 from Student’s t-test and data are means ± s.d.
The HD-ZIPIII transcription factors REVOLUTA (REV), PHB, and PHAVOLUTA are sufficient for xylem cell specification and secondary wall synthesis3. We found that VND7 bound REV and PHB promoters in yeast. VND7 has been to shown to act as a transcriptional activator9 or as a repressor when complexed with VNI224. With a dexamethasone-inducible version of VND725, transcript levels of REV and PHB were significantly decreased by 2.5-fold following induction (Fig. 3A). The REV transcription factor bound to the promoter of the lignin biosynthesis gene PHENYLALANINE AMMONIA LYASE4 (PAL4). In a rev-5 loss-of-function mutant, PAL4 significantly increased in transcript abundance (Fig. 3B) and transient induction of REV by a glucocorticoid receptor fusion26 resulted in a decrease of PAL4 expression (Fig. 3C). Taken together, these data suggest that E2Fc can activate VND7 expression in a dose-dependent manner, while VND7, possibly in concert with VNI2, can repress REV expression, and REV can repress expression of PAL4. This series of interactions predicted by the network model and tested by perturbation analyses ensures that activation of VND7 and coordination of lignin biosynthesis is tightly regulated.
Figure 3. Tissue-specific VND7 regulation and VND7 targets.

(A) REV and PHB expression relative to β-tubulin control following dexamethasone treatment of 35S::VND7:VP16:GR relative to untreated. n=4, a,b,c = p<0.01). (B) PAL4 expression relative to AT5G15710 control in rev-5 relative to wild-type. (C) PAL4 expression relative to UBC21 control following one hour dexamothasone treatment of 35S:REV:GR relative to untreated. *p<0.05 for panels B and C, n= 2 biological replicates with 3 technical replicates. All panels show data as means ± s.d, with p calculated from Student’s t-test.
We next sought to identify all transcription factors that potentially regulate secondary cell wall biosynthesis genes, not just in root xylem cells but also in aboveground cell types including xylary fibers, interfasicular fibers, and anthers. Many of the biosynthetic genes downstream of the key NAC domain transcription factors act in both the root and shoot9, To expand the network, we used Y1H to screen multiple smaller promoter fragments of a subset of promoters included in the root xylem network including genes associated with cellulose, hemicellulose, and lignin biosynthesis against a library of 1,664 full-length Arabidopsis transcription factors (Supplementary Table 5,6). We observed a total of 413 interactions that included proteins from 36 of the 75 protein families tested (Supplementary Table 7; Fig. 1D; http://gturco.github.io/trenzalore/secondary_cell_wall). We found an over-representation of AP2-EREBP, bZip, ZF-HD, MYB, and GeBP families (Supplementary Table 8). Each promoter interacted with an average of 38 different proteins, generating even more possibilities for combinatorial, redundant, or condition-specific gene regulation. Like the root-xylem network, previously reported protein-DNA interactions were observed in this screen including MYB46 and MYB83 binding the promoters of CESA genes (Supplementary Table 7)8,27. Since most of these interactions were novel, a subset was additionally validated. Transient expression of AIL1, MYB83, MYB54, NAC92, NST2, and SND1 caused a significant increase in CESA4::LUC activity in tobacco, indicating binding and activation of the CESA4 promoter (Fig. 4A). We further tested three regions of the CESA4 promoter with two NAC family proteins, SND1 and NST2 (Fig. 4B,C), using an in vitro electrophoretic mobility shift assay (EMSA). Extracts of Escherichia coli expressing either GST:NST2 or GST:SND1 in the presence of a CESA4-2pr promoter probe produced DNA species with retarded mobility (Fig. 4B,C). We also observed binding between CESA7, CESA8, and KOR promoter fragments with the NST2 protein and CESA8 with the SND1 protein (Extended Data Fig. 3). These interactions between NST2 and CESA4, CESA8, and KOR promoters were further confirmed in planta by ChIP. An antibody to GFP was used to immunoprecipitate NST2 protein from extracts of 35S::NST2:GFP plants. The complex was significantly enriched for fragments from the CESA4, CESA8 and KOR promoters (Fig 4D). The tracheary element-regulating cis-element (TERE = CTTNAAAGCNA) is a direct target of VND628,29. A perfect TERE is present in the CESA4 promoter (CTTGAAAGCTA) and TERE-like sequences are present in CESA8 (CTTCAATGTTA) and KOR (CTTGAAAATGA). Taken together, these data clearly demonstrate that the expression of CESA4 and other secondary cell wall genes is mediated by the direct binding of the NAC-domain binding transcription factors NST2 and SND1 to the target gene promoters via the TERE.
Figure 4. Multiple transcription factors bind the CESA4 promoter.
(A) Activation of CESA4::LUC by transcription factors in tobacco (n=5). *p<0.05 based on Student’s t-test. Data are means ± s.d. (B–C) EMSA with NST2 (B) and SND1 (C) with promoters. Arrowheads indicate protein-DNA complexes, arrows indicate free probe. (D) ChIP of NST2:GFP with CESA4, CESA7, CESA8, and KOR promoters.
Having generated a gene regulatory network supported by in vivo and in vitro approaches, we sought to test if the model could allow us to predict responses under abiotic stress perturbation. Co-opting a developmental regulatory network is likely a key mechanism to facilitate adaptation in response to stress. Thus, we hypothesized that stress responses are likely integrated into the gene regulatory network that determines xylem cell specification and differentiation and that we can predict the exact genes that these stresses manipulate within our network.
