Abstract
Celiac disease (CD) is a chronic inflammatory disease in the small intestine triggered by gluten uptake that occurs in genetically susceptible individuals. HLA-DQ2 protein encoded by HLA-DQA1*05 and DQB1*02 alleles is found in 90-95% of CD patients. All of the remaining patients carry HLA-DQ8 protein encoded by HLA-DQA1*03 and DQB1*03:02 alleles. Specific HLA-DQ genotypes define different risk for CD incidence. Presence of susceptible HLA-DQ genotypes does not predict certain disease development, but their absence makes CD very unlikely, close to 100%. Here we presented for the first time the distribution of HLA-DQ genotypes in the group of pediatric celiac patients from the University Children’s Hospital, Belgrade, Serbia and estimated risk for CD development that these genotypes confer. Seventy three celiac disease patients and 62 healthy individuals underwent genotyping for DQA1, DQB1 alleles and DRB1 allele. 94.5% of patients carried alleles that encode DQ2 protein variant and 2.7% carried alleles that encode DQ8 protein variant. Two patients carried single DQB1*02 allele. No patients were negative for all the alleles predisposing to CD. The highest HLA-DQ genotype risk for CD development was found in group of patients homozygous for DQ2.5 haplotype, followed by the group of heterozygous carriers of DQ2.5 haplotype in combination with DQB1*02 allele within the other haplotype. The lowest risk was observed in carriers of a single copy of DQB1*02 or DQA1*05 allele or other non-predisposing alleles. HLA genotyping, more informative than serological testing commonly used, proved to be a useful diagnostic tool for excluding CD development.
KEY WORDS: celiac disease, HLA-DQ alleles, HLA genotyping
INTRODUCTION
Celiac disease (CD) is a chronic inflammatory disease in the small intestine induced by the ingestion of gluten. The only effective treatment for CD consists of a strict lifelong gluten-free diet. Estimated prevalence of CD is approximately 1% in Caucasian population [1-3], but it is expected to be higher since the condition is greatly under-diagnosed. Many cases are subclinical. Earlier, celiac disease was recognized as a rare malabsorption syndrome in childhood, but taking into account the growing proportion of new cases diagnosed in adults, CD is now considered as common disorder that may arise at any age [4-6]. Celiac disease occurs more often in female than in male individuals, with a gender ratio of about 2:1 [7,8]. It is more frequent among first-degree relatives of patients (prevalence ranging from about 3% to 17%) [2,9]. Also, the high concordance rate (~75%) found among monozygotic twins further points out strong genetic contribution [10].
Celiac disease has a multifactorial nature. Gluten macromolecules, consisting of the gliadin and glutenin subcomponents present critical environmental factor, while both HLA and non-HLA genes are thought to be predisposing genetic factors. In CD patients, the integrity of the tight junction system is weakened [11], so poorly digestible gliadin can pass through the epithelial barrier, and interact with antigen-presenting cells in lamina propria [12,13]. Gluten-reactive CD4+ T cells of intestinal mucosa in celiac patients recognize gluten peptides attached to HLA-DQ2 and HLA-DQ8 molecules and induce immune reaction.
