Summary
A-type lamins are components of the nuclear lamina, a filamentous network of the nuclear envelope in metazoans that supports nuclear architecture. In addition, lamin A/C can also be found in the nuclear interior. This nucleoplasmic lamin pool is soluble in physiological buffer, depends on the presence of the lamin-binding protein, Lamina-associated polypeptide 2α (LAP2α) and regulates cell cycle progression in tissue progenitor cells. ΔK32 mutations in A-type lamins cause severe congenital muscle disease in humans and a muscle maturation defect in LmnaΔK32/ΔK32 knock-in mice. At molecular level, mutant ΔK32 lamin A/C protein levels were reduced and all mutant lamin A/C was soluble and mislocalized to the nucleoplasm. To test the role of LAP2α in nucleoplasmic ΔK32 lamin A/C regulation and functions, we deleted LAP2α in LmnaΔK32/ΔK32 knock-in mice. In double mutant mice the LmnaΔK32/ΔK32- linked muscle defect was unaffected. LAP2α interacted with mutant lamin A/C, but unlike wild-type lamin A/C, the intranuclear localization of ΔK32 lamin A/C was not affected by loss of LAP2α. In contrast, loss of LAP2α in LmnaΔK32/ΔK32 mice impaired the regulation of tissue progenitor cells like in lamin A/C wild type animals. These data indicate that a LAP2α-independent assembly defect of ΔK32 lamin A/C is predominant for the mouse pathology, while the LAP2α-linked functions of nucleoplasmic lamin A/C in the regulation of tissue progenitor cells are not affected in LmnaΔK32/ΔK32 mice.
Keywords: congenital muscular dystrophy, nuclear envelope, lamin A/C, lamina associated polypeptide 2α, nucleoplasmic lamins
Introduction
Lamins are intermediate filament proteins in metazoan cells that form the lamina, a scaffold structure tightly associated with the inner nuclear membrane (Dechat et al., 2008). They provide mechanical stability for the nuclear envelope and the nucleus and help to organize higher order chromatin structure. Lamins are grouped into A- and B-type, based on biochemical, structural and dynamic properties, sequence homologies and expression patterns. While B-type lamins are ubiquitously expressed throughout development, the major A-type lamins (lamin A and C) encoded by LMNA are expressed at later stages during development (Rober et al., 1989; Stewart and Burke, 1987). Lamin A and B-type lamins undergo posttranslational processing at their C-terminal CAAX motif, including farnesylation and carboxy-methylation (Rusinol and Sinensky, 2006). The hydrophobic farnesyl-group at the C-terminal cysteine facilitates tight interaction with the inner nuclear membrane. While mature B-type lamins remain farnesylated, lamin A undergoes an additional endoproteolytic processing step that removes 15 from the C-terminus including the farnesyl-group (Pendas et al., 2002). Thus mature lamin A and also lamin C, which is not processed post-translationally, lack a farnesyl group and are less tightly bound to membranes than B-type lamins. Consequently, a fraction of A-type lamins is also found in the nucleoplasma (Dechat et al., 2010).
The physiological relevance and functions of the nucleoplasmic lamin A/C pool are poorly understood, but they are likely involved in many of the reported functions of lamins in cell signaling and gene expression (Andres and Gonzalez, 2009; Dechat et al., 2010; Heessen and Fornerod, 2007; Prokocimer et al., 2009). Our recent studies showed that Lamina-associated polypeptide 2α (LAP2α) regulates the localization and functions of nucleoplasmic A-type lamins (Naetar et al., 2008). LAP2α is a unique member of the LAP2 protein family (Wilson and Foisner, 2010). While most LAP2 proteins are integral membrane proteins of the inner nuclear membrane and associate with lamins in the peripheral lamina, LAP2α localizes to the nuclear interior and interacts with nucleoplasmic lamins A and C (Dechat et al., 2000). Deletion of LAP2α in mice causes loss of lamins A and C in the nucleoplasm of dermal fibroblasts and proliferating tissue progenitor cells, while lamins at the peripheral lamina are unaffected (Naetar et al., 2008). Similarly, human fibroblasts lose nucleoplasmic lamins following RNA-interference-mediated knock-down of LAP2α (Pekovic et al., 2007). Moreover, during myoblast differentiation, LAP2α expression is downregulated and nucleoplasmic lamins are lost (Markiewicz et al., 2005). Nucleoplasmic lamins and LAP2α were shown to bind directly to the tumor suppressor retinoblastoma protein (pRb) in its active, hypo-phosphorylated form (Markiewicz et al., 2002) and to promote pRb repressor activity on pRb/E2F target gene promoters, mediating efficient cell cycle exit of proliferating cells (Dorner et al., 2006). Accordingly, LAP2α deletion in mice accompanied by loss of nucleoplasmic lamins results in hyperproliferation of tissue progenitor cells and tissue hyperplasia (Naetar et al., 2008).
Mutations in the LMNA gene and in several genes encoding lamin-associated proteins have been linked to phenotypically heterogenous diseases generally termed laminopathies. The disease variants range from muscular dystrophies over cardiomyopathies to lipodystrophies and systemic involvements of multiple tissues like the premature ageing disease Hutchinson-Gilford progeria syndrome (HGPS) (Worman and Bonne, 2007). The molecular disease mechanisms underlying the laminopathies are still poorly understood. While one disease model proposes defects in mechanical properties of the lamina in laminopathic cells, leading to increased fragility of nuclei, other models have proposed impaired functions of mutated lamins in chromatin regulation and gene expression (Gotzmann and Foisner, 2006).
In a recent study we described a novel mouse model for a severe, striated muscle–affecting laminopathy (Bertrand et al., 2012): LmnaΔK32/ΔK32 knock-in mice harbor a Lmna mutation that results in the loss of lysine 32 in the N-terminal domain of lamins A and C, and causes a severe form of Congenital Muscular Dystrophy (CMD) in humans (Quijano-Roy et al., 2008). Homozygous LmnaΔK32/ΔK32 mice were indistinguishable from their wild-type littermates at birth but soon exhibited striated muscle maturation delay and metabolic defects and died within 2-3 weeks (Bertrand et al., 2012). Interestingly, the ΔK32 mutation was previously proposed to impair the lateral assembly of lamin A/C head to tail polymers (Bank et al., 2011). In line with this observation, mutant lamins failed to assemble at the lamina and localized predominantly in the nucleoplasm in LmnaΔK32/ΔK32 mice. Taking into account our previous results on the important role of nucleoplasmic lamins in tissue progenitor cells (Dechat et al., 2010), we hypothesized that a deregulated nucleoplasmic lamin pool in LmnaΔK32/ΔK32 mice may contribute to the pathologies. Therefore, we set out to test whether LAP2α, a major regulator of nucleoplasmic lamins in Lmna+/+ mice (Naetar et al., 2008), is also involved in regulating nucleoplasmic ΔK32 lamin A/C, and if and how loss of LAP2α in LmnaΔK32/ΔK32 mice may affect mutant lamin A/C localization and function. Using LAP2α−/− LmnaΔK32/ΔK32 double mutant mice, we show in this manuscript that loss of LAP2α did not change the localization of mutant lamins, as shown for wild-type lamins. LAP2α and ΔK32 lamin A/C, however, formed “functional” complexes in the nucleoplasm, as loss of LAP2α in LmnaΔK32/ΔK32 mice caused hyperproliferation of epidermal progenitor cells and hyperplasia of epidermis, like in Lmna+/+ mice. These data indicate that a LAP2α-unrelated assembly defect of ΔK32 lamin may be the predominant molecular defect in LmnaΔK32/ΔK32 mice, while its LAP2α-dependent functions in the nucleoplasm are unaffected.