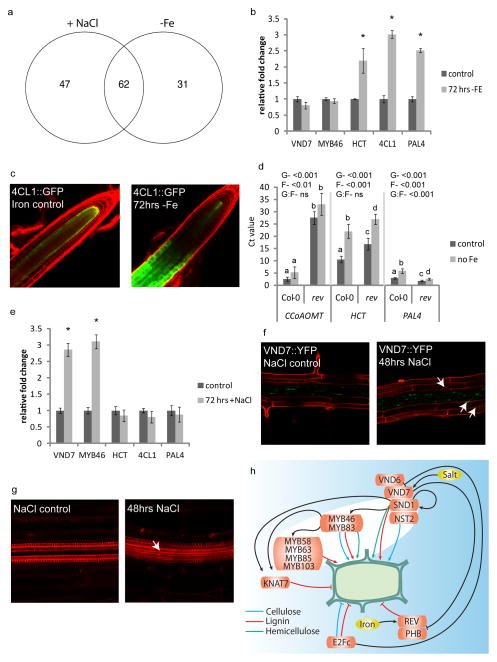
We first identified genes within the network whose expression was altered specifically in the root vasculature in response to salt, sulfur, iron and pH stress30,31 and nitrogen influx32. Genes within the root xylem secondary cell wall network were significantly differentially regulated in response to sulfur stress, salt stress and iron deprivation (Supplementary Table 9). Substantial overlap was observed between iron deprivation and salt stress gene responses and was further characterized (Fig. 5A). We filtered the xylem network to include only genes differentially expressed in salt or iron, creating stress-specific sub-networks (Extended Data Fig. 4). Previously, we determined that key developmental transcription factors have significantly more upstream regulators compared to other genes33. In response to iron deprivation, REV has the most upstream regulators, while in response to salt stress, VND7 and MYB46 have the most upstream regulators.
Figure 5. The xylem-specific gene regulatory network is responsive to high salinity and iron deprivation.
(A) Network genes responsive to high salinity and/or iron deprivation. (B) VND7, HCT, 4CL1, PAL4 expression after iron deprivation. (C) 4CL1::GFP expression after iron deprivation (representative images shown, n=4xline). (D) Lignin gene expression after iron deprivation in rev-5. G-genotype, F-Fe stress; p-values from ANOVA. (E) VND7, HCT, 4CL1, PAL4 expression after NaCl. (B,D,E) Expression relative to UBC10 and PP2AA3 controls. n= 2 biological replicates with 3 technical replicates. *p≤0.01based on Student’s t-test and data are means ± s.d. (F) Representative images of VND7::YFP (n=5) and (G) fuchsin-staining (n=5) after NaCl. Arrows- (F) non-stele cells and (G) extra metaxylem strand. (H) Proposed regulation of secondary wall biosynthesis.
Based on these data from the iron-deprivation sub-network, we hypothesized that REV plays a key role in regulating secondary cell wall development in response to iron deprivation. In order to additionally determine directionality and sign in the network, we constructed a network of 16 key nodes using the consensus network from four unsupervised and one supervised network inference method. REV was also predicted to be an important regulator of lignin biosynthesis gene expression in response to iron deprivation using these methods (Extended Data Fig. 5). First, to test the model-generated prediction that lignin biosynthetic gene expression is altered in response to iron deprivation, we measured phenylpropanoid-related gene expression. An increase in 4CL1, PAL4 and HCT gene expression was observed (Fig. 5B). Iron deprivation stress altered the timing and spatial distribution of the 4CL1 transcript (Fig. 5C; Extended Data Fig. 6A). These expression changes are accompanied by an increase in fuchsin staining indicative of increased phenylpropanoid deposition (Extended Data Fig. 6B). Expression in a rev-5 loss-of-function mutant in iron-deficient conditions revealed a REV- and stress-dependent influence on CCoAOMT1, PAL4 and HCT expression (Fig. 5D), thus validating our model predictions.
In the high-salinity sub-network VND7 and MYB46 contain the most upstream regulators (Extended Data Fig. 4). VND7 and MYB46 expression is greatly increased in roots in response to salt stress, but lignin biosynthetic gene expression is unaltered (Fig. 5E). In corroboration with this hypothesis, the network model constructed using the described in silico methods also predicts VND7 and MYB46 as main regulators in response to salt stress but not iron deprivation (Extended Data Figure 7), and indeed this was observed with an expansion of the domain of VND7 expression after salt treatment but not iron deprivation (Fig. 5E,F; Extended Data Fig. 6C). In conjunction with this ectopic increase, we observed an additional strand of metaxylem in roots exposed to high salinity (Fig. 5G).
Due to functional redundancy among regulators of secondary cell wall biosynthesis, transcription factors have largely eluded identification by loss-of-function genetic screens. Our network approach has identified hundreds of novel regulators and provided considerable insight into the developmental regulation of xylem cell differentiation. The network, which includes a cell cycle regulator, is comprised of many feed forward loops that likely ensure robust regulation of this process. Accordingly, we revealed that perturbation at distinct nodes changes the network subtly including phenylpropanoid biosynthesis in response to iron deprivation, and ectopic xylem cell differentiation in response to salt stress. We anticipate that these findings will be instrumental in biotechnology and in our understanding of cell fate acquisition.