Numerous studies have demonstrated that about 90-95% of CD patients express HLA-DQ2 protein, and nearly all of the remaining patients express DQ8 protein [14, 15]. These proteins are encoded by highly polymorphic HLA-DQ genes located at HLA class II loci on the short arm of chromosome 6 (6p21.3). HLA-DQA1 gene encodes α chain, while DQB1 encodes β chain of HLA-DQ protein. Among HLA-DQ alleles strong linkage disequilibrium exists. Variants of HLA-DRB1 gene, located in closely positioned HLA-DR locus, are in strong linkage disequilibrium with HLA-DQ variants and they form distinctive haplotypes [14]. DQ2 heterodimer can be encoded by DQA1*05 and DQB1*02 alleles present in cis on the DRB1*03-DQA1*05-DQB1*02 haplotype, or in trans with the DQA1*05 allele usually on DRB1*11/12/13-DQA1*05-DQB1*03:01 haplotype and DQB1*02 allele usually on DRB1*07-DQA1*02-DQB1*02 haplotype. DQ8 protein is encoded by DQA1*03 allele in cis position with DQB1*03:02 allele on DRB1*04-DQA1*03-DQB1*03:02 haplotype [14]. Among the DQ2 heterodimer carriers, the risk of CD development has been shown to be increased in individuals homozygous for DQB1*02 allele [16, 17]. Also, homozygosity of alleles that contribute to DQ2 genotype was associated with the development of serious complications of celiac disease, indicating gene dosage effect [18]. Because 25-40% of the general population carries these HLA genotypes without developing CD, their presence is necessary but not sufficient for the development of the disease [19]. Given the strong association, HLA genotyping is routinely used as a genetic test for CD. Presence of susceptible DQ variants does not predict certain disease development, but their absence makes CD very unlikely with a negative predictive value close to 100% [20,21].
In this study, we presented for the first time distribution of HLA-DQ genotypes in a group of pediatric celiac patients from the University Children’s Hospital, Belgrade, Serbia. Also, we estimated the disease risk associated with different HLA-DQ genotypes. The aim of the study was to determine the frequencies of at-risk HLA-DQ genotypes in our group of CD patients.
MATERIAL AND METHODS
Patients and controls
For this study, 73 blood samples were obtained from children who were previously diagnosed as positive for celiac disease at University Children’s Hospital, Belgrade, Serbia.
The diagnosis of CD was based on the criteria of the European Society for Pediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN) from 1989, i.e. on the characteristic pathohistological appearance of small bowel mucosa and clinical recovery on gluten-free diet, as well as on the confirmed clinical and/or morphological relapse during gluten challenge at age 5-7 years in those diagnosed before completed second year of life [22]. The patient group consisted of 54 females and 19 males and had median age of 12 (range 1-22) at sample collection, while the mean age was 10.62 (SD-4.87). Group of 62 healthy individuals randomly selected from the general population, matched for gender and ethnicity with CD group, was used as a control sample (median age was 11, range 1-53; and mean age was 18.70, SD-15.03).
This study was approved by the Ethics Committee of University Children’s Hospital, University of Belgrade, and informed consent was obtained from each participant. If participant was a minor, parental or guardian’s consent was taken.
HLA genotyping
Genomic DNA was extracted from peripheral blood samples using GeneJET Genomic DNA Purification Kit (Thermo Scientific). All individuals were typed for DQA1, DQB1 and DRB1 genes by sequence-specific primer-polymerase chain reaction (SSP-PCR) method developed by Lavant et al. [23]. Three different primer mixes were used, each containing at least one locus specific common primer labeled with a fluorescent dye (NED for DQA1, VIC for DQB1 and FAM for DRB1) and allele specific primers for detection of DQA1*01, *02, *03, *04, *05 and *06, DQB1*02, *03:01, *03:02, *03:03, *03:04, *04, *05 and *06 and DRB1*04, *03, *07 and *09 [23]. Each PCR reaction contained 4ng DNA, 0.25mM dNTPs (Thermo Scientific), 1xQiagen PCR Buffer, 1.25mM MgCl2, 0.25U HotStarTaq DNA polymerase (Qiagen) and 2µl primer mix 1 (DQA1, DQB1), primer mix 2 (DQB1), or primer mix 3 (DRB1) in a final volume of 6µl. The DNA was amplified following initial denaturation at 95°C for 10 min by 45 cycles at 94°C for 1 min, 66°C for 30 s and 72°C for 1 min, and a final 10 min at 72°C. PCR reactions were diluted in water, 1:10 for PCR reaction 1 (primer mix 1), 1:20 for PCR reaction 2 (primer mix 2) and 1:5 for PCR reaction 3 (primer mix 3). Then, 1µl of each diluted PCR reaction was mixed with 9µl of 2.5% GeneScan™ 500 LIZ™ standard (Applied Biosystems, USA) in HiDi Formamide. Further, mixtures were denatured by a single heating step at 95°C for 5 min prior to electrophoresis on 3130 Genetic Analyzer (Applied Biosystems), using Fragment Analysis Run Module. Detected fragment sizes were correlated to the LIZ-labeled internal standard peaks on the X-axis. Using GeneMapper Software v.4 (Applied Biosystems), alleles were easily identified due to their specific fluorescent dye and size. Because of the strong linkage disequilibrium between DQA1, DQB1 and DRB1 genes and fixed combinations, the HLA-DQ haplotypes can be deduced in individuals without family data with high probability [24,25].