Results
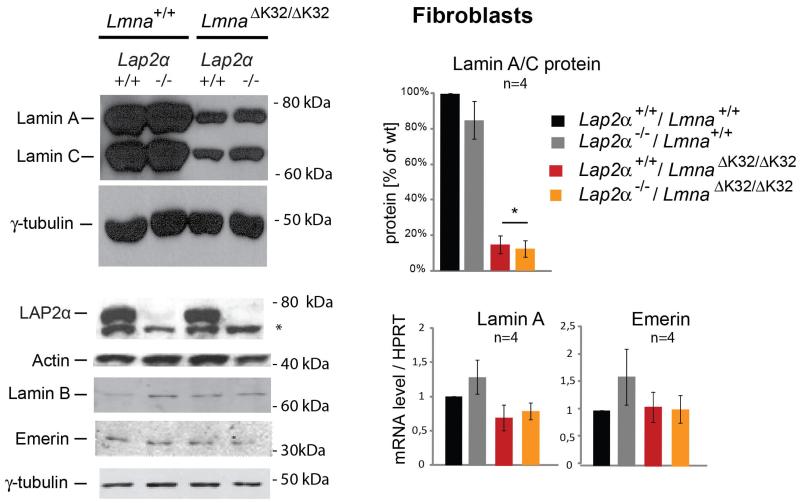
Loss of LAP2α does not affect protein levels and localization of ΔK32 lamin A/C
Mutant ΔK32 lamin A/C in LmnaΔK32/ΔK32 knock-in mice fails to assemble at the nuclear lamina and mislocalizes to the nucleoplasm (Bertrand et al., 2012). Since LAP2α has previously been found to regulate the nucleoplasmic pool of wild-type lamins A and C (Naetar et al., 2008), we wanted to examine the influence of LAP2α loss on ΔK32 lamin A/C expression and cellular distribution. We generated double mutant mice by crossing Lmna+/ΔK32 mice with heterozygous Lap2α-deficient mice and isolated fibroblasts and myoblasts from newborn littermates. Lamin A/C protein levels were massively reduced to 60% to 90% of wild-type lamin A/C levels in all LmnaΔK32/ΔK32 cells independent of the presence or absence of LAP2α (Fig. 1, Supplementary Fig. S1A). LAP2α and other nuclear envelope and/or lamina proteins, such as lamin B1 and emerin, were not affected in LmnaΔK32/ΔK32 cells. Similar results were obtained in lysates of liver and diaphragm derived from mice of the four genotypes (Supplementary Fig. S1B). Thus, LAP2α loss did not affect the expression level of mutant ΔK32 lamin A/C protein. Down-regulation of mutant lamin A/C likely occurrs post-transcriptionally, since lamin A mRNA levels in fibroblasts were similar in all genotypes (Fig. 1) in accordance with previous reports (Bertrand et al., 2012).
Fig. 1. Lamin A/C expression is significantly reduced in ΔK32 Lamin A/C expressing cells independent of the presence and absence of LAP2α.
Immunoblots of lysates of primary fibroblast derived from single and double mutant LmnaΔK32/ΔK32, and Lap2α−/− mice and wild-type control littermates probed for indicated proteins are shown. The star indicates an unspecific band produced by the LAP2α antibody. For quantification of lamin protein levels, band intensities of lamins A and C were combined and normalized to the band intensity of the actin loading control and presented as % of the wild-type control. Protein levels of single mutant LmnaΔK32/ΔK32 and double mutant LmnaΔK32/ΔK32 / Lap2α−/− fibroblasts were significantly reduced (n=4, P-values=0.04 and 0.03, respectively as determined by Student’s t-Test against wild-type). Lower right panels show mRNA levels of lamin A and emerin as determined by real-time PCR. mRNA levels were normalized to the corresponding wild-type levels. Means and s.e. of 3-4 independent experiments are shown. mRNA levels of lamin A or emerin are not altered in single and double mutant fibroblasts compared to wild type as determined by Student’s t-test: LmnaΔK32/ΔK32: n=4, P-value 0.2 (lamin A) and 0.8 (emerin); Lap2α−/−: n=4, P-value 0.4 (lamin) and 0.3 (emerin); LmnaΔK32/ΔK32 / Lap2α−/−: n=4, P-value 0.2 (lamin) and 0.9 (emerin).
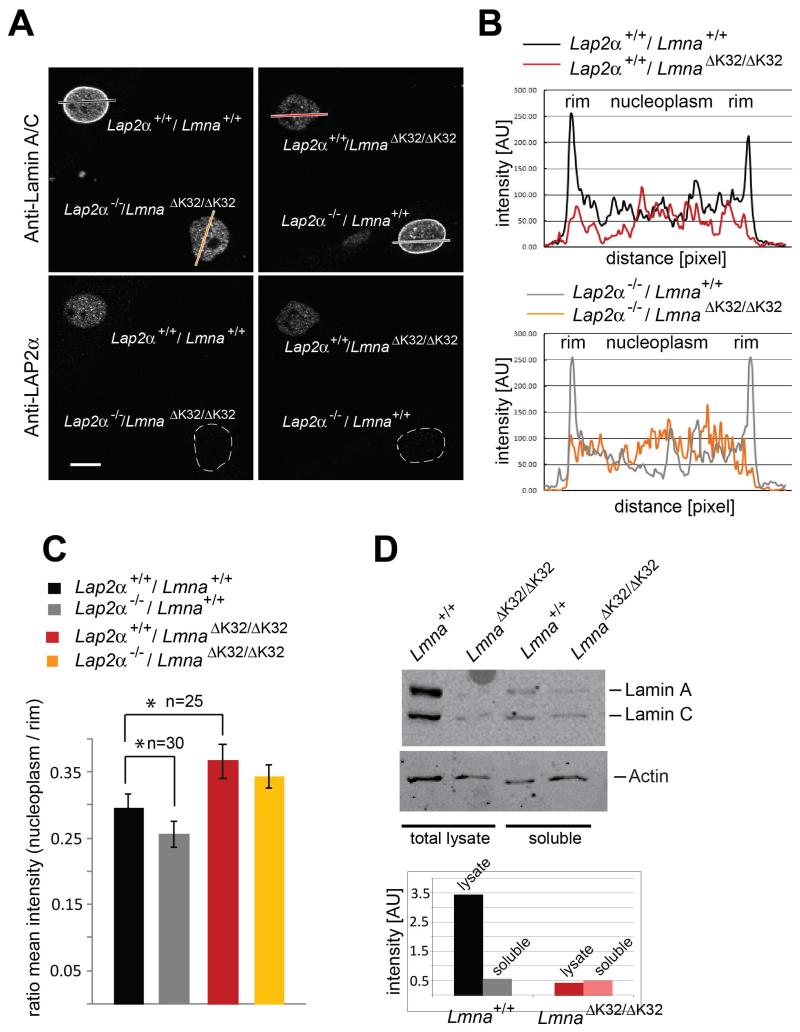
Immunofluorescence analyses of primary mouse fibroblasts confirmed the reduction of lamin A/C protein levels in single and double mutant LmnaΔK32/ΔK32 mice. Co-cultures of LAP2α-expressing and LAP2α-deficient Lmna+/+ and LmnaΔK32/ΔK32 cells allowed identifying LmnaΔK32/ΔK32 versus Lmna+/+cells in the co-culture (by the lack of LAP2α staining) and made it possible to compare lamin A/C levels and localization in the different genotypes under identical experimental conditions (Fig. 2A). While wild-type lamin A/C was predominantly found at the nuclear periphery with an additional weaker nucleoplasmic staining, the lowly expressed ΔK32 lamin A/C mutant was equally distributed throughout the nucleoplasm without any clear nuclear rim staining. Quantitative analyses of lamin A/C localization by plotting lamin A/C staining intensity profiles across the nuclear diameter in confocal immunofluorescence images confirmed the uniform distribution of the ΔK32 lamin A/C staining (Fig. 2B). LAP2α localization was unaffected in ΔK32 lamin A-expressing cells.
Fig. 2. Lamin A/C protein is redistributed to the nuclear interior in LmnaΔK32/ΔK32 fibroblasts in a LAP2α-independent manner.
(A) Co-cultures of primary dermal fibroblasts with indicated genotypes isolated from new-born littermates were processed for confocal immunofluorescence microscopy. Cells were stained with antibodies to lamin A/C and LAP2α, with the latter allowing identification of the genotype in mixed cultures. Scale bar denotes 10 μm. (B) Quantitation of intranuclear lamin staining was done by fluorescence intensity measurements along the dashed line shown in image using the profile tool in Zeiss LSM Image Browser. (C) Ratios of nucleoplasmic over peripheral mean A-type lamin fluorescence intensity were plotted in the histogram. In LmnaΔK32/ΔK32 fibroblasts, the ratios are significantly increased versus wild-type controls (n=25, P-value<0.05). In the Lmna+/+ background, nucleoplasmic lamins are lost and the ratio decreases significantly upon loss of LAP2α (n=30, P-value<0.05). (D) Immunoblots of total cell lysates and of soluble cell fractions following lysis of cells in Hepes buffer plus 0.5% Triton X-100 and 0.1% SDS, probed for lamin A/C and actin. Protein bands were quantified by ImageJ.
While loss of LAP2α in Lmna+/+ cells decreased lamin A/C levels in the nuclear interior (Naetar et al., 2008), the exclusive nucleoplasmic localization of ΔK32 lamin A/C mutant was unaffected by the absence of LAP2α as revealed in the staining intensity profiles (Fig. 2B). In addition we calculated the ratios of nucleoplasmic over peripheral mean lamin A/C fluorescence intensities for 25 to 30 fibroblasts for each genotype (Fig. 2C). In LmnaΔK32/ΔK32 fibroblasts, the ratios were significantly increased compared to Lmna+/+ cells (n=25, P-value< 0.05), reflecting the lack of accumulation of mutant lamin A/C at the periphery and its even distribution throughout the nucleus. Loss of LAP2α had no effect on the distribution of mutant lamin A/C (LmnaΔK32/ΔK32 background), while in the lamin A/C wild-type background loss of LAP2α caused a significant reduction of nucleoplasmic lamins (n=30, P-value< 0.05).
