Abstract
We studied the prevalence of polymorphisms in genes XRCC2 and XRCC3 in stomach cancer patients who lived in North Eastern Turkey. A total of 61 cancer patients and 78 controls were included in this study. Single nucleotide changes were studied in XRCC2 and XRCC3 genes at locus Arg188His and Thr241Met. Blood samples were taken from the patients and controls, and DNA was isolated. The regions of interest were amplified using a polymerase chain reaction method. After amplification, we used restriction enzymes (HphI and NcoI) to digest the amplified product. Digested product was then run through gel electrophoresis. We identified changes in the nucleotides in these specific regions. It was found that the Arg188His polymorphism of the XRCC2 gene was about 39% (24 out of the 61) among cancer patients. However, only 15% (12 out of 78) of the control group indicated this polymorphism. We also observed that 18 of the 61 cancer patients (29%) carried the Thr241Met polymorphism of the XRCC3 gene whereas 11 of the 78 (14%) individuals in the control group had the polymorphism. Our results showed a significant difference in polymorphism ratios between the cancer patients and health control group for the regions of interest. This result clearly showed that these polymorphisms increase the risk of stomach cancer and might be a strong marker for early diagnosis of gastric cancer.
KEY WORDS: gastric cancer, XRCC2 and XRCC3 genes, polymorphism, Turkey
INTRODUCTION
Nowadays, cancer threatens the public health substantially and is accepted as the plague of our century. The number of people who lose their lives annually in the world due to cancer is about 7.6 million [1]. This includes approximately 170,000 to 175,000 solely in Turkey [2]. It is estimated that the number of annual deaths in the world linked with cancer of the digestive system is approximately 920,000 [1]. In Turkey, the number of deaths due to gastric cancer is estimated to be about 64,000 [2].
It is estimated that the human genome includes between 23,000 and 25,000 genes [3]. More than 400 genes are found to be related to cancer formation. The proto-oncogenes responsible for cancer in the human genome can be classified as tumor suppressor genes and DNA repair genes [4]. The connections between genes, the environment, and cancer formation are an attractive research venue [5]. In all organisms, DNA repair mechanisms work to prevent the changes in the genome during the replication of the DNA with an aim to prevent unintended changes. The DNA repair is also realized via the enzymes produced from the genes [5,6]. These enzymes that prevent DNA damage perform the enzymatic operations that decrease cell death rate, mutations occurring in DNA, replication errors, and genomic instability. In this biological process, any change in the enzymes responsible for the DNA repair mechanism may cause the acceleration of the cell death and thus a formation of a cancer. Some genes that influence DNA repair are XRCC1, XRCC2, XRCC3, XPC, XPD/ERCC2, XPF and RAD51 [7]. Narter et al. (2009) conducted a study on SF mice, and observed that the XRCC2 and XRCC3 genes might have a vital importance in DNA repair and chromosome arrangements [8]. The chromosomal region that carries the XRCC2 and XRCC3 genes in the human genome was transformed to the mice cells, which was deformed by the mutation and a repair of the region was observed proving the role of XRCC2 and XRCC3 genes [9]. In the cell, the DNA repair genes can direct the functions of the proteins and can manage repair of the damaged DNA in the cell. The lack of repairing capacity and genetic instability in different cells can cause a cancer formation [9].
Gastric cancer is known as the third most common cancer type in the world among the known cancer types [10,11]. It represents approximately 10% of all cancers encountered worldwide. Especially in Japan, the disease is considered at epidemic dimensions by scientists. The regions with a high risk of cancer are Asia, Southern America, and Western Europe, and the low-risk regions are North America, northern Europe, and some African countries [12]. There are 10 to 20 times difference between the low and high-risk regions in terms of encountering of gastric cancer [13]. Such a big difference among the regions might provide strong clues regarding the fact that this disease is linked with genetics. According to the Lauran criteria revised in 1997, many gastric cancers are classified as intestinal and diffuse system [14]. In reality depicted in genome studies, it is observed that genetic polymorphisms and mutations that occur are linked with the geographic origin of patients and could be important triggers for cancer [15]. The general acceptance is that if the frequency of the allele which is less encountered in the population is less than one percent and if the polymorphism is more rare, it can be interpreted as a mutation [16]. In addition to the formation of cancer in the cell with the delay of the repair duties of the DNA repair proteins, the other genes and proteins which have close relations are also affected, the mechanisms repairing the DNA can be impaired [17]. In this study we have investigated the polymorphism variations in DNA repair genes XRCC2 and XRCC3 in patients suffering from gastric cancer and in healthy control group.
