Abstract
The mammalian cerebral cortex is responsible for the highest-levels of associative, cognitive and motor functions. In the CNS, the cortex stands as a prime example of extreme neuronal diversity, broadly classified into excitatory projection neurons and inhibitory interneurons. Here, we review recent progress made in understanding the strategies and mechanisms that shape projection neuron diversity during embryogenesis and discuss how projection neuron classes may be maintained, postnatally, for the life of the organism. In addition, we consider the intriguing possibility that projection neurons may be amenable to directed reprogramming of their class specific features to allow enhanced cortical plasticity in the adult.
The cerebral cortex: master of cellular complexity
Over a century ago neuroscientists generated the first depictions of the neuronal and non-neuronal structures they observed within the Central Nervous System (CNS) [1–3]. Collective efforts in the field have since demonstrated the great cellular complexity of the brain and highlighted how the mammalian cerebral cortex in particular stands uncontested as the most heterogeneous region of the CNS, composed of billions of neuron and glia whose subtype- specific classification remains to this day incomplete.
The neocortex processes information that regulates high-level functions including cognition, sensory perception, regulation of fine motor skills and, in humans, articulate language. These complex behaviors are centrally executed by two major groups of neurons: the excitatory projection neurons (PNs) and the inhibitory interneurons (INs), both present in a plethora of different subtypes (reviewed in [4, 5]). Excitatory PNs are born from neural progenitors located in the developing proliferative zones of the dorsal telencephalon; they are glutamatergic and send long-distance axons to targets within and outside of the cortex [4]. The activity of PNs is finely modulated by cortical INs, which are instead generated from neural progenitors residing in the ventral telencephalon [6], and display a great diversity of molecular signatures, electrophysiological properties, connectivity and synaptic dynamics; they are GABAergic and connect locally within the cortical microcircuitry [5].
The development and classification of cortical INs has been reviewed elsewhere [5, 7, 8]. Here, we will focus exclusively on the establishment of PN diversity and its maintenance. We will first briefly cover the classification of projection neurons. We will then review the strategies employed during development to achieve the generation of PN diversity and discuss its effect on the behavior of other cell types of the cortex. Finally, we will consider strategies to maintain PN diversity unchanged in the adult and touch upon the idea that despite the known immutability of postmitotic neuronal identity in the mammalian CNS projection neurons may retain the ability to reprogram their class-specific features in vivo, potentially providing a new substrate for cortical plasticity.
Achieving Cortical Pyramidal Neuron Diversity
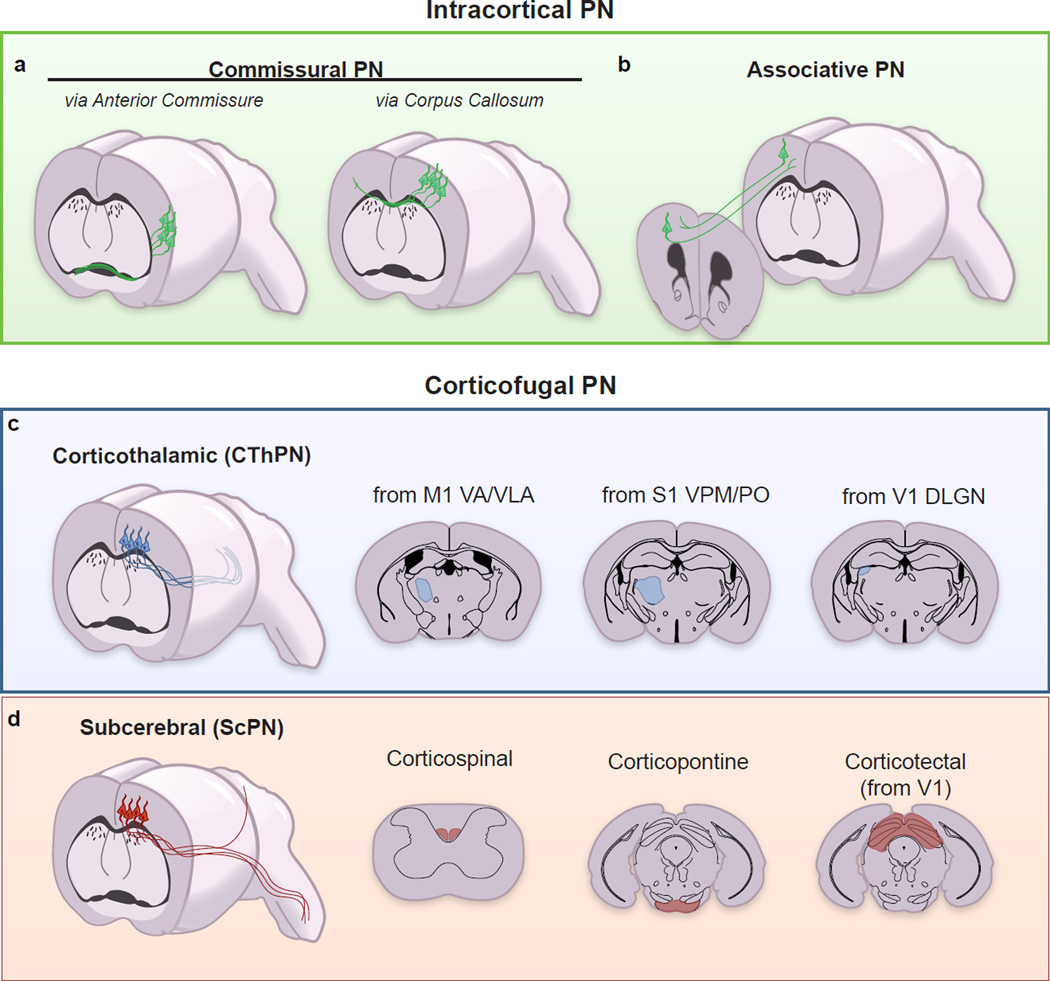