Notation
The subjects were classified in genotype groups depending on the presence of at-risk HLA-DQ haplotypes. Classification method was accepted from previous studies [26,27]. If the alleles DQA1*05 and DQB1*02 were present on a single chromosome (in cis configuration), haplotype was indicated as DQ2.5. If the DQA1*05 and DQB1*02 were inherited on separate chromosomes (in trans configuration), those were marked as α5/β2. In case that DQB1*02 allele was present in absence of DQA1*05 allele, haplotype was named β2. Also, DQA1*05 positive/DQB1*02 negative haplotype was labeled as α5. In individuals who lacked both DQA1*05 and DQB1*02 alleles, haplotypes were marked as X/X. Combination of DQA1*03 and DQB1*03:02 alleles on a single chromosome was indicated as DQ8 haplotype. Individuals with HLA-DQ2 encoding genotypes (DQ2.5/DQ2.5, DQ2.5/β2, DQ2.5/X, α5/β2) were denoted as DQ2 positive, while carriers of HLA-DQ8 encoding genotypes (DQ8/DQ8, DQ8/X) were denoted as DQ8 positive. For developing CD risk assessment, CD patients and controls were split into risk groups presented in Table 1, depending on the presence of specific HLA-DQ genotypes [15].
Statistical analysis
The distribution of HLA-DQ genotypes in patients and controls were analyzed by Fisher’s exact test using 2x2 contingency tables. All of the tests were performed as two-tailed and differences were considered statistically significant when p< 0.05. Risk of developing disease for specific genotype group was calculated as previously described [15] and presented relatively to the group with the highest risk, which value was denoted as 1.
RESULTS
In 73 children who were diagnosed with celiac disease and 62 controls, HLA variants were genotyped.
Frequency of each HLA-DQ genotype in CD and control group is presented in Table 2A. In particular, 19.2% of patients were homozygous for DQ2.5 haplotype (DQ2.5/DQ2.5), 16.4% carried single DQ2.5 haplotype with DQB1*02 allele within the other haplotype (DQ2.5/β2) and 52.1% were heterozygous for DQ2.5 haplotype and any haplotype unrelated to CD (DQ2.5/X). In control group, 3.2% of healthy individuals were carriers of DQ2.5/DQ2.5, 3.2% carried DQ2.5/β2 and 14.5% carried DQ2.5/X genotype. Positive association of those three genotypes with CD development was found (p=0.006, p=0.02 and p=4.5x10-6, respectively). The great majority of DQ2 positive patients carried at least one DQ2.5 haplotype (87.7%), and the rest were α5/β2 positive (6.8%). In CD group, two patients (2.7%) were negative for alleles that contribute to both DQ2 and DQ8 genotypes and carried only DQB1*02 allele (β2). No patient was negative for all the alleles predisposing to celiac disease. In the control group, among individuals lacking alleles that contribute to DQ2/DQ8 variants, 4.8% of subjects were β2 positive, 27.4% were α5 positive, and 24.2% lacked all the susceptible alleles. Negative association between CD development and α5 haplotype was observed (p=4.6x10-7). Proportion of haplotypes non-related to CD was significantly higher in healthy in comparison with patient group (p=3x10-6).