Our data suggest that ΔK32 lamin A/C has a LAP2α-independent assembly defect, preventing it from assembly at the nuclear lamina, which is in line with previous reports in C. elegans (Bank et al., 2011). To test this hypothesis, we performed cell lysis in physiological buffer plus 0-5% Triton X-100 / 0.1% SDS and determined the soluble, unassembled pool of lamin A/C by immunoblotting and densitometric analyses. While only around 10% of total wild-type lamin A/C were soluble, all of ΔK32 lamin A/C was solubilized under these conditions (Fig. 2D).
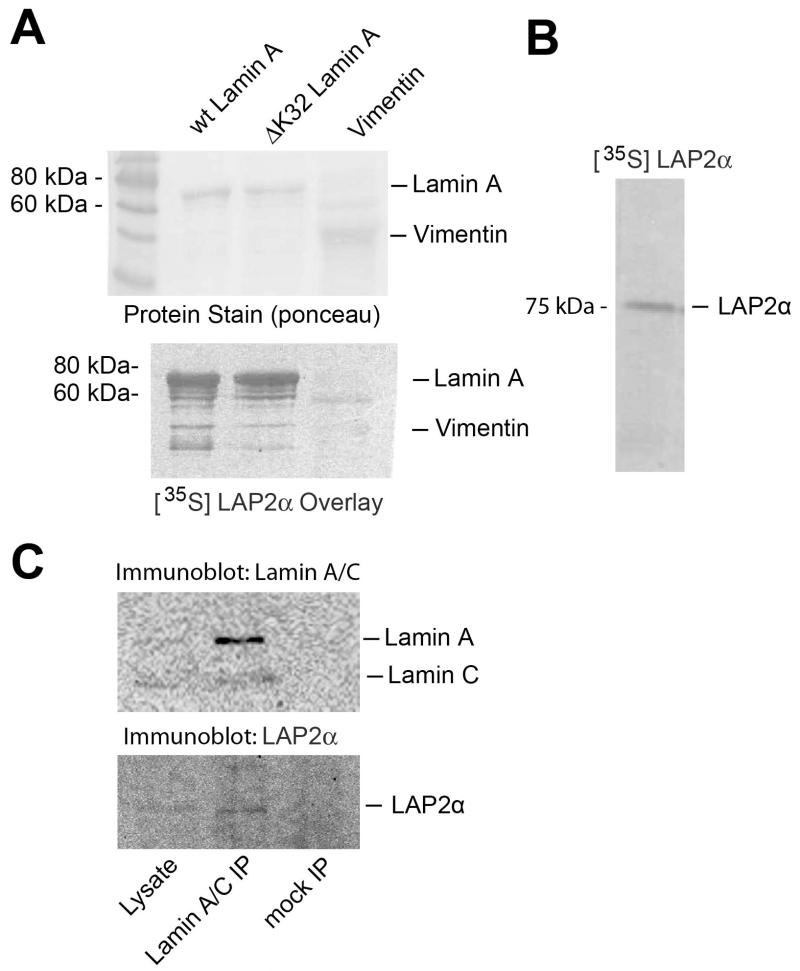
ΔK32 lamin A interacts with LAP2α like wild-type lamin A
Next we tested whether the mutated ΔK32 lamin A is able to directly interact with LAP2α. Bacterially expressed ΔK32 and wild type pre-lamin A were transferred to nitrocellulose and probed with in vitro translated, [35S]-labeled LAP2α. Autoradiography of the blot revealed binding of LAP2α to both wild-type and ΔK32 lamin A, while binding to a related, cytoplasmic intermediate filament protein, vimentin was not detectable (Fig. 3). To test whether ΔK32 lamin A - LAP2α complexes also exist in vivo we lysed ΔK32 lamin A-expressing cells in physiological buffer containing 0-5% Triton X-100 / 0.1% SDS, and immunoprecipitated ΔK32 lamin A using specific antibodies. Unlike in mock precipitations using empty beads, LAP2α was co-precipitated with ΔK32 lamin A (Figure 3C).
Fig. 3. Wild-type and ΔK32 lamin A bind LAP2α in vitro and in vivo.
(A) Ponceau protein staining of bacterially expressed and blotted recombinant wild-type prelamin A, ΔK32-prelamin A, or vimentin on nitrocellulose, and autoradiogram of the same blot after probing with [35S] labelled LAP2α are shown. (B) Autoradiography of in vitro-translated [35S} labelled LAP2α separated by SDS-PAGE. (C) Immunoblots of total cell lysates of ΔK32-lamin A/C fibroblasts (5% of input) and immunoprecipitates obtained with lamin A/C antibodies or empty beads (mock).
The lethal postnatal phenotype of LmnaΔK32/ΔK32 mice is not affected by loss of LAP2α
In order to test the effect of loss of LAP2α on the organismal and tissue phenotype of LmnaΔK32/ΔK32 mice, we analyzed litters of mice heterozygous for both LAP2α and ΔK32 lamin A/C (Lap2α+/−/Lmna+/ΔK32). The breeding produced genotypes according to Mendelian ratios (n=384; Lmna+/+/Lap2α+/+ = 4.7%; Lmna+/+/Lap2α−/− = 7.5%; LmnaΔK32/ΔK32/Lap2α+/+ = 4.9%; LmnaΔK32/ΔK32/LAP2α−/− = 8.4%). All genotypes were indistinguishable from wild-type littermates at birth. From postnatal day six on, all mutant mice homozygous for LmnaΔK32/ΔK32, independent of the Lap2α genotype, started to present a generally smaller appearance, a kinked tail, progressive growth retardation and stagnation in weight gain, as well as atrophied muscles and reduced mobility. By post-natal day 15 for single LmnaΔK32/ΔK32 mutants and day 17 for double LmnaΔK32/ΔK32/Lap2α−/− mutants, only 50% of mice were alive, none survived longer than post-natal day 21 (Supplementary Fig. S2). Thus, double mutants for LmnaΔK32/ΔK32 and Lap2α−/− showed a slightly prolonged, though statistically insignificant survival in comparison to the single LmnaΔK32/ΔK32 littermates, indicating that the LmnaΔK32/ΔK32 – linked phenotype was prominent.
LAP2α−/− specific epidermal paw hyperplasia is not counteracted by ΔK32 lamin A/C
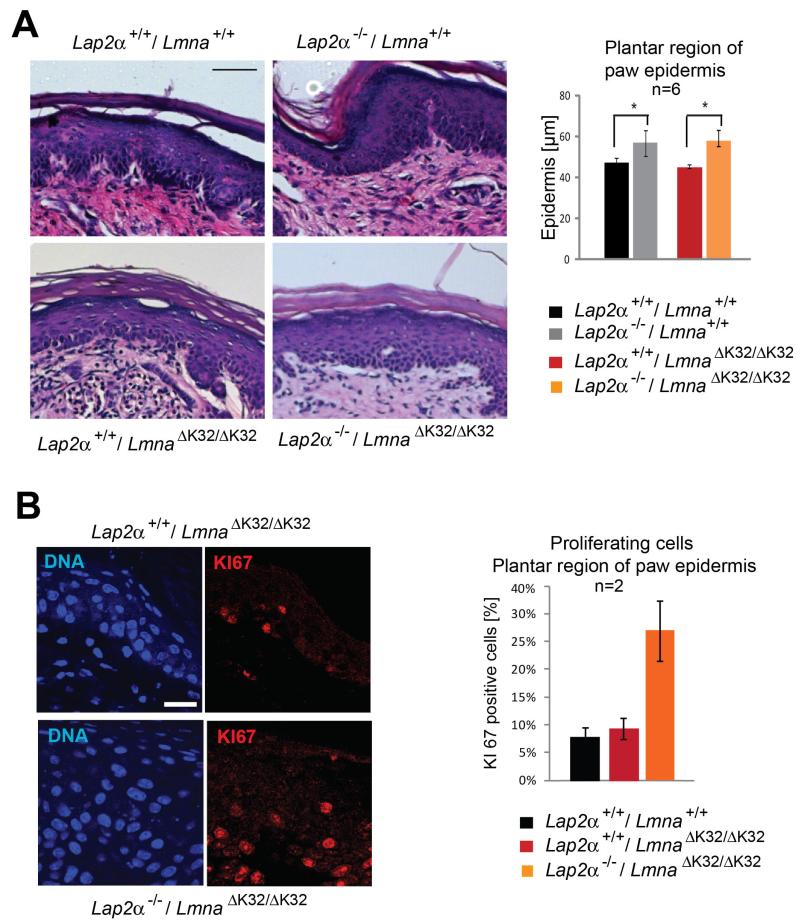
Having shown that ΔK32 lamin A/C fails to form a lamina at the nuclear periphery, but was still able to interact with LAP2α in the nucleoplasm, we sought to test whether ΔK32 lamin A – LAP2α complexes can still function in tissue progenitor cell regulation. Loss of nucleoplasmic lamin A/C - LAP2α complexes by either deletion of Lap2α or deletion of Lmna (causing loss of nucleoplasmic and peripheral lamina) was shown to cause hyperproliferation of epidermal progenitor cells and progressive hyperplasia of the paw epidermis during post-natal life (Naetar et al., 2008). If the nucleoplasmic ΔK32 lamin A/C – LAP2α complex is still functional in this pathway, LAP2α knock-down in LmnaΔK32/ΔK32 mice is expected to have a similar effect on epidermal progenitor cells and epidermal thickness as in wild type mice. Despite the young age of LmnaΔK32/ΔK32 single and double mutant mice, the paw epidermis was ~20% thicker in LmnaΔK32/ΔK32 / Lap2α−/− versus LmnaΔK32/ΔK32 / Lap2α+/+ littermates (Fig. 4A). Moreover, and in line with our previous findings, a higher number of proliferating (KI67 positive) cells was detected in double mutant LmnaΔK32/ΔK32 / Lap2α−/− versus LmnaΔK32/ΔK32 / Lap2α+/+ mice (Fig. 4B), pointing towards an increased proliferation of epidermal progenitor cells. Thus, we concluded that the ΔK32 lamin A/C mutant is still active in regulating progenitor cells in conjunction with LAP2α.