Methods Section
Yeast one-hybrid (Y1H) protein-DNA interaction assays
The root vascular-expressed transcription factor collection is described in Gaudinier et al. 7. The 1,663 transcription factor collection was assembled primarily from clones deposited in the Arabidopsis Biological Resource Center by various collaborative projects including the Peking-Yale Consortium34, REGIA35, TIGR36, and the SSP Consortium37. Translational fusions to the GAL4 activation domain were generated essentially as described by Pruneda-Paz et al.38. A total of 1,663 E. coli strains harboring different Arabidopsis transcription factors (Supplementary Table 5) were arrayed in 96-well plates and plasmids were prepared using the Promega Wizard SV 96 plasmid purification DNA system according to manufacturer recommendations.
Root secondary cell wall gene promoters (2–3 kb of upstream regulatory region from the gene’s translational start site, or the next gene, whichever comes first) were cloned and recombined with reporter genes according to Brady et al.33. Promoter sequences and primers used are described in Supplementary Table 1. AT1G30490, AT5G60690, AT2G34710, AT1G71930, AT1G62990 promoter sequences and primers are described in Brady et al 2011, while the promoter sequences and primers for AT5G15630 are described in Brady et al 2007. For dissection of cell wall biosynthesis promoters, approximately 1,000 bp of sequence upstream of the translational start site was tested for interactions with the transcription factor library. Three overlapping fragments of approximately equal and average size of 419 bp were independently cloned for each promoter according to Pruneda-Paz et al.38. The oligonucleotides used to amplify promoter fragments and details of their coordinates for 4CL1 (At1g51680), CESA4/IRX5 (At5g44030), CESA7/IRX3 (At5g17420), CESA8/IRX1 (At4g18780), COBL4/IRX6 (At5g15630), HCT (At5g48930), IRX9 (At1g27600), IRX14 (At4g36890), KOR/IRX2 (At5g49720), LAC4/IRX12 (At2g38080), and REF8 (At2g40890) are described in Supplementary Table 6.
Root bait promoters were screened against the stele-expressed transcription factor collection using the Y1H protocol as previously described7. The 1,663 transcription factor library was transformed into each yeast strain and the β-galactosidase activity was determined as described by Pruneda-Paz et al.38, but in 384-well plates. Positive interactions were visually identified as incidence of yellow caused by the presence of ortho-nitrophenyl cleavage from colorless ortho-nitrophenyl-β-D-galactoside by β-galactosidase. The DNA bait strains were similarly tested for self-activation prior to screening by not transforming with prey vectors in the presence of thiamine. All interacting transcription factors were assembled into a cell wall interaction library and the screen was repeated to confirm the results and each clone was sequenced to reconfirm identity.
Statistical analysis for protein family enrichment
Enrichment was determined using the hypergeometric distribution online tool (stattrek.com). The population size is the number of transcription factors in the xylem transcription factor collection while the successes within the population is the number of transcription factors within that transcription factor family in the xylem. The number of successes in the sample was the number of proteins belonging to that family, and the number in the sample is the total number of transcription factors within the network. The A. thaliana transcription factor list is as described in Gaudinier et al.7.
Power graph compression approach
The power graph compression was performed using the algorithm as previously described14.
Plant material
The E2Fc RNAi line is described by del Pozo et al.23 and was verified by quantifying E2Fc transcript abundance relative to the Col-0 control using an E2Fc primer compared to an ACTIN control primer (Supplementary Table 1). VND7::YFP lines are described in Kubo et al.39. The VND7 glucocorticoid induction line is described in Yamaguchi et al.9. The rev-5 loss-of-function mutant was described in et al.40.
Cloning and insertion of the 4CL1 promoter into a pENTR p4-p1R donor vector was performed according to Brady et al.33 (for sequence, see Supplementary Table 1). The promoter was then recombined into binary vector pK7m24GW,3 along with pENTR 221 ER-GFP:NOS. The resulting 4CL1::GFP vector was transformed into Agrobacterium strain GB3101. Col-0 plants were then transformed using the floral dip method.
Plant growth conditions
All plants were grown vertically on plates containing 1X Murashige and Skoog salt mixture, 1% sucrose, and 2.3 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.8) in 1% agar. NaCl plates were made by adding 140mM NaCl to this standard media. Iron control and deprivation media were made according to Dinneny et al.30. Plants grown on stress media (iron or salt) were first germinated on nylon mesh placed over control media for four days before transferring mesh with seedlings to iron deprivation or NaCl plates. Plants used for RNA isolation were also grown on nylon mesh placed over the agar to facilitate the collection of root material5.
Determination of crystalline cellulose
Roots of 7-day-old plants were harvested and lyophilized. Six to ten plates of seedlings grown at the same time on the same media were pooled to make a single biological replicate. Crystalline cellulose was measured according to Updegraff41. After hydrolysis of non-cellulosic polysaccharides from an alcohol insoluble residue wall preparation with the Updegraff reagent (acetic acid : nitric acids : water, 8:1:2 v/v ), the remaining pellet was hydrolyzed in 72% sulfuric acid. The resulting glucose quantity was determined by the anthrone method42.
Phloroglucinol staining
Five day after imbibition seedlings to be stained with phloroglucinol were fixed in a 3:1 95% EtOH:glacial acetic acid solution for 5 minutes. Samples were then transferred to a solution of 1% phloroglucinol in 50% HCl for 1–2 minutes. Whole seedlings were then mounted in 50% glycerol on slides and viewed using an Olympus Vanox microscope. Images were captured with a PIXERA Pro-600ES camera.