TABLE 2.
Distribution of HLA-DQ genotypes in patients with celiac disease and control group

Frequencies of alleles that contribute to DQ2 and DQ8 genotypes are summarized in Table 2B. 97.3% of patients were identified as carriers of alleles that contribute to DQ2 and/or DQ8 variants, while in control group 43.5% carried those alleles. DQ2 variants were found in 94.5% of patients and 25.8% of controls, showing significantly higher proportion in patient group in comparison with the control group (p=9.6x10-18). DQ8 variants were detected in 2.7% of patients, and in 17.7% of healthy individuals. Difference in frequencies of DQ8 carriers between CD and control group showed to be of statistical significance (p= 0.004).
Carriers of susceptible HLA-DQ genotypes were divided into genotype groups (Table 1) and risk for disease development was calculated (Table 3). The gradient of genotype relative risk (GRR) was observed, being the highest in group 1 (DQ2.5/DQ2.5 genotype) (GRR=1.000), and followed by group 2 (DQ2.5/β2 genotype) (GRR=0.857). In group 3 (DQ2.5/X genotype) relative risk was 1.6 times lower than in group 1 (GRR=0.603). For patients in group 4 (α5/β2) relative risk was 4 times lower than for patients in group 1 (GRR=0.238). Estimated relative risk in group 5 (DQ8/DQ8, DQ8/X) was almost 40 times and in group 6 (β2, α5, X/X) even 125 times lower than in group 1 (GRR=0.026 and GRR=0.008, respectively).
TABLE 1.
HLA-DQ genotype groups
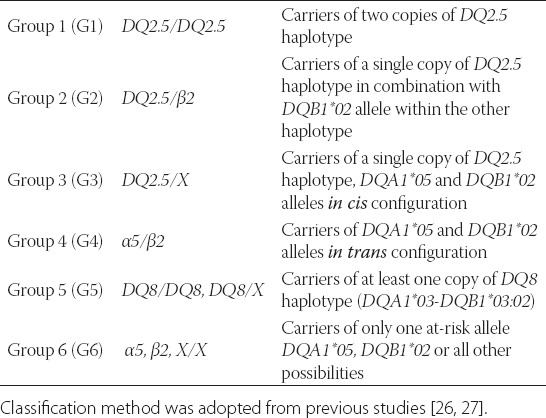
TABLE 3.
Estimated HLA-DQ genotype group relative risks (GRR) for CD patients

DISCUSSION
Celiac disease is a chronic inflammatory disorder triggered by gluten uptake that occurs in genetically susceptible individuals. Genetic elements in CD predisposition have been well elucidated, and it has been shown by numerous studies that the disease rarely develops in the absence of specific HLA-DQ alleles. It is reported that vast majority of patients with celiac disease carried HLA-DQA1*05 and DQB1*02 alleles, both encoding HLA-DQ2 protein. In nearly all of the remaining cases DQA1*03 and DQB1*03:02 alleles were found, both encoding HLA-DQ8 protein. Very rarely, CD patients carried genotypes that encode different HLA-DQ proteins [15].