Fig. 4. Lap2α−/− -specific epidermal paw hyperplasia is not affected in LmnaΔK32/ΔK32 mice.
(A) Paraffin-embedded paw sections of 18 day old single and double mutant LmnaΔK32/ΔK32 and Lap2α−/− mice and respective wild-type control littermates were stained with hematoxylin/eosin (HE). Scale bar denotes 50 μm. Epidermal thickness of the plantar region of the paw is shown. Upon loss of LAP2α, the thickness of the epidermis is increased irrespective of LmnaΔK32/ΔK32 (n=6, P-value<0.05). (B) Paraffin embedded paw sections were processed for immunofluorescence microscopy and stained for proliferation marker KI67. Scale bar is 20 μm. KI67-positive nuclei, were quantified by counting and found to be increased upon loss of LAP2α (n=2).
Loss of LAP2α in LmnaΔK32/ΔK32 mice increassd number of skeletal muscle progenitor cells
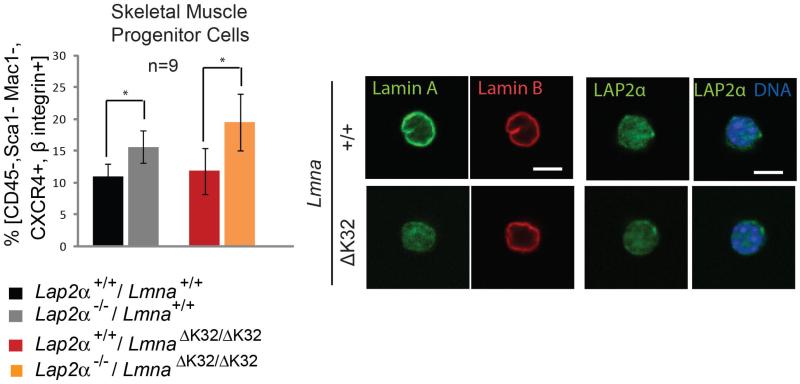
LAP2α loss was previously also shown to increase the number of skeletal muscle progenitor cells (Gotic et al., 2010). To test the number of satellite cells in LmnaΔK32/ΔK32 mice, we enriched skeletal muscle progenitor cells (SMPC) from an isolated pool of muscle fiber associated cells by flow cytometry, based on the expression of CXCR4 and β1-integrin and lack of expression of CD45, Sca1 or Mac1. Immunofluorescence microscopy of these cells confirmed mislocalization of ΔK32 lamin A/C and unaltererd lamin B distribution in lamin mutant versus wild-type cells (Fig. 5). Importantly, in both Lmna+/+ and LmnaΔK32/ΔK32 mice the SMPC population was increased in Lap2α−/− versus Lap2α+/+ background (Fig. 5), supporting the hypothesis that ΔK32 lamin A/C in conjunction with LAP2α is equally capable of regulating muscle progenitor cells like the wild-type lamin A/C. The increase in SMCP cells in LAP2α-deficient versus LAP2α-expressing LmnaΔK32/ΔK32 mice may slightly improve growth capability of muscle, which may also contribute to the subtle increase in survival of LmnaΔK32/ΔK32 / Lap2α−/− versus LmnaΔK32/ΔK32 / Lap2α+/+ mice (see supplementary Fig. S2).
Fig. 5. Lap2α−/− -specific increase in SMPCs is not affected in LmnaΔK32/ΔK32 mice.
Skeletal muscle progenitor cells (SMPCs) (CD45-/Sca1-/Mac1-/CXCR4+/β1-integrin+) were obtained from skeletal muscles (Gastrocnemius, Soleus, Tibialis anterior, Quadriceps, Triceps) and analyzed by flow cytometry or processed for confocal immunofluorescence microscopy. The number of CD45-Sca1-Mac1-CXCR4+β1-integrin+ (SMPC) cells within the parent population (CD45-Sca1-Mac1-) is presented in left panel. Lap2α−/− mice show a significant increase in SMPCs in comparison to their wild-type littermates (n=9, P-value=0.03 as determined by Student’s t-test). Similarly double mutant LmnaΔK32/ΔK32 / Lap2α−/− mice show an increase in SMPCs in comparison to their single mutant LmnaΔK32/ΔK32 littermates (n=9, P-value=0.02 as determined by Student’s t-test). (Right panel shows immunofluorescence microscopic confocal images of isolated SMPCs stained for lamin A/C, lamin B and LAP2α. Scale bar denotes 5 μm.
Based on the reported muscle phenotype of LmnaΔK32/ΔK32 mice (Bertrand et al., 2012) we also compared SMPC cell numbers in LmnaΔK32/ΔK32 versus Lmna+/+ littermates. The number of muscle fiber associated SMPC cells was indistinguishable in these genotypes (Fig. 5), indicating that an exhaustion of the SMPC pool is unlikely to contribute to muscle disease in LmnaΔK32/ΔK32 mice.
Muscular atrophy in LmnaΔK32/ΔK32 mice is not affected by loss of LAP2α
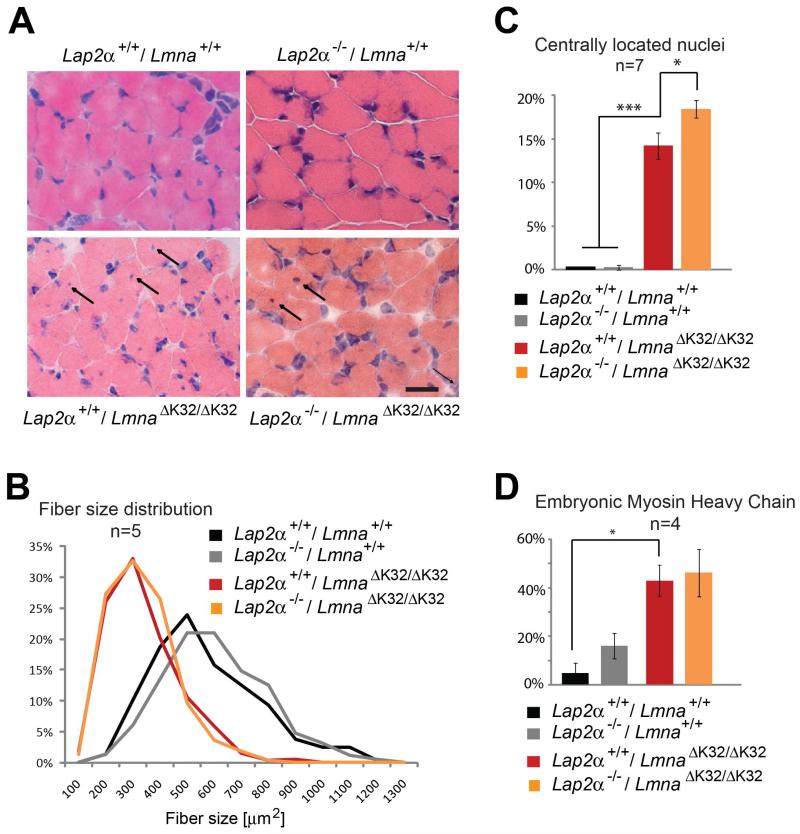
Histological haematoxylin/eosin staining of gastrocnemius and soleus muscle sections of 16 day old LmnaΔK32/ΔK32 mice revealed a generally atrophied muscle, decreased fiber cross sectional area, variability in fiber size and a significantly increased proportion of muscle fibers with centrally located or internalized nuclei (Fig. 6). Other dystrophic changes like replacement of functional muscle fibers by connective tissue or fat, cellular infiltrates, increased endomysial space, ruptured fibers or an increase in serum levels of creatine kinase (CK) levels as reviewed in (Costanza and Moggio, 2010) were not observed. This phenotype was predominantly caused by the lamin A/C mutant, as the presence or absence of LAP2α did not grossly affect this phenotype. However, one striking phenotype upon loss of LAP2α in LmnaΔK32/ΔK32 mice was the further increase in the number of fibers with centrally located nuclei (14.2% to 18.4%, n=7, P-value= 0.019) (Fig. 6C). This observation is consistent with a subtle increase in muscle growth in LAP2α-deficient background probably due to the higher number of SMPCs (Fig. 5). The presence of central nuclei within muscle fibers may also be an indicator for a muscle maturation defect as reported (Bertrand et al., 2012). Indeed, the proportion of muscle fibers expressing the embryonic form of myosin heavy chain was significantly increased in LmnaΔK32/ΔK32 versus Lmna+/+ mice (Fig. 6D, n=4, P-value= 0.04), but was independent of LAP2α expression.