MATERIAL AND METHODS
Subjects
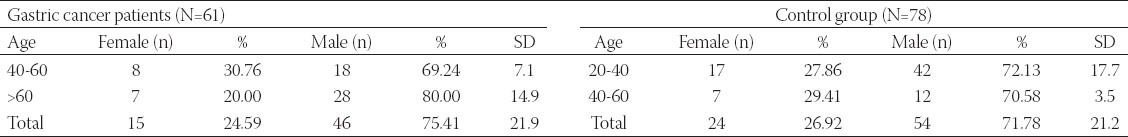
In this study we have researched the polymorphism prevalence of the XRCC2 and XRCC3 genes in patients diagnosed with gastric cancer who are admitted to Kafkas University Medicine Faculty Surgery Polyclinics and Adana Numune Oncology Hospital between March 2011 and May 2013. The clinical diagnosis of the gastric cancer was officially set using histopathological, biochemical, and radiological methods, and 61 such individual was included in a gastric cancer group. About 78 people, that were tested similar to the cancer patients and declared as healthy, were included in control group. The gastric cancer patients were selected from the second and third phase patients, and their previous gastric disorders were not considered. From each of the study's participants, both patients and controls, 8 ml of blood was taken into tubes with EDTA. The demographical structure of the examination groups are given in Table 1.
TABLE 1.
Gender and age distribution of gastric cancer patients and control group
Genotyping
DNA isolation was performed according to the Salting-Out method from all the individuals [18]. The isolated DNA samples were measured at the Nanodrop Spectrophotometer (Thermo ND1000) and kept at -80 degrees Celsius. The primer pairs selected were as follows: XRCC2 gene: F5’-TGTAGTCACCCATCTCTCTGC-3’andR 5’- AG TTGCTGCCATGCCTTACA-3’; XRCC3 gene: F 5’-G CCTGGTGGTCATCGACTC-3’ and R 5’-AC AGGG CTCT GGAAGGCACTGCTCAGCTCAC GCACC-3’ [19]. A final volume 25 µl PCR protocol that included 2.5 µl 10X Taq polymerase buffer solution, 1µl magnesium chloride (2 mM), 2 µl dNTP mix (0.2 mM), 2.5 µl forward primer (10 pmol), 2.5 µl reverse primer (10 pmol), 1µl genomic DNA (100ng/µl), 0.2 µl DNA taq polymerase enzyme (5u/µl), and 8.3 µl distilled water; a total volume of 25 µl was used [20]. PCR conditions were as follows: an initial denaturation for 3 min at 95°C, then 35 cycles at 94°C for 30 s, at 57°C for 30 s, at 72°C for 45 s, and a final extension at 1 cycle 72°C for 7 min. The PCR products were detected by agarose gel electrophoresis (at 90V, 300 A for 1.5 h) on 2% agarose gel containing ethidium bromide, and the fluorescent intensity of each band was evaluated with a UV transilluminator (Gel Logic Pro 2200, Montreal, Canada). For the XRCC2 (Arg188His) polymorphism, the PCR amplification bands were observed as 290 bp, and the XRCC3 (Thr241Met) amplification bands as 136bp. Amplified products were digested: XRCC2 (Arg188His) with 3U Haemophilus parahaemolyticus (HphI), and XRCC3 (Thr241Met) was digested with 3U Nocardia corallina (NcoI) (New England Biolabs, INC UK) [21]. Digestion products were visualized, and resulting fragments were separated on 2.5% agarose gels and with ethidium bromide staining under ultraviolet illumination (Gel Logic Pro 2200, Canada).
The single amplicon of 290 bp as a result of the section of the XRCC2 (Arg188His) polymorphism with the restriction enzyme was separated into two DNA fragments as 148 bp and 142 bp. In the evaluation, XRCC2 (Arg188His) was analyzed as having the HphI enzyme (290 bp bands Arg/Arg [homozygote wild tip] genotype; 148 bp+142 bp bands as having the Arg/His [heterozygote] genotype; 290 bp + 148 bp + 142 bp bands as having the His/His [homozygote mutant genotype]). As a result of the section of XRCC3 (Thr241Met) with NcoI enzyme, the 136 bp bands were observed as having 97 bp + 39 bpThr/Thr (homozygote genotype), 139 bp + 97 bp + 36 bp bands as having Thr/Met (heterozygote genotype), and single band 136 bp as having Met/Met (homozygote [mutant] genotype) [19].