The neocortex presents a high-degree of neuronal diversity, which is organized in six layers and multiple functional areas (reviewed in [4]). Distinct PN subtypes can be recognized and canonically classified based on the laminar position of their cell bodies, soma and dendritic morphology, electrophysiological properties and, above all, axonal connectivity [9, 10]. Indeed, PNs derive their classic nomenclature from their axonal targets and can be broadly classified into intracortical projection neurons (commissural and associative PNs) and corticofugal projection neurons (subcortical and subcerebral PNs (Figure 1). Intracortical neurons, although present in all six cortical layers, reside in larger numbers in the upper cortical layers (L2/3), and extend axons across the midline to the opposite hemisphere. The majority of intracortical neurons project to contralateral targets via the corpus callosum, and are thus coined callosal projection neurons (CPNs), whereas a small percentage projects via the anterior commissure, the most ancient commissure of the brain (Figure 1a). Commissural neurons have been identified in all areas of the neocortex, where they are responsible for integrating bilateral information between homologous areas of the two cerebral hemispheres [10]. Neurons projecting contralaterally through the anterior commissure are mainly located in the most lateral cortical areas, which are part of the olfactory-limbic system [11] (Figure 1a). Associative PNs extend axons within the same cortical hemisphere. They can project to either short-distance targets (such as layer IV granular neurons) or long-distance targets in the frontal cortex, for example (Figure 1b).
Figure 1. Cortical Projection Neuron Classification by Connectivity.
PNs are broadly divided into two groups: Intracortical PNs and Corticofugal PNs. Intracortical PNs are further subdivided into Commissural PNs (a), which project to the contralateral hemisphere, and Associative PNs (b), which project to cortical areas within the same hemisphere (e.g. ipsilateral forward and backward projecting neurons). Some commissural PN connect through the corpus callosum (Callosal PNs, CPNs) while others, residing within the lateral cortex project via the anterior commissure (a). Corticofugal PNs project to subcortical targets and are further divided into Corticothalamic PNs (CThPNs) (c) and Subcerebral PN (ScPNs) (d). CThPNs are located in L6 and project to various nuclei of the thalamus in an area-dependent manner (c). From the primary motor cortex (M1), the majority of CThPNs project to the Ventral Anterior (VA) and Anterior Ventral Lateral (VLA) nuclei. From the somatosensory cortex (S1), the majority of CthPNs project to the Ventral Posterior Medial nucleus (VPM) and the Posterior nucleus (PO). From the visual cortex (V1), the majority of CFuPNs project to the Dorsal Lateral Geniculate Nucleus (dLGN). ScPNs are also further divided based on their axonal targets (d). Corticospinal motor neurons send primary axons to the spinal cord. Corticopontine neurons extend axons to the pontine nuclei within the brainstem, and Corticotectal neurons have axon projections to the optic tectum in the midbrain.
Corticofugal projection neurons (CFuPNs), mainly located in the deep layers of the cortex (L5 and L6), send axons to distal targets outside of the cortex. Corticothalamic projection neurons (CThPNs) are a heterogeneous group of neurons that target different nuclei of the thalamus, while subcerebral projection neurons (ScPNs) extend axons to multiple targets below the brain, most prominently connecting the cortex to the nuclei of the brainstem and the spinal cord (Figure 1c–d). ScPNs are also highly diverse. Their somas are in L5b (across different cortical areas) and different subgroups of ScPNs extend axons to distinct anatomical and functional targets. ScPNs include the corticospinal motor neurons (CSMNs) that connect to the spinal cord, the cortico-pontine PNs that connect to the brainstem motor nuclei and the corticotectal PNs that project to the superior colliculus (Figure 1d)[10].