Fig. 6. Skeletal muscle phenotype of double mutant LmnaΔK32/ΔK32 / Lap2α−/− mice.
(A) Cross-sectional sections of cryo-preserved gastrocnemius muscles of 18 day old wild-type, and single and double mutant LmnaΔK32/ΔK32 I Lap2α−/− mice were stained with hematoxylin/eosin (HE). Arrows denote fibers exhibiting a central nucleus. Scale bar is 50 μm. (B) Fiber cross-sectional area (n=5) was measured. (C) Quantification of fibers with centrally located nuclei is shown (n=7, P-value= 9.9 E-05 for Lmna+/+ against LmnaΔK32/ΔK32 / Lap2α+/+ and P-value=0.019 for LmnaΔK32/ΔK32 / Lap2α+/+ against LmnaΔK32/ΔK32 / Lap2α−/−) (D) Quantification of embryonic myosin heavy chain (eMHC) positive muscle fibers (n=4, P-value = 0.04 for Lmna+/+ against LmnaΔK32/ΔK32 / Lap2α+/+).
In vitro differentiation of LmnaΔK32/ΔK32 myoblasts is massively delayed and insufficient
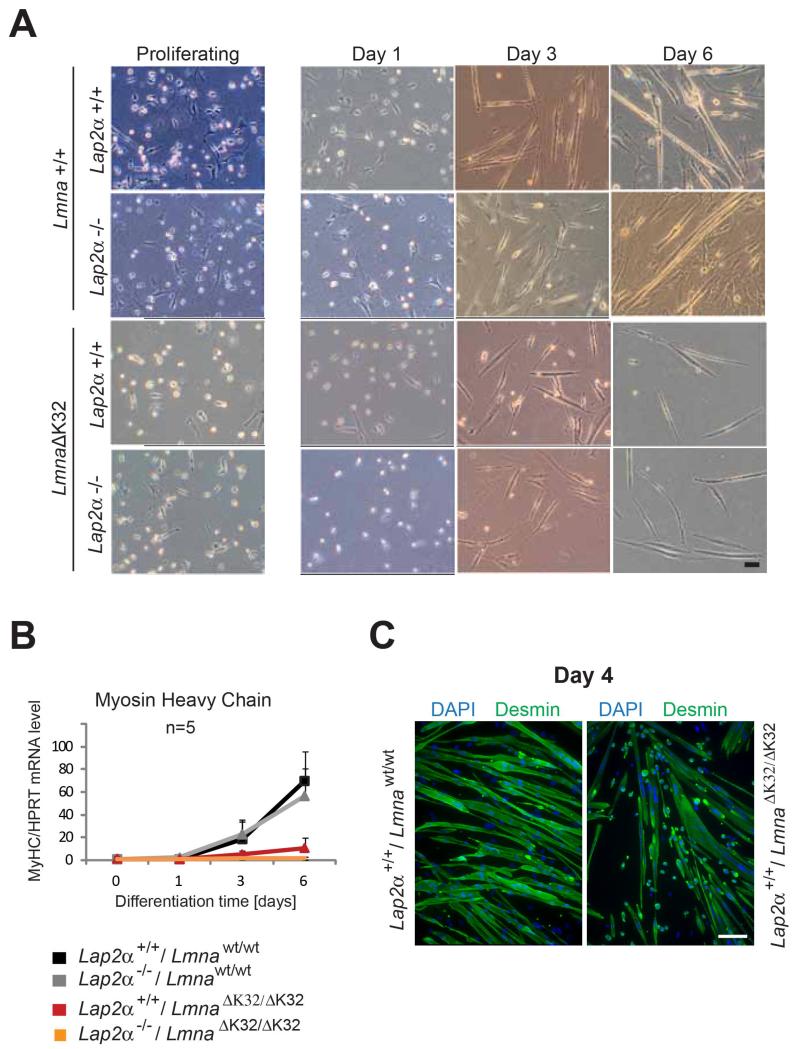
When LmnaΔK32/ΔK32 myoblasts were cultivated in vitro and induced to differentiate by withdrawal of serum, we observed a delayed onset of differentiation, an insufficient formation of myotubes and failure to upregulate MyHC compared to the wild-type littermates, pointing towards an impairment of the differentiation potential of LmnaΔK32/ΔK32 myoblasts. Additional loss of LAP2α did not noticeably aggravate or ameliorate this phenotype. Fig. 7A shows bright field images revealing a lag of LmnaΔK32/ΔK32 myoblast differentiation at day 3, irrespective of LAP2α expression and a massive reduction of myotube formation at day 6 of in vitro muscle differentiation, as confirmed also by immunofluorescence microscopy of myotubes at day 4 of differentiation (Fig. 7C). In line with this, qRT PCR analyses revealed a failure of MyHC upregulation during differentiation (Fig. 7B).
Fig. 7. Primary LmnaΔK32/ΔK32 myoblasts exhibit delayed and insufficient in vitro differentiation irrespective of presence or absence of LAP2α.
Primary murine myoblasts were isolated from newborn littermates and expanded and differentiated on collagen-coated dishes. At 1, 3 and 6 days after induction of differentiation, cultures were analyzed by various assays: (A) bright field images in the left column show proliferating myoblasts, those in other columns show myoblasts that have been plated at the same densities and induced to differentiate for 1, 3 and 6 days. A lag of LmnaΔK32/ΔK32 myoblast differentiation irrespective of LAP2α at day 3 and massive reduction of myotube formation at day 6 of in vitro muscle differentiation is detectable (bar = 100 μm). (B) Real time PCR analyses of myosin heavy chain (MyHC) normalized to endogenous levels of Hprt showing an absence of MyHC upregulation upon LmnaΔK32/ΔK32 myoblast differentiation. Means of 5 independent experiments are shown and only positive standard errors are shown as error bars. (C) Confocal immunofluorescence microscopic analyses of differentiating myoblasts at day 4 of differentiation stained with antibodies to desmin, showing impaired fusion of LmnaΔK32/ΔK32 myoblasts. DNA was stained with DAPI. Scale bar denotes 50 μm.
Discussion
In this manuscript we confirm and extend our previous findings showing that CMD-linked ΔK32 lamin A/C mutants fail to accumulate at the nuclear lamina in primary fibroblasts of LmnaΔK32/ΔK32 knock-in mice. Mutant lamin A/C is expressed at significantly reduced protein level and localizes uniformly throughout the entire nucleus. Also in wild-type cells and tissues, a highly dynamic and cell cycle-dependent pool of lamin A/C has been described in the nuclear interior (Moir et al., 2000; Naetar et al., 2008). This nucleoplasmic pool of lamin A/C has been found to associate with LAP2α and has been implicated in the regulation of proliferation and differentiation of tissue progenitor cells during tissue homeostasis (Naetar et al., 2008). It was thus tempting to speculate that an abnormally regulated pool of nucloplasmic lamins or a misbalance between lamina-associated and lamina-independent lamins were responsible for some of the pathologies described in the LmnaΔK32/ΔK32 mice (Bertrand et al., 2012). To test this hypothesis we investigated, whether any of the previously described functions and regulation mechanisms of nucleoplasmic lamins - LAP2α complexes are impaired in ΔK32 lamin A/C knock-in mice.