Statistical Analysis
In this study, statistical analyses were made with the GraphPad Prism 6 package program. In the evaluation of the data, in addition to the descriptive statistical methods (mean, standard deviation), the χ2 goodness of fit test was used in the comparison of the patient and control groups, and the Fisher exact test was used in the comparisons of the qualitative data. The significance level of α=0.05 was used for significance level. In the calculation of the genotypes and allele frequencies, Hardy-Weinberg equality was tested.
RESULTS
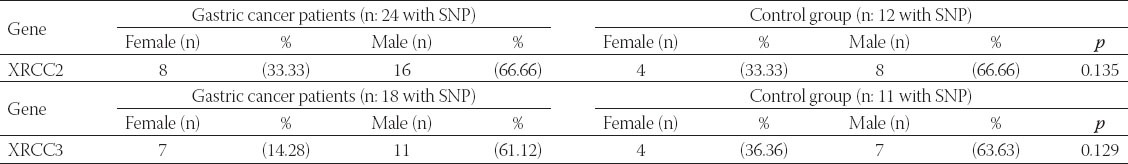
The sampled population used in this study was composed of 61 gastric cancer patients (15 females and 46 males) and 78 healthy individuals (24 females and 54 males). When our findings were evaluated in terms of the XRCC2 gene, in the gastric cancer patients, the Arg188His polymorphism was observed at the rate of 39% (24 of 61 patients), and the nucleotide change was observed in the control group at the rate of 15% (12 of 78 controls). The change rate of the XRCC3 gene in the Thr241Met polymorphism was observed in the rate of 29% (18 of 61) in patients, and the nucleotide change was observed in the rate of 14% (11 of 78) in the controls. When evaluated in terms of the general population, the rate of encounter in females was observed to be less frequent than in males in terms of the polymorphism prevalence of the XRCC2 and XRCC3 genes. The general distribution of the XRCC2 and XRCC3 gene polymorphisms in the patients and controls is given in Table 2.
TABLE 2.
Prevalence of XRCC2 and XRCC3 gene polymorphisms in the patients and controls (With SNP: Expresses a nucleotide change. Without SNP: Expresses no nucleotide change)

The overall difference in the frequency of the polymorphisms in the patients and controls as a result of the genomic evaluations are given in Table 3.
TABLE 3.
Frequency of SNP of XRCC2 and XRCC3 genes according to the gender in the patients and controls (With SNP: Explains a nucleotide change)
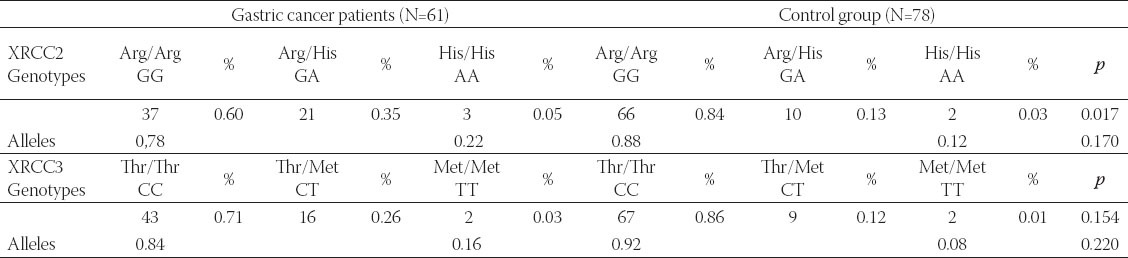
The genotypic and allelic frequencies of the patients and control group are provided in Table 4.
TABLE 4.