Confocal laser scanning microscopy
Confocal laser scanning microscopy was carried out on a Zeiss LSM700. Cell walls were stained using propidium iodide as previously described30.
Transient protein-DNA interaction detection in tobacco
B-GLUCURONIDASE
For transient transactivation expression assays, the VND7, GAL4, and/or CyclinB1 promoters were cloned into pGWB3 to generate GUS (β-glucoronidase gene) fusion reporters for E2Fc transcriptional activity. The E2Fc effector vector43 (in PYL436) was kindly provided by Savithramma Dinesh Kumar (UC Davis, CA). The effector and reporter constructs were transformed into Agrobacterium tumefaciens strain GV3101 and co-infiltrated with the p19 silencing inhibitor into 3-weeks-old Nicotiana benthamiana leaves at OD600 0.6:0:6:1 respectively. Leaves were harvested 3 days after agro-infiltration and homogenized in GUS extraction buffer (50 mM Na2PO4 pH:7, 10 mM Na2-EDTA, 0.1% SDS, 0.1% Triton TX-100 and 10 mM β-mercaptoethanol). Quantitative MUG fluorescent assay for GUS determination was performed using 100 μg of protein/sample in 500 μL of GUS assay buffer (1 mM 4-Methyl umbelliferyl β-D-glucuronide –SIGMA- in Extraction Buffer). Samples were covered in aluminum foil and incubated at 37°C. Reaction was stopped at different time points by transferring 50 μL to a tube with 450 μL of Stop Buffer (0.2 M Na2CO3). 4-methylumbelliferone fluorescence was determined using a Infinite® 200 Pro-series reader (excitation at 365 nm, emission at 455 nm).
LUCIFERASE (Figure 2)
Over-night cultures of Agrobacterium (GV3101, OD=0.6) carrying VND7 promoter fused to luciferase (LUC) and 35S::E2Fc were prepared in infiltration medium (2 mM Na3PO4, 50 mM MES, 0.5% glucose, 100 μM acetosyringone) at OD600=0.1. Subsequently, cultures containing VND7::LUC and 35S::E2Fc at respective ratios of 1:0, 1:0.5, 1:1, 1:2, 1:5, or 1:10 were spot-infiltrated into 6–7 weeks old Nicotiana benthamiana leaves. To prevent gene silencing, Agrobacterium strain carrying the pBIN19 suppressor from tomato bushy stunt virus was included in each of the combinations44. The LUC activity was inspected at 72 to 96 hours post infiltration using CCD camera (Andor Technology).
Luciferase imaging of VND7::LUC was performed as previously described with modifications45. Briefly, tobacco leaves were cut off after 3-d of transient transformation and sprayed with 1 mM luciferin (Promega) in 0.01% Tween-80, then were imaged using an Andor DU434-BV CCD camera (Andor Technology). Images were acquired every 10 min for 12 pictures. Luciferase activity was quantified for a defined area as mean counts pixel−1 exposure time−1 using Andor Solis image analysis software (Andor Technology). Statistical analyses were performed using two-tailed Student’s t-tests. The difference was considered significant if p<0.05.
LUCIFERASE (Figure 4)
A vector system was created to generate a single vector with the CaMV 35S constitutive promoter (35S) fused to a transcription factor, a promoter fragment fused to the firefly luciferase reporter gene, and 35S fused to the Renilla luciferase reporter gene. The constitutively expressed Renilla gene served as a control to normalize for transformation efficiency. This system includes one destination vector pLAH-LARm and three entry vectors pLAH-TF, pLAH-PROM and pLAH-VP6435T using MultiSite Gateway Pro Technology (Invitrogen) to simultaneously clone three DNA fragments (Extended Data Fig. 8). To develop the expression vector, promoter fragments and transcription factors were BP cloned into pDONR-P3-P2 and pDONR-P1-P4 to create pLAH-TF and pLAH-Prom, respectively. PacI digested pMDC32 was ligated with the 2.427 kb pFLASH fragment following HindIII and SacI digestion to yield pLAR-L with the firefly luciferase (LUC) reporter gene. The 3 kb pRTL2-Renilla HindIII digested fragment was inserted into SacI digested pLAH-L to create pLAR-LR with both firefly LUC and Renllia luciferase (REN) genes. To generate pLAH-LAR, a SpeI digested PCR fragment containing the AmpR gene amplified from pDEST22 was ligated with SpeI digested pLAR-LR. To add the minimal CaMV 35S fragment (Mini35S) before the LUC reporter gene, the gateway cassette ccdB/CmR of pLAR-LAR was replaced by a HindIII digested PCR fragment Mini35S-ccdB-CmR amplified from pMDC32 using specific primer pHindIII-Rv and primer Mini35S-attR2. The final destination vector is referred to as pLAH-LARm.
The protein coding regions of select transcription factor genes were amplified. Each amplified fragment was recombined with pDONR-P1-P4 vector by performing BP reactions to produce pLAH-TF. Target promoter fragments were amplified from A. thaliana genomic DNA using appropriate primers with attB3 and attB2 sites (Supplementary Table 10). Each amplified fragment was cloned into pDONR-P3-P2 vector by performing BP reactions to produce pLAH-PROM. A third pDONR vector (pLAH-VP64Ter) was designed to create a C-terminal fusion of the strong transcription activation domain VP64 to the transcription factor followed by the 35S transcription terminator (35St). A PCR fragments containing VP64 region and 35S terminator was amplified from pB7-VP64 using specific primers with attB4r and attB3r sites (Supplementary Table 10) into pDONR P4r-P3r to produce pLAH-VP6435T. Finally, the fully functional expression vector was generated by Gateway LR cloning of destination vector and the three entry clones: pLAH-LARm, pLAH-TF, and pLAH-VP64Ter (Extended Data Fig. 7).