Although the regulation of the intranuclear, nucleoplasmic lamin A/C pool is not completely understood yet, our previous studies revealed one direct regulator of nucleoplasmic lamins A and C, a nucleoplasmic isoform of the Lamina-associated polypeptide 2 family, termed LAP2α. While most other LAP2 isoforms are transmembrane proteins of the inner nuclear membrane and bind lamins in the nuclear lamina (Foisner and Gerace, 1993), LAP2α lacks a transmembrane domain and localizes to the nuclear interior (Dechat et al., 1998) and binds specifically A-type lamins (Dechat et al., 2000). We showed that loss of LAP2α in LAP2α knock-out mice reduced the levels of lamins A and C in the nuclear interior in proliferating epidermal progenitor cells and primary fibroblasts, while re-expression of LAP2α into LAP2α-deficient fibroblasts rescued the nucleoplasmic lamin A/C pool (Naetar et al., 2008). These data suggested that LAP2α is essential and sufficient for targeting and/or stabilizing nucleoplasmic lamins A and C. Since this activity of LAP2α required its C-terminal lamin A/C-interaction domain, it was assumed that LAP2α regulates intranuclear lamin A/C by direct binding. Since we found here that LAP2α also interacted with mutant ΔK32 lamin A/C, we reasoned that loss of LAP2α in LmnaΔK32/ΔK32 mice may reduce the potentially abnormal nucleoplasmic ΔK32 lamin A/C pool in mutant mice and may allow mutant lamin to associate with the peripheral lamina. However, loss of LAP2α did neither affect the levels nor localization of ΔK32 lamin A/C. Therefore we concluded that mutant lamin A/C is incapable of assembling at the nuclear lamina even in the absence of LAP2α, supporting previous studies that suggested that the ΔK32 mutation in lamin A/C impairs the lateral association of head-to-tail dimer protofilaments into anti-parallel tetrameric filaments (Bank et al., 2011). These studies, which were performed in C. elegans did however not reveal a uniform nucleoplasmic distribution of mutant lamin but rather nucleoplasmic aggregates. This difference may be due to the fact that C. elegans contains only one lamin gene, which encodes a farnesylated B-type lamin. In any case, our studies reveal that mouse ΔK32 lamin A shows a LAP2α-independent assembly defect.
Our binding analyses show that LAP2α and ΔK32 lamin A/C can form complexes in the nucleus. Are these complexes functional? While lamins A and C at the nuclear lamina have been implicated in a number of functions, including nuclear architecture (Sullivan et al., 1999), (hetero-)chromatin organization (Guelen et al., 2008) and signaling (reviewed in (Andres and Gonzalez, 2009; Heessen and Fornerod, 2007)), we have previously shown that LAP2α and nucleoplasmic lamin A/C function in the regulation of the pRb-pathway (Dorner et al., 2006; Naetar et al., 2008). This function, which is likely independent of the peripheral lamina, has been proposed to regulate the proliferation and differentiation of tissue progenitor cells during tissue homeostasis. Since classical gene knock-out or knock-in approaches in mouse by targeting the Lmna gene affect both the peripheral lamina and the nucleoplasmic lamin A/C pool, it has been difficult to distinguish and specifically test the activities of peripheral versus nucleoplasmic lamins. The LAP2α knock-out mouse is currently the only model that selectively affects the nucleoplasmic pool of lamin A/C, while the peripheral lamina remains unaffected (Naetar et al., 2008). The fact that both the selective loss of nucleoplasmic lamins (by LAP2α deletion) and the complete loss of lamin A/C in Lmna−/− mice (affecting nucleoplasmic and peripheral lamins) show a similar hyperproliferation of progenitor cells in the paw epidermis and epidermal hyperplasia (Naetar et al., 2008) indicates that the phenotype is directly linked to the loss of nucleoplasmic lamin-LAP2α complexes rather than the loss of lamin-independent functions of LAP2α. If the nucleoplasmic ΔK32 lamin A/C - LAP2α complexes were still functional, we would expect similar consequences on tissue progenitor cells upon LAP2α loss in wild type and LmnaΔK32/ΔK32 mice. Indeed, we observed an increase in proliferating cells in paw epidermis and a thickening of the epidermis upon knocking out LAP2α in LmnaΔK32/ΔK32 mice. These data show i) that nucleoplasmic ΔK32 lamin A/C still functions in the regulation of tissue progenitor cells and ii) that the nucleoplasmic lamins require LAP2α for this activity. Forcing lamin A/C into the nuclear interior by, for instance interfering with their assembly at the lamina (as done in LmnaΔK32/ΔK32 mice) is insufficient to generate “active” nucleoplasmic lamin complexes.
Based on our results, it is likely that the pathologies described in the LmnaΔK32/ΔK32 mice are primarily caused by loss of peripheral lamins and/or the downregulation of lamin protein levels. Can loss of LAP2α affect the mutant lamin A/C-linked phenotype? One of the most prominent phenotype described in the LmnaΔK32/ΔK32 mice is an impaired peri- and postnatal muscle maturation, reflected by an increased number of muscle fibers with centrally located nuclei and increased embryonic myosin heavy chain expression. Since LAP2α loss has been shown to increase the number of fiber-associated progenitor cells, we speculated that the larger pool of skeletal muscle progenitor cells might partially rescue the mutant lamin A/C-mediated muscle defect. Although we saw a LAP2α loss-mediated increase in muscle progenitor cells in LmnaΔK32/ΔK32 mice, which may theoretically contribute to regeneration of defective muscle, we did not see any significant rescue of muscle morphology and maturation in double mutant LmnaΔK32/ΔK32 / Lap2α−/− versus LmnaΔK32/ΔK32 mice. The slight increase in muscle fibers with centrally located nuclei in muscle of LmnaΔK32/ΔK32 / Lap2α−/− versus LmnaΔK32/ΔK32 mice would be consistent with an increased regeneration/growth activity. However, the prominent lamin mutant-linked defect in myoblast differentiation in vitro may preclude any further improvement of the double mutant phenotype.
Overall our studies show that a LAP2α-independent defect of the assembly and stability of ΔK32 lamin A/C and the accompanied loss of lamin A/C at the peripheral lamina are prominent in LmnaΔK32/ΔK32 mice and responsible for the pathologies. In contrast, the nucleoplasmic ΔK32 lamin A/C is still able to bind LAP2α and function in the regulation of tissue progenitor cells.
Materials and Methods
Mice
Mice were maintained in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. Animal experiments were performed according to permissions from Austrian authorities. Data acquisition was done by observers blinded for the genotype of the animal. Lap2α-deficient mice were generated by deleting the Lap2α-specific exon 4 in the Lap2 gene (also known as thymopoietin, Tmpo) using the Cre/loxP system (Naetar et al., 2008). LmnaΔK32/ΔK32 mice were generated by a knock-in strategy replacing the wild-type Lmna exon 1 with an exon 1 where the lysine at position 32 (delAAG) is deleted (Bertrand et al., 2012). In order to obtain double mutants and littermate controls, mice heterozygous for both Lap2α and Lmna ΔK32 (Lap2α+/−, Lmna+/ΔK32) were crossed. All experiments were performed in a mixed genetic background (C57BL/6, B6129F1, BALB/c) on postnatal day 15 to 18, following cervical dislocation. For genotyping genomic DNA was prepared from tail tips and PCR analyses were performed using puRETaq Ready-To-Go PCR beads (GE Healthcare Biosciences, NJ, USA) in a PTC-200 Peltier Thermo Cycler (TJ Research). The used primers were:
Lap2α: Exon 4: 5′-CACAATCCCTAGAGGACTTCACTT-3′, Intron 4: 5′-CTGTGACTTTGCTGGCCTTCCAGTCTA-3′ and Exon 3: 5′-CAGGGAACTGAATCGAGATCCTCTAC-3′; Lmna: intron 2 forward: 5′-CAAAGTGCGTGAGGAGTTCA-3′ and intron 2 reverse in: 5′-TGACAGCATAGGCCCTGTCAC-3′
Tissue sections, histology, immunohistochemistry and immunofluorescence
Following isolation, organs were immediately dipped into pre-chilled 2-methyl-butane and snap frozen in liquid nitrogen, embedded in TBS™ Tissue freezing medium (Triangle Biomedical Sciences, Durham, NC, USA) and 5 μm sections were cut using a Cryostat HM500 OM at −23°C. Alternatively, tissues were fixed in 4% formaldehyde (Rotifix from Roth, Karlsruhe, Germany), dehydrated, cleared, embedded in paraffin and sectioned using a Leica RM 2155 microtome. Haematoxylin and eosin (H & E) staining was done according to standard protocol using an automated Ass-1 staining unit. Immunostainings for embryonic myosin heavy chain were performed using mouse monoclonal antibody against embryonic myosin (F1.652; DSHB, University of Iowa, Iowa City, USA) and biotinylated anti-mouse antibody. Stainings were developed using DAB (Vector Laboratories, Burlingame, CA, USA), nuclei were counterstained with Haematoxylin. The sections were dehydrated, mounted in Entellan (Merck, Darmstadt, Germany), and analyzed using a Zeiss Axio Imager.M1 microscope equipped with a Zeiss AxioCam MRc5 and images processed by AxioVision Rel. 4.5 software.