Genotype and allele frequency of XRCC2 and XRCC3 gene polymorphisms in the patients and controls
DISCUSSION
In the present study, we researched the differences in the polymorphism rates of the XRCC2 and XRCC3 genes in gastric cancer patients in the northeastern population of Turkey. Linking variation in polymorphic sites of the XRCC2 and XRCC3 has been conducted for various cancer types [22-24]. It was found that the XRCC3 Thr241Met polymorphism was associated with the risk of breast cancer [25]. In a study conducted in Turkey in 2009, variations in the XRCC3 gene were estimated for 85 bladder cancer patients and 45 controls and compared. The frequency of T allele was reported to be five times more prevalent in the controls compared to the cancer patients [21,26-27]. In this study, we found that the frequency of the T allele was twofold higher in gastric cancer patients. Also, the G allele of the APE gene family was observed more frequently in patients with common digestive system cancer types [24,28]. In the present study, the frequency of the G allele was 0.78 and therefore high. Zhao et al. (2012) argued that the XRCC1 and XRCC3 polymorphisms affected the tumor formation in the nerve system, and the XRCC1 and XRCC3 polymorphisms were not related to the digestive system cancers [6,29,30]. Our research indicated that the XRCC3 polymorphism may be sensitive in cancer cases linked to the digestive system. On the other hand, Zhang et al. (2005) determined that there were polymorphic regions that were more frequently associated with melanoma skin cancer than cancer occurring in head, neck, bladder, and breast cancer. Therefore, the effect of the XRCC3 gene varies in different cancer types [31-35]. A recent study conducted in Poland focused on same genomic regions and cancer type, and has shown that the regions of interest are associated with cancer [19,20]. Concordance between our research and Krupa et al. study (2011) [19], provides grounds for discussion that polymorphic variations are not necessarily population dependent or local, the results are rather consistent and should be further evaluated across other populations.
CONCLUSION
In this study, we found that there were significant variations in certain polymorphic regions of two DNA repair genes XRCC2 and XRCC3 between gastric cancer patients and control group among Turkish population. Results obtained from this study were concordant with the previous reports in other populations implying a true causative polymorphism, and more importantly, a possibility to be used in early diagnosis of the cancer. In order to extrapolate findings of our study to a global pattern, further research should be taken in different populations with a larger sample size.
ACKNOWLEDGEMENTS
We are grateful to the Kafkas University Scientific Research Project unit (Kars, Turkey, Grant No: MMF: 2011-60) for financially supporting this study.
DECLARATION OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- [1].Jing-Jing Jing J, Hui-Yuan Liu HY, Jin-Kuan Hao JK, Li-Na Wang LN, Yun-Ping Wang YP, Li-Hua Sun LH, et al. Gastric cancer incidence and mortality in Zhuanghe, China, between 2005 and 2010. World J Gastroenterol. 2012;18:1262–1269. doi: 10.3748/wjg.v18.i11.1262. http://dx.doi.org/10.3748/wjg.v18.i11.1262 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yücetürk G, Sabah D, Keçeci B, Kara AD, Yalçinkaya S. Prevalence of bone and soft tissue tumors. Acta Orthop Traumatol Turc. 2011;45(3):135–43. doi: 10.3944/AOTT.2011.2504. http://dx.doi.org/10.3944/AOTT.2011.2504 . [DOI] [PubMed] [Google Scholar]
- [3].Vineis P, Manuguerra M, Kavvoura FK. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101(1):24–36. doi: 10.1093/jnci/djn437. http://dx.doi.org/10.1093/jnci/djn437 . [DOI] [PubMed] [Google Scholar]
- [4].Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68(10):3551–7. doi: 10.1158/0008-5472.CAN-07-5835. http://dx.doi.org/10.1158/0008-5472.CAN-07-5835 . [DOI] [PubMed] [Google Scholar]