Our study presented results of HLA genotyping in group of 73 celiac disease pediatric patients from the University Children’s Hospital, Belgrade, Serbia and 63 healthy individuals. We found that in our cohort 94.5% of CD patients carried DQA1*05 and DQB1*02 alleles, either in cis or in trans configuration. Results from this case-control study confirmed strong association of the presence of alleles that contribute to HLA-DQ2 genotype with CD development. This data is in concordance with the results obtained for CD patients from several European countries regarding HLA-DQ2 variants: in Scandinavian probands - 92%; French - 87%; Italian - 84%; and in UK - 88% [14, 15]. Two other Italian studies reported that 80.8% and 64% of Italian CD cohort carried HLA-DQ2 variants [26, 27]. Dissimilarity in the frequencies of carriers of HLA-DQ2 variants was observed, with the higher proportion of DQ2 positive patients in northern than in the southern European populations. Furthermore, it is noticed that proportions of patients who had DQA1*05 and DQB1*02 alleles in cis and the ones with the alleles in trans position were different, with distinct north-south gradient [15]. Precisely, in southern (France and Italy) populations, frequency of patients carrying the DQ2 in trans genotype was higher than in northern populations. Also, relative risk for DQ2 in trans genotype was higher compared to DQ2 in cis genotype. Proportion of carriers of HLA-DQ2 variants in our patient group, as well as frequency of DQ2 in cis (DQ2.5/X) and in trans (α5/β2) carriers was the most similar to the Scandinavian group [14, 15]. The observed differences could be explained by involvement of another genetic factor linked with HLA complex (MICA, TNF) subdividing the DQA1-DQB1 haplotypes in different at-risk subhaplotypes, some being more at risk than others [28-30].
The highest HLA-DQ genotype relative risk (GRR) for CD in our cohort was associated with DQ2.5/DQ2.5 genotype carriers. Also, carriers of DQ2.5 haplotype with two DQB1*02 alleles (DQ2.5/β2 genotype) were in group with high relative risk. Recent large prospective study also confirmed that DQ2.5 haplotype homozygosity conferred the highest risk for CD and was associated with the earliest onset [31]. In our study, group of DQ2/non-DQ2 carriers (DQ2.5, α5/β2 genotypes) showed medium relative risk for CD. The lowest risk was detected in group of non-DQ2/non-DQ2 genotype carriers (DQ8, β2, α5, X/X genotypes). This data indicated dose effect for DQB1*02 allele, since groups with two DQB1*02 alleles showed higher GRR than groups with single DQB1*02 allele (Table 3). Gene dosage effect for DQB1*02 allele has already been shown before [17].
In Serbia, so far, involvement of specific DQ variants in CD susceptibility has been studied by serologic techniques only [32]. It was confirmed that the most frequent HLA protein variants in patients with CD in the northern Serbian region, Vojvodina, were DR3 and DQ2. However, serological HLA typing method could not reveal HLA haplotypes.
When presented results were compared with the data obtained from the groups closely related to our group (Croatian and Slovenian group), we noticed that proportions of at-risk HLA-DQ genotypes are similar. In Croatian CD group, frequency of patients positive for alleles that contribute to DQ2 genotype were 93.7%, similar to our group (94.5%) and slightly higher than in Slovenian CD group (88.8%) [33, 34]. Further, prevalence of carriers of alleles that contribute to DQ8 genotype were lower in our and Croatian CD group (2.7% and 4.8% respectively) compared with Slovenian CD group (8.8%). Also, Slovenian study identified one patient with proven CD lacking all susceptible HLA-DQ alleles, emphasizing the fact that CD patients completely negative for at-risk HLA-DQ alleles can be found as well.
Considering its multifactorial nature, CD is a rare example in which a genetic test is of great importance in clinical practice. It is recommended that clinicians should not classify patients only as DQ2 and DQ8 positive or negative, but must also consider the presence of DQB1*02 and DQA1*05 alleles alone [14], regardless of their relative low risk for CD development. Recent study presented that even 5.8% of patients lacking HLA-DQ2 and HLA-DQ8 variants carried DQB1*02 allele, therefore this allele should be recognized as risk allele for disease occurrence [35]. It may be useful to consider HLA-DQ genotype gradient risk in selecting individuals who must undergo recurrent clinical and serologic follow-ups, especially in high-risk groups. As recent study suggested, screening of genetically susceptible infants can lead to a diagnosis of CD at a very early age [31]. Moreover, in estimating the CD risk, the role of non-HLA genes should be considered. Several genes in HLA non related loci, in particular genes included in the control of the adaptive immune response, have been identified as potential contributors to the CD [36-39]. Also, it is important to note that in celiac disease risk estimation, apart from genotype data, records related to family history of this disease, along with gender and country of residence should not be disregarded [31].