For immunofluorescence microscopy, cryosections were fixed either in 3.7% formaldehyde (Merck, Darmstadt, Germany) in PBS or in ice-cold acetone. Paraffin sections were incubated in xylene for 20 min, in isopropanol for 10 min, in 96%, 80%, 70% and 60% ethanol for 2 min each, and in ddH2O for 5 min. Rehydrated sections were incubated for 60 min in citrate buffer (1.8 mM citric acid and 8.2 mM sodium citrate) at 100°C. After washing in PBS, sections were incubated in 0.1% Triton-X-100 / PBS for 30 min, blocked with goat serum (Vectastain; Vector Labs, Burlingame, CA, USA), and incubated with antibodies and Hoechst-dye as described (Naetar et al., 2007). Samples were viewed in a Zeiss Axiovert 200M microscope equipped with a Zeiss LSM510META confocal laser-scanning unit, an alpha Plan-Fluor 100x/1.45 Oil and a Plan-Apochromat 63x/1.40 Oil DIC MC27 objective (Zeiss). Images were prepared with Adobe Photoshop software.
The following antibodies were used: goat polyclonal anti Lamin A/C antibody N18 (Santa Cruz Biotecj Inc., Heidelberg, Germany), rabbit antiserum to LAP2α (Vlcek et al., 2002), mouse monoclonal antibody to LAP2α (15/2) (Dechat et al., 1998), rabbit serum NCL-KI67p (Novocastra Lab., New Castle UK) and rabbit antiserum to desmin (ab8592; Abcam, Cambridge, UK). DNA was stained with DAPI: (#32670, Sigma, St Loius, MO, USA).
Primary cells, isolation and analyses
Primary fibroblasts were isolated 1-3 days after birth from back skin of newborn mice as described (Andra et al., 1998). Cells were cultivated in high glucose DMEM, 10% fetal calf serum (FCS), 50 U/ml Penicillin, 50 μg/ml Streptomycin and 0.2 μM L-Glutamine (all from Invitrogen, Carlsbad, CA, USA) at 37°C and 5% CO2. Experiments were performed between passage 1 and 4. For immunofluorescence, cells were seeded on coverslips and processed as previously described using following antibodies: goat polyclonal anti Lamin A/C antibody N18 (Santa Cruz), mouse monoclonal anti lamin A/C antibody, clone 4C11, provided by E. Ogris (Roblek et al., 2010), rabbit antiserum to LAP2α (Vlcek et al., 2002), goat polyclonal anti lamin B antibody (C20, Santa Cruz)
Fluorescence intensity measurements were done using the profile tool in Zeiss LSM Image Browser version 4.2.0.121 software. Ratios of nucleoplasmic to peripheral mean A-type lamin fluorescence intensities were calculated for 25 or 30 fibroblasts of each genotype. Primary myoblasts were obtained from de-skinned front and hind limbs of neonatal mice (2 days old) as described (Gotic et al., 2010). To induce muscle differentiation, proliferation medium (20% FCS / 2.5 ng/ml basic FGF / Hams’ F-10 / Penicillin and Streptomycin) was substituted by DMEM containing 5% horse serum containing penicillin and streptomycin. All cells were kept on collagen coated dishes in a humidified atmosphere at 37°C and 5% CO2.
Isolation of myofiber-associated satellite cells
Mice were euthanized and single fibers from particular muscles (tibialis anterior extensor digitorum longus, soleus, gastrocnemius, quadriceps, triceps and biceps brachii) were isolated according to (Shefer and Yablonka-Reuveni, 2005) and modified as described in (Gotic et al., 2010). In brief, muscles were collected in PBS and subsequently incubated in sterile 0.2% collagenase I (Gibco Life Technol., Carlsbad, CA, USA) / DMEM solution (3 ml/50 mg of tissue) for 1.5 – 2 hours in a shaking water bath at 37°C. Muscle digestion was stopped by transferring samples to a series of DMEM containing Petri dishes coated with filtered horse serum. Single fibers were released by gentle trituration, collected in DMEM and pelleted by centrifugation at 17 g for 5 min. After washing twice in PBS, fibers were resuspended in 0.01% collagenase II (Gibco Life Technol., Carlsbad, CA, USA) / 0.15 U/ml dispase II (Roche Applied Science, Mannheim, Germany) / PBS and incubated for 30 min at 37°C by shaking. Samples were filtered through 40 μm pore cell strainers and cells were pelleted by 5 min centrifugation at 210 g. After two washes in PBS, samples were stained on ice for 30 min in 2% FCS/PBS containing following antibody cocktail: anti-CD45, APC conjugated anti mouse CD45 (Ly-5); anti-Sca1, PerCP-Cy5.5 conjugated anti mouse Sca-1 (Ly-6A/E); anti-Mac1, APC conjugated anti mouse CD11b,; and anti-β1-integrin, PE anti mouse/rat CD29, (all from eBioscience, Frankfurt, Germany; and anti-CXCR4, FITCS rat-anti mouse CD184 (CXCR4) (BD Pharmingen™ Heidelberg, Germany) .
Cells were washed and analyzed using a FASCAria equipped with DIVA acquisition software (BD Biosciences) as described in (Cerletti et al., 2008). In brief, viable cells were first analyzed on the basis of size and granulation. Subsequently, a population of cells homogenous in size was tested for the expression of CD45, Mac1 and Sca1 surface markers and a subpopulation of CD45-/Sca1-/Mac1- cells was selected for CXCR4 and β1-integrin expression analysis. The number of CD45-/Sca1-/Mac1-/CXCR4+/β1-integrin+ cells in each mouse sample was presented as percentage within the parent CD45-/Sca1-/Mac1- population.
Immunoblot analyses
Cells and tissues were lysed and analyzed by SDS-PAGE and immunoblotting as previously described (Naetar et al., 2008) unsing the following antibodies: antiserum to LAP2α (Vlcek et al., 2002), goat polyclonal anti-lamin A/C serum N18 (Santa Cruz), anti lamin B (C-20, Santa Cruz), anti-emerin (NCL-Emerin, Novoscastra), rabbit polyclonal actin antiserum A-2066 (Sigma), anti γ-tubulin (B-5-1-2, Sigma) and rabbit polyclonal antiserum to histone 3, (Abcam, Cambridge, MA, USA). Quantitation of protein levels was performed with LICOR Odyssee Infrared Imaging System, application software version 2.1.12. Band intensities of lamins A and C were combined and normalized to the band intensity of the actin or γ-tubulin or histone 3 as loading control.
Quantitative real time PCR
Total RNA was isolated from muscle tissue and cultured cells using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) or RNeasy® Plus Micro Kit (Qiagen, Hilden, Germany). cDNA was synthesized by First Strand cDNA Synthesis Kit for RT-PCR (Roche Applied Science, Mannheim, Germany) according to manufacturers’ instructions and specific sequences were subsequently amplified on an Mastercycler® ep realplex (Eppendorf, Hamburg, Germany) using MESA GREEN qPCR MasterMix Plus for SYBR Assay I TTP (Eurogentec, Liege, Belgium) for quantitative PCR. Specific primers are listed in Supplementary Table 1 (see also Gotic et al., 2010; Ozawa et al., 2006; Usami et al., 2003). Data were documented using Mastercycler® ep realplex software (Eppendorf, Hamburg, Germany) and processed by Microsoft Excel XP. Endogenous levels of Hprt (Hypoxanthine-Guanine Phosphoribosyltransferase) in quantitative PCR were used for data normalization according to the Pfaffl method (Pfaffl, 2001).
In vitro binding assay and immunoprecipitation
The ΔK32 mutation was introduced into prelamin A cDNA, in the pET24-LA construct (Goldman et al., 2004) by in vitro mutagenesis using a QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA), using the following primers:
FOR: 5′-AGGAGGAGGACCTGCAGGAGCTCAATG-3′, REV: 5′-AGGTCCTCCTCCTGCAGCCGGGTGA-3′. The construct was sequenced before use. Wild-type and ΔK32 prelamin A were expressed in bacteria as described in (Dechat et al., 2000) and resolved on a 10% SDS-PAGE and transferred to a nitrocellulose membrane. Purified rat Vimentin was used as a negative control (Foisner et al., 1988). Nitrocellulose membranes were stained with PonceauS, washed in PBST (PBS, 0.05% Tween 20) and incubated in overlay buffer (10 mM Hepes, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, 1 mM DTT) for 1 hour with three changes. After blocking with 2% BSA in overlay buffer, membranes were probed overnight at 4°C with in vitro-translated, radioactively labeled FLAG - tagged LAP2α, diluted 1:50 in overlay buffer plus 1% BSA (w/v) and 1 mM PMSF. For this, a plasmid containing FLAG - tagged LAP2α cDNA (pSV5) (Vlcek et al., 1999) was in vitro translated using the TnT® T7 Quick Coupled Transcription/Translation System (Promega, Mannheim, Germany) according to the manufacturer’s instructions using 35S-labelled Methionine (Hartmann Analytic, Braunschweig, Germany). After extensive washing in overlay buffer, nitrocellulose was air dried, and bound proteins were detected by autoradiography.