- [5].Varki A, Geschwind DH, Eichhler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat Rev Gen. 2008;(10):749–63. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhao Y, Deng X, Wang Z, Wang Q, Liu Y. Genetic polymorphisms of DNA repair genes XRCC1 and XRCC3 and risk of colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13(2):665–9. doi: 10.7314/apjcp.2012.13.2.665. http://dx.doi.org/10.7314/APJCP.2012.13.2.665 . [DOI] [PubMed] [Google Scholar]
- [7].Shin WG, Kim HU, Song HJ, Hong SJ, Shim KN, Sung IK, et al. Surveillance strategy of atrophic gastritis and intestinal metaplasia in a country with a high prevalence of gastric cancer. Dig Dis Sci. 2012;57(3):746–520. doi: 10.1007/s10620-011-1919-0. http://dx.doi.org/10.1007/s10620-011-1919-0 . [DOI] [PubMed] [Google Scholar]
- [8].Narter KF, Ergen A, Agaçhan B, Görmüs U, Timirci O, Isbir T. Bladder cancer and polymorphisms of DNA repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1) Anticancer Res. 2009;29(4):1389–93. [PubMed] [Google Scholar]
- [9].Silva SN, Tomar M, Paulo C. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol. 2010;34(1):85–92. doi: 10.1016/j.canep.2009.11.002. http://dx.doi.org/10.1016/j.canep.2009.11.002 . [DOI] [PubMed] [Google Scholar]
- [10].Benhamou S, Tuimala J, Bouchardy C, Dayer P, Sarasin A, Hirvonen A. DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aero digestive tract. Int J Cancer 2. 2004;112(5):901–4. doi: 10.1002/ijc.20474. 8. [DOI] [PubMed] [Google Scholar]
- [11].Yen CY, Liu SY, Chen CH. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37(5):271–7. doi: 10.1111/j.1600-0714.2007.00608.x. http://dx.doi.org/10.1111/j.1600-0714.2007.00608.x . [DOI] [PubMed] [Google Scholar]
- [12].Inoue M, Tsugane S. Epidemiology of gastric cancer in Japan. Postgrad Med J. 2005;81(957):419–24. doi: 10.1136/pgmj.2004.029330. http://dx.doi.org/10.1136/pgmj.2004.029330 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Loizidou MA, Michael T, Neuhausen SL. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res Treat. 2008;112(3):575–9. doi: 10.1007/s10549-007-9881-4. http://dx.doi.org/10.1007/s10549-007-9881-4 . [DOI] [PubMed] [Google Scholar]
- [14].Popanda O, Tan XL, Ambrosone CB. Genetic polymorphisms in the DNA double-strand break repair genes XRCC3, XRCC2, and NBS1 are not associated with acute side effects of radiotherapy in breast cancer patients. Cancer Epidemiol Biomarkers Prev. 2006;15(5):1048–50. doi: 10.1158/1055-9965.EPI-06-0046. http://dx.doi.org/10.1158/1055-9965.EPI-06-0046 . [DOI] [PubMed] [Google Scholar]
- [15].Hussain S, Wilson JB, Blom E. Tetratricopeptide-motif-mediated interaction of FANCG with recombination proteins XRCC3 and BRCA2. DNA Repair (Amst) 2006;5(5):629–40. doi: 10.1016/j.dnarep.2006.02.007. http://dx.doi.org/10.1016/j.dnarep.2006.02.007 . [DOI] [PubMed] [Google Scholar]
- [16].García-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet. 2006;119(4):376–88. doi: 10.1007/s00439-006-0135-z. http://dx.doi.org/10.1007/s00439-006-0135-z . [DOI] [PubMed] [Google Scholar]
- [17].Tan P. Germline polymorphisms as modulators of cancer phenotypes. BMC Med. 2008;6(27):6–12. doi: 10.1186/1741-7015-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rapley R, Walker JM. 1-2. Ottowa, New Jersey: Humana Press; 2008. The Nucleic Acid Protocol Handbook; pp. 3–29. [Google Scholar]
- [19].Krupa R, Sliwinski T, Wisniewska-Jarosinska M, Chojnacki J, Wasylecka M, Dziki L, et al. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer a case control study. MolBiol Rep. 2011;38(4):2849–54. doi: 10.1007/s11033-010-0430-6. http://dx.doi.org/10.1007/s11033-010-0430-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sliwinski T, Krupa R, Majsterek I. Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res Treat. 2005;94(2):105–9. doi: 10.1007/s10549-005-0672-5. http://dx.doi.org/10.1007/s10549-005-0672-5 . [DOI] [PubMed] [Google Scholar]