Agrobacterium tumefaciens strain GV3103 (MP90) carrying expression constructs were grown in Luria-Bertani media with rifampiycin and ampicillin and suspended in infiltration buffer 10 mM MES, pH5.7, containing 10 mM MgCl2 and 150 μM acetosyringone. The cultures were adjusted to an OD600 of 0.8 and incubated at room temperature for at least 3 h prior to infiltration. The cultures were hand infiltrated using a 1 mL syringe into 3- to 4-week-old N. benthamiana leaves. Leaf samples were harvested 36 h after infiltration and assayed for luciferase activity according to manufacturer instructions using the Dual-Luciferase Reporter Assay Systems (Promega). Approximately 100 mg of tissue was frozen in liquid N and homogenized using a Retsch Mixer Mill MM400 for 1 min at 30 Hz. Ground tissue was then thawed in lysis buffer (0.1 M HEPES, pH7.8, 1% Triton X-100, 1 mM CaCl2 and 1 mM MgCl2) at 25°C for 15 min. Then 50 μL of Luciferase Assay Reagent II was added to 10 uL aliquots of the lysates to measure firefly luciferase activity, 1000 ms intergration time, using a Spectra Max M5/M5e plate reader to measure total light emission. Firefly luciferase activity was quenched with 50 μL of Stop & Glo Reagent, which contains Renilla luciferin substrate, also measured, 100 ms integration time, as total light emission. An expression vector containing part of the coding sequence (+X/+Y) of the β-glucuronidase reporter gene rather than a transcription factor gene was used for baseline measurement of firefly luciferase activity. To estimate relative transcription factor affinity with each promoter fragment, three biological replicates of transcription factor expressing vectors were compared to the average results for the GUS expression vector. First, dividing firefly luciferase activity by Renilla luciferase activity normalized the transformation efficiency of each infiltrated leaf sample. Relative binding of the transcription factors to the promoter bait sequences was determined relative to the GUS control using a Student’s t-test in R v2.11.0.
Electrophoretic mobility shift assays
To express recombinant NST2 or SND1 protein, coding sequence was cloned and fused to glutathione S-transferase tag in the pDONR211 vector and then transferred into pDEST15 (Invitrogen). E. coli strain BL21-AI (Invitrogen) transformed with pDEST15-GST:NST2 were grown in liquid media to an OD600 of 0.4, treated with 0.2% L-arabinose to induce expression overnight and harvested by centrifugation the following day. Cells were treated with 1mg/mL lysozyme on ice for 30 min in minimal volume of 1X PBS buffer and lysed by sonication. Cell lysates were clarified by centrifugation and incubated with 100 μL of glutathione sepharose beads (GE Healthcare) for 30 min at 4 °C with rotation. The beads were transferred to a column, washed with 10 volumes of 1X PBS. Protein was eluted in 100 mM Tris-HCl pH8.0, 100 mM NaCl and 3 mg/mL glutathione buffer and purified protein was resuspended in 50% glycerol and stored at −80 °C.
Three overlapping probes were generated for CESA7, CESA8 and KOR promoters using the same oligonucleatides described in Hazen Table S1, whereas three probes were generated for CESA4 using the following primers: CESA4pr-1fwd, CACCGGGCCTTTGTGAAATTGATTTTGGGC; CESA4pr-1rev, TGTATTTCTACTTTAGTCTTAC; CESA4pr-2fwd, CCAGATTTGGTAAAGTTTATAAG; CESA4pr-2rev, GTGTCATAAGAAAGCTTCAAG; CESA4pr-3fwd, TCTTATGACACAAACCTTAGAC; CESA4pr-3rev, ACACTGAGCTCTCGGAAGCAGAGCAG. Reactions were carried out in binding buffer (10 mM Tris, pH7.5, 50 mM KCl, 1mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.1% IGEPAL CA-630, and 0.05 ug/ul calf thymus DNA). Following the addition of 150 ng of protein from the GST purification eluate, reactions were incubated at room temperature for 30 min. Protein-DNA complexes were separated from the free DNA on 1% agarose/1X TAE gels at 4 °C. The agarose gels were stained with ethidium bromide and bands visualized under UV light. For the titration of promoter DNA with NST2 protein, CESA4 promoter fragment-2 DNA and KOR promoter fragment-1 DNA in 30 ng were titrated with increasing amounts of NST2 protein: 25, 50, 150, 300, and 600 ng. Binding reaction and the separation of protein-DNA complexes were carried out as described above.