CONCLUSION
We presented for the first time the distribution of HLA-DQ genotypes in the group of pediatric celiac patients from the University Children’s Hospital, Belgrade, Serbia and estimated risk for CD development that these genotypes confer. New ESPGHAN guidelines for the diagnosis of CD confirmed that HLA genotyping can be used as an efficient adjunct in the CD diagnostic algorithm [40]. Negative predictive value of HLA genotyping proved itself as a useful tool for avoiding additional invasive diagnostic testing or to exclude risk in relatives and patients in high-risk groups.
ACKNOWLEDGEMENTS
This work was supported by Ministry of Education, Science and Technological Development, Republic of Serbia (Grant No. III41004).
DECLARATION OF INTEREST
The authors report no conflict of interests.
REFERENCES
- [1].Fasano A, Berti I, Gerarduzzi T, Not T, Colletti RB, Drago S, et al. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: a large multicenter study. Arch Intern Med. 2003;163(3):286–292. doi: 10.1001/archinte.163.3.286. [DOI] [PubMed] [Google Scholar]
- [2].Dubé C, Rostom A, Sy R, Cranney A, Saloojee N, Garritty C, et al. The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology. 2005;128(4 Suppl 1):S57–67. doi: 10.1053/j.gastro.2005.02.014. [DOI] [PubMed] [Google Scholar]
- [3].Mustalahti K, Catassi C, Reunanen A, Fabiani E, Heier M, McMillan S, et al. The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann Med. 2010;42(8):587–595. doi: 10.3109/07853890.2010.505931. [DOI] [PubMed] [Google Scholar]
- [4].Sanders DS, Hurlstone DP, Stokes RO, Rashid F, Milford-Ward A, Hadjivassiliou M, et al. Changing face of adult coeliac disease: experience of a single university hospital in South Yorkshire. Postgrad Med J. 2002;78(915):31–33. doi: 10.1136/pmj.78.915.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Heel DA, West J. Recent advances in coeliac disease. Gut. 2006;55(7):1037–1046. doi: 10.1136/gut.2005.075119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Leeds JS, Hopper AD, Sanders DS. Coeliac disease. Br Med Bull. 2008;88(1):157–170. doi: 10.1093/bmb/ldn044. [DOI] [PubMed] [Google Scholar]
- [7].Llorente-Alonso MJ, Fernández-Acenero MJ, Sebastián M. Gluten intolerance: Sex-and age-related features. Can J Gastroenterol. 2006;20(11):719–722. doi: 10.1155/2006/470273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Megiorni F, Mora B, Bonamico M, Barbato M, Montuori M, Viola F, et al. HLA-DQ and susceptibility to celiac disease: evidence for gender differences and parent-of-origin effects. Am J Gastroenterol. 2008;103(4):997–1003. doi: 10.1111/j.1572-0241.2007.01716.x. [DOI] [PubMed] [Google Scholar]
- [9].Esteve M, Rosinach M, Fernández-Bañares F, Farré C, Salas A, Alsina M, et al. Spectrum of gluten-sensitive enteropathy in first-degree relatives of patients with coeliac disease: clinical relevance of lymphocytic enteritis. Gut. 2006;55(12):1739–1745. doi: 10.1136/gut.2006.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Greco L, Romino R, Coto I, Di Cosmo N, Percopo S, Maglio M, et al. The first large population based twin study of coeliac disease. Gut. 2002;50(5):624–628. doi: 10.1136/gut.50.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lionetti E, Catassi C. New clues in celiac disease epidemiology, pathogenesis, clinical manifestations, and treatment. Int Rev Immunol. 2011;30(4):219–231. doi: 10.3109/08830185.2011.602443. [DOI] [PubMed] [Google Scholar]