- [21].Brooks J, Shore RE, Zeleniuch-Jacquotte A. Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(4):1016–9. doi: 10.1158/1055-9965.EPI-08-0065. http://dx.doi.org/10.1158/1055-9965.EPI-08-0065 . [DOI] [PubMed] [Google Scholar]
- [22].Romanowicz-Makowska H, Smolarz B, Zadrozny M, Westfal B, Baszczynski J, Polac I, et al. Single nucleotide polymorphisms in the homologous recombination repair genes and breast cancer risk in Polish women. Tohoku J Exp Med. 2011;224(3):201–8. doi: 10.1620/tjem.224.201. http://dx.doi.org/10.1620/tjem.224.20 . [DOI] [PubMed] [Google Scholar]
- [23].Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol. 2000;2(10):757–61. doi: 10.1038/35036399. http://dx.doi.org/10.1038/35036399 . [DOI] [PubMed] [Google Scholar]
- [24].Zhang Z, Wan J, Jin X, Jin T, Shen H, Lu D, et al. Genetic polymorphisms in XRCC1, APE1, ADPRT, XRCC2, and XRCC3 and risk of chronic benzene poisoning in a Chinese occupational population. Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2614–9. doi: 10.1158/1055-9965.EPI-05-0143. http://dx.doi.org/10.1158/1055-9965.EPI-05-014.3 . [DOI] [PubMed] [Google Scholar]
- [25].Jacobsen NR, Nexøt BA, Olsen A, Overvad K, Wallin H, Tjønneland A, Vogel U, et al. No association between the DNA repair gene XRCC3 T241M polymorphism and risk of skin cancer and breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12(6):584–5. [PubMed] [Google Scholar]
- [26].Bignell GR, Greenman CD, Davies H. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–8. doi: 10.1038/nature08768. http://dx.doi.org/10.1038/naturne08768 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013 Mar 29;339(6127):1567–70. doi: 10.1126/science.1230184. http://dx.doi.org/10.1126/science.1230184 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kilpivaara O, Aaltonen LA. Diagnostic cancer genome sequencing and the contribution of germlinevariants. Science. 2013;339:1559–62. doi: 10.1126/science.1233899. http://dx.doi.org/10.1126/science.1233899 . [DOI] [PubMed] [Google Scholar]
- [29].Larrea AA, Lujan SA, Kunkel TA. SnapShot: DNA mismatch repair. Cell. 2010;14(4):141, 4730e1. doi: 10.1016/j.cell.2010.05.002. [DOI] [PubMed] [Google Scholar]
- [30].Sjödahl K, Lu Y, Nilsen TI, Ye W, Hveem K, Vatten L, et al. Smoking and alcohol drinking in relation to risk of gastric cancer: a population-based, prospective cohort study. Int J Cancer. 2007;120(1):128–32. doi: 10.1002/ijc.22157. http://dx.doi.org/10.1002/ijc.22157 . [DOI] [PubMed] [Google Scholar]
- [31].Zhang Z, Wan J, Jin X, Jin T, Shen H, Lu D, et al. Genetic polymorphisms in XRCC1, APE1, ADPRT, XRCC2, and XRCC3 and risk of chronic benzene poisoning in a Chinese occupational population. CancerEpidemiol Biomarkers Prev. 2005 Nov;14(11 Pt 1):2614–9. doi: 10.1158/1055-9965.EPI-05-0143. http://dx.doi.org/10.1158/1055-9965.EPI-05-0143 . [DOI] [PubMed] [Google Scholar]
- [32].Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carciogenesis. 2001;22(4):593–7. doi: 10.1093/carcin/22.4.593. http://dx.doi.org/10.1093/carcin/22.4.593 . [DOI] [PubMed] [Google Scholar]
- [33].Pérez LO, Crivaro A, Barbisan G, Poleri L, Golijow CD. XRCC2 R188H (rs3218536), XRCC3 T241M (rs861539) and R243H (rs77381814) single nucleotide polymorphisms in cervical cancer risk. Pathol Oncol Res. 2013;19(3):553–8. doi: 10.1007/s12253-013-9616-2. http://dx.doi.org/10.1007/s12253-013-9616-2 . [DOI] [PubMed] [Google Scholar]
- [34].Mates IN, Jinga V, Csiki IE, Mates D, Dinu D, Constantin A, et al. Single nucleotide polymorphisms in colorectal cancer: associations with tumor site and TNM stage. J Gastrointestin Liver Dis. 2012;21(1):45–52. [PubMed] [Google Scholar]
- [35].Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K. Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancerpatients. Mutat Res. 2008;648(1-2):65–72. doi: 10.1016/j.mrfmmm.2008.09.014. http://dx.doi.org/10.1016/j.mrfmmm.2008.09.014 . [DOI] [PubMed] [Google Scholar]