Chromatin immunoprecipitation of NST2
Chromatin immunoprecipitation was conducted as described by Nusinow et al.46 with the following modifications. Roughly 5 g (fresh weight) whole stems from six-week-old Arabidopsis were harvested and crosslinked for 15 min under vacuum in crosslinking buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 250 mM sucrose, 1 mM PMSF and 1% formaldehyde). Technical replicates containing approximately 1.5 mg DNA were resuspended in 800 μl SII buffer, incubated with 2 μg anti-GFP antibody (ab290, Abcam) bound to Protein G Dynabeads (Invitrogen) for 1.5 h at 4 °C and then washed five times with SII buffer. Chromatin was eluted from the beads twice at 65 °C with Stop buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 20 mM EDTA and 1% SDS). RNase- and DNase-free glycogen (2 μg) (Boehringer Mannheim) was added to the input and eluted chromatin before they were incubated with DNase- and RNase-free proteinase K (Invitrogen) at 65 °C overnight and then treated with 2 μg RNase A (Qiagen) for 1 h at 37 °C. DNA was purified by using Qiagen PCR Purification kit and resuspended in 100 μl H20. Quantitative PCR reactions of the technical replicates were performed using Quantifast SYBR Green PCR Kit (Qiagen), with the following PCR conditions: 2 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 15s at 55 °C and 20 s at 68 °C. Primers used in this study are listed in Hazen Table S4. Results were normalized to the input DNA, using the following equation:100 × 2(Ct input-3.32--Ct ChIP).
Quantitative RT-PCR
Primers for QRT-PCR were designed to amplify a 100 bp region (or a 400 bp region for REV, PHB, and PHV transcripts due to sequence similarity) on the 3′ end of each transcript33. Primer sets used for QRT-PCR are listed in Supplementary Table 1. Each plate was considered a biological replicate and Columbian and reference genotypes were plated on the same plate. Five days after imbibition, total RNA was extracted from seedling roots using an RNeasy Kit (QIAGEN). cDNA was synthesized by treatment with reverse transcriptase and oligo(dT) primer (SuperScript III First-Strand Synthesis System; Invitrogen). QRT-PCR was performed in an iCycler iQ Real-Time PCR Detection System (Bio-rad) using the Bio-rad iQ SYBR green Supermix. Gene expression was measured between wild-type and mutant pairs across at least two biological replicates with three technical replicates using the Δ-ΔCT method30.
VND7 induction experiments
VND7-VP16-GR9 plants were grown vertically on sterile mesh placed on top of MS media with sucrose. Five days after imbibition, seedlings were transferred, with the mesh, to MS media containing 10μM dexamethasone and roots were collected for QRT-PCR (RNeasy Kit; Qiagen) after 0, 1, 2, 3, or 4 hours on dexamethasone (n=3). As a positive control, upregulation of MYB46 expression was confirmed using QRT-PCR.
Nitrogen influx, salt stress, iron deprivation, sulfur stress, pH stress analysis
The datasets used contained mean expression values for each gene in both control and treatment, and a q-value for each gene indicating the significance of the hypothesis that the expression values of control and treatment are drawn from distributions with the same means. These data sets were filtered to extract only those genes whose q-value was ≤0.01 and whose fold change between mean expression values was ≥1.5 in either direction. Fisher’s exact test was used to test whether the number of such genes is overrepresented in the xylem cell specification and differentiation gene regulatory network.
Gene regulatory network inference
Expression data30 were used, after normalization with the mmgMOS method used in the PUMA R package47. The supervised regulatory interactions network was constructed using SIRENE48. The directionality of the interactions is defined by the protein-DNA interactions from Y1H data. The interaction sign is derived by Pearson’s correlation coefficient for each protein-DNA interaction. The analysis performed was categorized as a) Supervised Tier Ia network inferred with SIRENE with the provided Y1H gene regulatory connections and the corresponding gene expression profiles (16 genes - 4 TFs), b) Supervised Tier Ib: an additional three verified connections from the supervised Tier Ia and unsupervised Tier I were considered in the inference. The unsupervised regulatory interaction network was constructed using the consensus from four different gene regulatory network inference methods, GENIE349, Inferelator50, TIGRESS51 and ANOVerence52. The data used were the same as the supervised TIERIa network. The default parameters were used in all methods and a rank-based method was used to build the consensus network as in Marbach et al.53.
Extended Data
Extended Data Figure 1. Number of novel and previously described protein-DNA interactions and transcription factors involved in secondary cell wall biosynthesis and xylem development.
Venn diagrams of overlap between previously reported19 (A) interactions or (B) transcription factors and those of the xylem-specific gene regulatory network. *=includes genes that were not included in yeast one hybrid screen.
Extended Data Figure 2. Activation or repression of VND7 by E2Fc is dynamic and dose-dependent.
(A) Intensity of LUC bioluminescence quantified using Andor Solis image analysis software. Data are means ± s.d. (n=20). Asterisks denote significance at p<0.05 determined by Student’s t-test. (B) Quantitative real-time PCR of E2Fc and VND7 transcripts in ΔN-E2Fc (E2Fc overexpressor line lacking the N-terminal domain) expressing plants versus Col-0 control. Red dashed line marks the point at which VND7 is unchanged compared to control. Each data point is an individual biological replicate with 3 technical replicates. (C) 3-week old tobacco leaves were infiltrated with the p19 silencing inhibitor and either the reporter VND7::GUS or VND7::GUS and either 35S::E2Fc:MYC or 35S::RBR:GFP, or both. Extracted protein was then used in a quantitative MUG fluorescent assay, where relative fluorescence was measured 60 min after incubation with substrate. Data are means ± s.d., n=3.
Extended Data Figure 3. Binding of NST2 and SND1 to fragments of CESA7, CESA8, and KOR promoters.