- [12].Matysiak-Budnik T, Moura IC, Arcos-Fajardo M, Lebreton C, Ménard S, Candalh C, et al. Secretory IgA mediates retrotranscytosis of intact gliadin peptides via the transferrin receptor in celiac disease. J Exp Med. 2008;205(1):143–154. doi: 10.1084/jem.20071204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Caillat-Zucman S. Molecular mechanisms of HLA association with autoimmune diseases. Tissue Antigens. 2009;73(1):1–8. doi: 10.1111/j.1399-0039.2008.01167.x. [DOI] [PubMed] [Google Scholar]
- [14].Karell K, Louka AS, Moodie SJ, Ascher H, Clot F, Greco L, et al. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol. 2003;64(4):469–477. doi: 10.1016/s0198-8859(03)00027-2. [DOI] [PubMed] [Google Scholar]
- [15].Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, et al. HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens. 2004;63(6):562–567. doi: 10.1111/j.0001-2815.2004.00237.x. [DOI] [PubMed] [Google Scholar]
- [16].Clerget-Darpoux F, Bouguerra F, Kastally R, Semana G, Babron MC, Debbabi A, et al. High risk genotypes for celiac disease. C R Acad Sci III. 1994;317(10):931–936. [PubMed] [Google Scholar]
- [17].Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, et al. The HLA-DQ2 gene dose effect in coeliac disease is directly related to the magnitude and breadth of the gluten-specific T cell responses. Proc Natl Acad Sci USA. 2003;100(21):12390–12395. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Al-Toma A, Goerres MS, Meijer JW, Peña AS, Crusius JB, Mulder CJ. Human leukocyte antigen-DQ2 homozygosity and the development of refractory celiac disease and enteropathy-associated T–cell lymphoma. Clin Gastroenterol Hepatol. 2006;4(3):315–319. doi: 10.1016/j.cgh.2005.12.011. [DOI] [PubMed] [Google Scholar]
- [19].Green P, Cellier C. Celiac disease. N Engl J Med. 2007;357(17):1731–1743. doi: 10.1056/NEJMra071600. [DOI] [PubMed] [Google Scholar]
- [20].Kaukinen K, Partanen J, Mäki M, Collin P. HLA-DQ typing in the diagnosis of celiac disease. Am J Gastroenterol. 2002;97(3):695–699. doi: 10.1111/j.1572-0241.2002.05471.x. [DOI] [PubMed] [Google Scholar]
- [21].Wolters VM, Wijmenga C. Genetic background of celiac disease and its clinical implications. Am J Gastroenterol. 2008;103(1):190–195. doi: 10.1111/j.1572-0241.2007.01471.x. [DOI] [PubMed] [Google Scholar]
- [22].Walker-Smith JA, Guandalini S, Schmitz J, Shmerling DH, Visakorpi JK. Revised criteria for diagnosis of coeliac disease. Report of working group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child. 1990;65(8):909–911. doi: 10.1136/adc.65.8.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lavant EH, Agardh DJ, Nilsson A, Carlson JA. A new PCR-SSP method for HLA DR-DQ risk assessment for celiac disease. Clin Chim Acta. 2011;412(9-10):782–784. doi: 10.1016/j.cca.2010.12.033. [DOI] [PubMed] [Google Scholar]
- [24].Allele Frequency Net Database. [accessed: 20 February 2014]. http://www.allelefrequencies.net .
- [25].National Marrow Donor Program Bioinformatics. [accessed: 18 February 2014]. http://bioinformatics.nmdp.org/HLA/HLA_Resources.aspx .