For solubility assays, 1 × 107 mutant or wild-type fibroblasts were lysed by sonication in 3 ml lysis buffer (20 mM Hepes, pH 7.4, 78 mM KCl, 42 mM NaCl, 10 mM EGTA, 8.4 mM CaCl2, 4 mM MgCl2, 1 mM DTT) supplemented with protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Following addition of 0.5% Triton X-100 and 0.1% SDS, soluble fractions were obtained by centrifugation at 1700 × g for 10 min. Total cell lysates and soluble fractions were used for immunoblotting and protein amounts were quantified by ImageJ.
Immunoprecipitation of ΔK32 lamin A/C was done from soluble cell fractions (see, above) as described (Dechat et al., 200) using mouse monoclonal 3A6-4C11 anti Lamin A/C antibody (Active Motif, Carlsbad, CA) and protein A Sepharose beads (Sigma).
Statistical analysis
Data are presented as the mean of n individual experiments (n being indicated in each figure), the error bars denote standard errors. Log-Rank tests for survival curves, Chi-square test, Student’s t-test and one-way ANOVA were applied using Microsoft Excel HP. Statistical significance was assumed at a P-value < 0.05 and is highlighted in graphs using a star. Three stars are indicating a P-value < 0.01.
Supplementary Material
Acknowledgements
We acknowledge grant support from the Austrian Science Research Fund (grant number FWF P22043) to RF and from the European Union Sixth Framework Programmes [Eurolaminopathies #018690) to RF and GB. and support from COST action BM1002 (Nanonet) We thank Thomas Sauer, MFPL Vienna, for valuable help in FACS analysis of SMPCs. The monoclonal antibody F1.652 developed by Helen Blau was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. We thank Bob Goldman, Northwestern University Chicago for generous gift of lamin A plasmids and Egon Ogris, Medical University Vienna for anti lamin A/C monoclonal antibody.
References
- Andra K, Nikolic B, Stocher M, Drenckhahn D, Wiche G. Not just scaffolding: plectin regulates actin dynamics in cultured cells. Genes Dev. 1998;12:3442–3451. doi: 10.1101/gad.12.21.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres V, Gonzalez JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187:945–957. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank EM, Ben-Harush K, Wiesel-Motiuk N, Barkan R, Feinstein N, Lotan O, Medalia O, Gruenbaum Y. A laminopathic mutation disrupting lamin filament assembly causes disease-like phenotypes in C. elegans. Mol Biol Cell. 2011;22:2716–2728. doi: 10.1091/mbc.E11-01-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand AT, Renou L, Papadopoulos A, Beuvin M, Lacene E, Massart C, Ottolenghi C, Decostre V, Maron S, Schlossarek S, et al. DelK32-lamin A/C has abnormal location and induces incomplete tissue maturation and severe metabolic defects leading to premature death. Hum Mol. Gen. 2012;21:1037–1048. doi: 10.1093/hmg/ddr534. [DOI] [PubMed] [Google Scholar]
- Brachner A, Foisner R. Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem Soc Trans. 2011;39:1735–1741. doi: 10.1042/BST20110724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanza L, Moggio M. Muscular dystrophies: histology, immunohistochemistry, molecular genetics and management. Curr Pharm Des. 2010;16:978–987. doi: 10.2174/138161210790883471. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gesson K, Foisner R. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harbor symposia on quantitative biology. 2010;75:533–543. doi: 10.1101/sqb.2010.75.018. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gotzmann J, Stockinger A, Harris CA, Talle MA, Siekierka JJ, Foisner R. Detergent-salt resistance of LAP2alpha in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J. 1998;17:4887–4902. doi: 10.1093/emboj/17.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J Cell Sci. 2000;113:3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner D, Vlcek S, Foeger N, Gajewski A, Makolm C, Gotzmann J, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha regulates cell cycle progression and differentiation via the retinoblastoma-E2F pathway. J Cell Biol. 2006;173:83–93. doi: 10.1083/jcb.200511149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–79. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Foisner R, Leichtfried FE, Herrmann H, Small JV, Lawson D, Wiche G. Cytoskeleton-associated plectin: in situ localization, in vitro reconstitution, and binding to immobilized intermediate filament proteins. J Cell Biol. 1988;106:723–733. doi: 10.1083/jcb.106.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotic I, Schmidt WM, Biadasiewicz K, Leschnik M, Spilka R, Braun J, Stewart CL, Foisner R. Loss of LAP2 alpha delays satellite cell differentiation and affects postnatal fiber-type determination. Stem Cells. 2010;28:480–488. doi: 10.1002/stem.292. [DOI] [PubMed] [Google Scholar]
- Gotzmann J, Foisner R. A-type lamin complexes and regenerative potential: a step towards understanding laminopathic diseases? Histochem Cell Biol. 2006;125:33–41. doi: 10.1007/s00418-005-0050-8. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Heessen S, Fornerod M. The inner nuclear envelope as a transcription factor resting place. EMBO reports. 2007;8:914–919. doi: 10.1038/sj.embor.7401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E, Dechat T, Foisner R, Quinlan RA, Hutchison CJ. Lamin A/C binding protein LAP2alpha is required for nuclear anchorage of retinoblastoma protein. Mol Biol Cell. 2002;13:4401–4413. doi: 10.1091/mbc.E02-07-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E, Ledran M, Hutchison CJ. Remodelling of the nuclear lamina and nucleoskeleton is required for skeletal muscle differentiation in vitro. J Cell Sci. 2005;118:409–420. doi: 10.1242/jcs.01630. [DOI] [PubMed] [Google Scholar]
- Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naetar N, Hutter S, Dorner D, Dechat T, Korbei B, Gotzmann J, Beug H, Foisner R. LAP2alpha-binding protein LINT-25 is a novel chromatin-associated protein involved in cell cycle exit. J Cell Sci. 2007;120:737–747. doi: 10.1242/jcs.03390. [DOI] [PubMed] [Google Scholar]
- Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, Gotic I, Fuchs P, Cohen TV, Bittner R, et al. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol. 2008;10:1341–1348. doi: 10.1038/ncb1793. [DOI] [PubMed] [Google Scholar]
- Ozawa R, Hayashi YK, Ogawa M, Kurokawa R, Matsumoto H, Noguchi S, Nonaka I, Nishino I. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. AmerJ Pathol. 2006;168:907–917. doi: 10.2353/ajpath.2006.050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekovic V, Harborth J, Broers JL, Ramaekers FC, van Engelen B, Lammens M, von Zglinicki T, Foisner R, Hutchison C, Markiewicz E. Nucleoplasmic LAP2alpha-lamin A complexes are required to maintain a proliferative state in human fibroblasts. J Cell Biol. 2007;176:163–172. doi: 10.1083/jcb.200606139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–1085. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano-Roy S, Mbieleu B, Bonnemann CG, Jeannet PY, Colomer J, Clarke NF, Cuisset JM, Roper H, De Meirleir L, D’Amico A, et al. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann. Neur. 2008;64:177–186. doi: 10.1002/ana.21417. [DOI] [PubMed] [Google Scholar]
- Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Roblek M, Schuchner S, Huber V, Ollram K, Vlcek-Vesely S, Foisner R, Wehnert M, Ogris E. Monoclonal antibodies specific for disease-associated point-mutants: lamin A/C R453W and R482W. PLoS One. 2010;5:e10604. doi: 10.1371/journal.pone.0010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinol AE, Sinensky MS. Farnesylated lamins, progeroid syndromes and farnesyl transferase inhibitors. J Cell Sci. 2006;119:3265–3272. doi: 10.1242/jcs.03156. [DOI] [PubMed] [Google Scholar]
- Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol Bio.l. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami A, Abe S, Ide Y. Myosin heavy chain isoforms of the murine masseter muscle during pre- and post-natal development. Anat Histol Embryol. 2003;32:244–248. doi: 10.1046/j.1439-0264.2003.00481.x. [DOI] [PubMed] [Google Scholar]
- Vlcek S, Just H, Dechat T, Foisner R. Functional diversity of LAP2alpha and LAP2beta in postmitotic chromosome association is caused by an alpha-specific nuclear targeting domain. EMBO J. 1999;18:6370–6384. doi: 10.1093/emboj/18.22.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlcek S, Korbei B, Foisner R. Distinct functions of the unique C terminus of LAP2alpha in cell proliferation and nuclear assembly. J Biol Chem. 2002;277:18898–188907. doi: 10.1074/jbc.M200048200. [DOI] [PubMed] [Google Scholar]
- Wilson KL, Foisner R. Lamin-binding Proteins. Cold Spring Harb Perspect Biol. 2010;2:a000554. doi: 10.1101/cshperspect.a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–333. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.