Electrophoretic mobility shift assays showing NST2 (A–D) and SND1 (E–F) protein specifically binds the promoters of cellulose-associated genes. Probe was incubated in the absence or presence of GST or GST:SND1 protein extracts. The arrowheads indicate the specific protein-DNA complexes, while arrows indicate free probe.
Extended Data Figure 4. Sub-networks of network genes differentially expressed in response to iron deprivation of high salinity.
Sub-network of genes with q-values of ≤0.01 and whose fold change between mean expression values was ≥1.5 in either direction in iron deprivation (A) or high NaCl (B) stress microarray dataset. Nodes are colored according in in-degree as shown on scale bars below sub-networks. Transcription factors with the highest in-degree are labeled and indicated with a black circle.
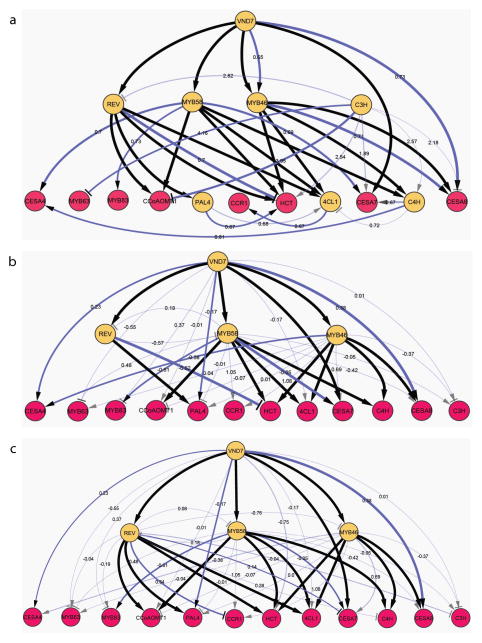
Extended Data Figure 5. The reconstructed gene regulatory consensus network based on analysis of the iron-deprivation expression dataset by different network inference methods.
(A) Unsupervised, (B) supervised in the first pass, (C) Supervised after the validated two connections have been added in the training set. Edge transparency denote p ≤ 0.06 for the Pearson Correlation Coefficient (PCC); edge width is proportional to PCC; edge value correspond to the total edge score; a greater value corresponds to more significant score. Yellow and red nodes correspond to transcription factor and target gene nodes, respectively; black and blue edges denote Y1H-derived and inferred interactions, respectively.
Extended Data Figure 6. Iron deprivation and NaCl stress influences lignin and phenylpropanoid biosynthesis associated gene expression.
(A) No change was observed in the expression of 4CL1::GFP in 4 DAI roots transferred to a control media (left, n=4) or media with 140 mM NaCl for 48 hours (right, n=4). (B) Increased fuchsin staining of xylem cells as well as of cell walls of non-vascular cells in 4 DAI roots transferred to a control media (left) or media with an iron chelator for 72 h (right). (C) No change was observed in the expression of VND7::YFP in 4 DAI roots transferred to a control media (left, n=4) or media with an iron chelator for 72 h (right, n=5).
Extended Data Figure 7. Schematic diagram of dual-luciferase reporter vector development.
(A) Three distinct donor vectors harboring either the transcription factor, VP64 activation domain fused to the 35S minimal promoter, or a promoter fragment. (B) The dual reporter vector, pLAH-LARm, is then recombined with the three donor vectors to generate the (C) single reporter vector.
Supplementary Material
Acknowledgments
We thank M. Tierney (University of Vermont) for 35S::GFP seeds, T. Demura for VND7 resources, M.K. Barton for REV:GR seeds, E.P. Spalding for advice on manuscript revision, and C. Gutierrez for E2Fc RNAi and E2Fc N-terminal deletion overexpressor seeds and useful discussion. This research was supported by the Office of Science (BER) Department of Energy Grant DE-FG02-08ER64700DE (to S.P.H. and S.A.K.), National Institute of General Medical Sciences of the National Institutes of Health under award numbers RO1GM056006 and RC2GM092412 (to S.A.K), National Institute of Health (R01GM107311) and National Science Foundation (IOS-1036491 and IOS-1352478) to K.D., USDA CRIS 1907-21000-030 to D.W. and L.F. and UC Davis Startup Funds and a Hellman Fellowship (to S.M.B).
Footnotes
The authors declare no competing financial interests.
Author Contributions. M.T.-T., L.L. and M.dL. contributed equally to this work, T.T.W. and A.G. contributed equally to this work. S.M.B. and S.P.H. contributed equally to this work. M.T.-T, L.L., M.dL., S.M.B, S.P.H. designed the research. M.T.-T., L.L., M.dL., A.G., G.X., N.F.Y., G.M.T., M.T.V., R.L., P.P.H, C.W., and K.D. performed the research. M.T.-T., L.L., G.T., T.W.T., N.T., J.C., M.P., D.K., I.T., S.A., S.M.B. and S.P.H. analyzed the data. L.Z., D.W., G.B., J.L.P.-P., and S.A.K. contributed new reagents/analytic tools. M.T.-T., L.L., G.M.T., S.M.B. and S.P.H. wrote the article. All authors discussed the results and commented on the manuscript.