- [26].Megiorni F, Mora B, Bonamico M, Barbato M, Nenna R, Maiella G, et al. HLA-DQ and risk gradient for celiac disease. Hum Immunol. 2009;70(1):55–59. doi: 10.1016/j.humimm.2008.10.018. [DOI] [PubMed] [Google Scholar]
- [27].Piccini B, Vascotto M, Serracca L, Luddi A, Margollicci MA, Balestri P, et al. HLA-DQ typing in the diagnostic algorithm of celiac disease. Rev Esp Enferm Dig. 2012;104(5):248–254. doi: 10.4321/s1130-01082012000500005. [DOI] [PubMed] [Google Scholar]
- [28].Louka AS, Moodie SJ, Karell K, Bolognesi E, Ascher H, Greco L, et al. European Genetics Cluster on Celiac Disease. A collaborative European search for non-DQA1*05-DQB1*02 celiac disease loci on HLA-DR3 haplotypes: analysis of transmission from homozygous parents. Hum Immunol. 2003;64(3):350–358. doi: 10.1016/s0198-8859(02)00822-4. [DOI] [PubMed] [Google Scholar]
- [29].Louka AS, Lie BA, Talseth B, Ascher H, Ek J, Gudjónsdóttir AH, et al. Coeliac disease patients carry conserved HLA-DR3-DQ2 haplotypes revealed by association of TNF alleles. Immunogenetics. 2003;55(5):339–343. doi: 10.1007/s00251-003-0586-5. [DOI] [PubMed] [Google Scholar]
- [30].Bolognesi E, Karell K, Percopo S, Coto I, Greco L, Mantovani V, et al. Additional factor in some HLA DR3/DQ2 haplotypes confers a fourfold increased genetic risk of celiac disease. Tissue Antigens. 2003;61(4):308–316. doi: 10.1034/j.1399-0039.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- [31].Liu E, Lee HS, Aronsson CA, Hagopian WA, Koletzko S, Rewers MJ, et al. Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014;371(1):42–9. doi: 10.1056/NEJMoa1313977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vojvodić S, Ademović-Sazdanić D. HLA II class antigens and susceptibility to coeliac disease. Genetika. 2011;43(3):517–526. [Google Scholar]
- [33].Žunec R, Grubić Z, Jurčić Z, Peršić M, Kaštelan A, Kerhin-Brkljačić V. Hla-DQ2 heterodimer in the diagnosis of celiac disease. Biochemia Medica. 2004;14:3–4. [Google Scholar]
- [34].Dolinšek J, Micetik-turk D, Urlep-Žužej D, Zagradišnik B, Haimilia K, Holopainen P. Importance of Celiac Disease Patients Lacking Hla Dq2 or Dq8 Heterodimer in Slovenia. J Pediatr Gastroenterol Nutr. 2006;42(5):18–19. [Google Scholar]
- [35].Mubarak A, Spierings E, Wolters V, van Hoogstraten I, Kneepkens CM, Houwen R. Human Leukocyte Antigen DQ2.2 and Celiac Disease. J Pediatr Gastroenterol Nutr. 2013;56(4):428–430. doi: 10.1097/MPG.0b013e31827913f9. [DOI] [PubMed] [Google Scholar]
- [36].Sollid LM, Lie BA. Celiac disease genetics: current concepts and practical applications. Clin Gastroenterol Hepatol. 2005;3(9):843–851. doi: 10.1016/s1542-3565(05)00532-x. [DOI] [PubMed] [Google Scholar]
- [37].Garner CP, Murray JA, Ding YC, Tien Z, van Heel DA, Neuhausen SL. Replication of celiac disease UK genome-wide association study results in a US population. Hum Mol Genet. 2009;18(21):4219–4225. doi: 10.1093/hmg/ddp364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40(4):395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Romanos J, van Diemen CC, Nolte IM, Trynka G, Zhernakova A, Fu J, et al. Analysis of HLA and non-HLA alleles can identify individuals at high risk for celiac disease. Gastroenterology. 2009;137(3):834–840. doi: 10.1053/j.gastro.2009.05.040. [DOI] [PubMed] [Google Scholar]
- [40].Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci. 2012;19:88. doi: 10.1186/1423-0127-19-88. [DOI] [PMC free article] [PubMed] [Google Scholar]