References
- 1.Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 6.Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudinier A, et al. Enhanced Y1H assays for Arabidopsis. Nat Meth. 2011;8:1053–1055. doi: 10.1038/nmeth.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim WC, Ko JH, Han KH. Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol Biol. 2012;78:489–501. doi: 10.1007/s11103-012-9880-7. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi M, et al. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front Plant Sci. 2013;4:325. doi: 10.3389/fpls.2013.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walhout A. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 2011;12:109. doi: 10.1186/gb-2011-12-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim WC, Kim JY, Ko JH, Kang H, Han KH. Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol Biol. 2014;85:589–599. doi: 10.1007/s11103-014-0205-x. [DOI] [PubMed] [Google Scholar]
- 14.Ahnert SE. Power graph compression reveals dominant relationships in genetic transcription networks. Mol BioSyst. 2013;9:2681–2685. doi: 10.1039/c3mb70236g. [DOI] [PubMed] [Google Scholar]
- 15.Kim WC, et al. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 2013;73:26–36. doi: 10.1111/j.1365-313x.2012.05124.x. [DOI] [PubMed] [Google Scholar]
- 16.del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the Ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell. 2006;18:2224–2235. doi: 10.1105/tpc.105.039651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Pozo JC, Boniotti MB, Gutierrez C. Arabidopsis E2Fc functions in cell division and Is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell. 2002;14:3057–3071. doi: 10.1105/tpc.006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jager SM, Menges M, Bauer UM, Murray JAH. Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol Biol. 2001;47:555–568. doi: 10.1023/a:1011848528377. [DOI] [PubMed] [Google Scholar]
- 19.Mariconti L, et al. The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem. 2002;277:9911–9919. doi: 10.1074/jbc.M110616200. [DOI] [PubMed] [Google Scholar]
- 20.Kosugi S, Ohashi Y. Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 2002;128:833–843. doi: 10.1104/pp.010642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jager S, et al. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol. 2009;71:345–365. doi: 10.1007/s11103-009-9527-5. [DOI] [PubMed] [Google Scholar]
- 22.Heckmann S, et al. The E2F transcription factor family regulates CENH3 expression in Arabidopsis thaliana. Plant J. 2011;68:646–656. doi: 10.1111/j.1365-313X.2011.04715.x. [DOI] [PubMed] [Google Scholar]
- 23.del Pozo Jc, Diaz-Trivino S, Cisneros N, Gutierrez C. The E2FC-DPB transcription factor controls cell division, endoreplication and lateral root formation in a SCFSKP2A- dependent manner. Plant Signal Behav. 2007;2:273–274. doi: 10.4161/psb.2.4.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi M, et al. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell. 2010;22:1249–1263. doi: 10.1105/tpc.108.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi M, et al. VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010:906–914. doi: 10.1104/pp.110.154013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wenkel S, Emery J, Hou BH, Evans MMS, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell. 2007;19:3379–3390. doi: 10.1105/tpc.107.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong R, Ye ZH. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012;53:368–380. doi: 10.1093/pcp/pcr185. [DOI] [PubMed] [Google Scholar]
- 28.Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010;22:3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyo H, Demura T, Fukuda H. TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J. 2007;51:955–965. doi: 10.1111/j.1365-313X.2007.03180.x. [DOI] [PubMed] [Google Scholar]
- 30.Dinneny JR, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 31.Iyer-Pascuzzi AS, et al. Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell. 2011;21:770–782. doi: 10.1016/j.devcel.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Nat Acad Sci. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady SM, et al. A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol. 2011;7 doi: 10.1038/msb.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong W, et al. Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 2004;135:773–782. doi: 10.1104/pp.104.042176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paz-Ares J The REGIA Consortium. REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genom. 2002;3:102–108. doi: 10.1002/cfg.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Underwood BA, Vanderhaeghen R, Whitford R, Town CD, Hilson P. Simultaneous high-throughput recombinational cloning of open reading frames in closed and open configurations. Plant Biotech J. 2006;4:317–324. doi: 10.1111/j.1467-7652.2006.00183.x. [DOI] [PubMed] [Google Scholar]
- 37.Yamada K, Lim J, Dale J, Chen H. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 38.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawker NP, Bowman JL. Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 2004;135:2261–2270. doi: 10.1104/pp.104.040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Updegraff D. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 42.Scott TA, Melvin EH. Determination of dextran with anthrone. Anal Chem. 1953;25:1656–1661. [Google Scholar]
- 43.Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem. 2004;279:2101–2108. doi: 10.1074/jbc.M310029200. [DOI] [PubMed] [Google Scholar]
- 44.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 45.Walley J, et al. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007;3:1800–1812. doi: 10.1371/journal.pgen.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearson R, et al. puma: a Bioconductor package for propagating uncertainty in microarray analysis. BMC Bioinformatics. 2009;10:211. doi: 10.1186/1471-2105-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mordelet F, Vert JP. SIRENE: supervised inference of regulatory networks. Bioinformatics. 2008;24:i76–i82. doi: 10.1093/bioinformatics/btn273. [DOI] [PubMed] [Google Scholar]
- 49.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE. 2010;5:e12776. doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenfield A, Madar A, Ostrer H, Bonneau R. DREAM4: Combining genetic and dynamic information to identify biological networks and dynamical models. PLoS ONE. 2010;5:e13397. doi: 10.1371/journal.pone.0013397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haury AC, Mordelet F, Vera-Licona P, Vert JP. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst Biol. 2012;6:145. doi: 10.1186/1752-0509-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Küffner R, Petri T, Tavakkolkhah P, Windhager L, Zimmer R. Inferring gene regulatory networks by ANOVA. Bioinformatics. 2012;28:1376–1382. doi: 10.1093/bioinformatics/bts143. [DOI] [PubMed] [Google Scholar]
- 53.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nat Meth. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.