Of note, some PNs send axons to multiple targets and they cannot be easily ascribed to one neuronal subtype. Among those are the ScPNs with backward projections, which extend axons to both subcerebral targets and to ipsilateral caudal cortex [12]; and the corticostriatal PNs, which are mainly present in L5 and project to the ipsilateral and contralateral striatum and also innervate the contralateral cortex (CStrPN IT-type, intratelencephalic type) [13].
Although classical schemes of nomenclature for PN classes directly build on anatomical parameters, like laminar location and axonal connectivity, it is clear that these only provide a basic framework to begin to classify PN diversity. PNs are distinct also by molecular identity, the presence of primary and collateral axonal connections, somatodendritic morphology, and electrophysiological properties. The molecular classification of anatomically identified PN classes is only beginning to be known. Several studies have purified and transcriptionally compared distinct PN subtypes, providing the first sets of class-specific genes [14–20]. To date, CSMNs and CPNs are amongst the neurons best defined at the molecular level. For example, Fezf2, Cntn6, Cad13, Bcl11b, Cry-mu, Ldb2 among others can be used to label CSMNs (and other ScPNs), while they are excluded from CPNs [14, 21]. Conversely, Cux2, Inhba, Btg1, Lpl, Cited2, PlexinD1 are among genes expressed in CPNs but not in CSMNs [17].
A few important lessons have emerged from these molecular studies. First, that each and every one of these markers present different degrees of restricted expression in any given projection neuron class and thus that only the combinatorial use of multiple genes can identify, specifically, one PN population versus the others. Second, the combination of genes that distinguish one PN class at a given point in time may not do so at another, indicating that signature profiles of gene expression for individual classes of PNs are temporally dynamic. Third, perhaps not surprisingly, transcript expression for marker genes does not always reflect protein distribution. For example, CTIP2 (Bcl11b), a commonly used marker for CFuPNs shows protein expression exclusively in postmitotic neurons, while its RNA is expressed in progenitors too ([14, 22] and unpublished data). This suggests the existence of regulatory mechanisms, possibly non-coding RNAs, that prevent transcript translation until progenitors give rise to neurons and it may reflect a strategy for generating large amount of proteins in a very short time. Finally, many of these new molecular markers label only subsets of the current classes of anatomically-defined neurons. This indicates that canonical classes of PNs are likely further subdivided into subclasses and that each neuronal population is per se heterogeneous. Single-cell transcriptional profiling of individual populations should in the near future help to clarify the measure of diversity within each class of PNs and, further, define the functional meaning of intra-population heterogeneity. This is a budding field of research enabled by the latest technology for molecular profiling of small population of cells, down to single neurons [23].
The elusive strategies employed to generate projection neuron diversity
Great research effort has been focused on determining the molecular regulatory grammar that orchestrates the generation of PN diversity in the embryo and on defining the cellular context where key molecular decisions of lineage fate specification take place. Classic [3H] thymidine labeling [24, 25] and more recent genetic studies (reviewed in [26]) have shown that cortical PNs are born in a specific temporal sequence from a pool of neural progenitor cells in the dorsal telencephalon. However, the strategy employed by progenitors to achieve this daunting task, together with the molecular nature of the decisions made specifically at the progenitor stage, remain a matter of debate. At the core of the problem lays the long-standing question of whether this stereotypic production of neurons is due to (i) a progressive, temporal restriction of progenitor fate, such that at any given point in time the choice of neurons that a pool of progenitors can generate is restricted and/or (ii) the existence of classes of progenitors pre-fated to generate specific neuronal subtypes.
Pioneering heterochronic transplantation studies demonstrated that early cortical progenitors are multipotent while late progenitors are unable to produce the earlier fates [27–29]. The work provided clear evidence that progenitor potential is progressively, temporally restricted. In agreement, lineage fate mapping- using retroviruses- showed that when a single progenitor is labeled early in corticogenesis it can give rise to neurons of all layers [30, 31]. Ex vivo studies by Sally Temple’s group further credited this model by showing that multipotent progenitors sequentially give rise to deep layer neurons first and upper layer neurons later, although observing the birth of all lineages in vitro from the same single progenitor has been challenging [32]. Similarly, directed differentiation of murine embryonic stem (ES) cells into cortical projection neuron-like cells points at least partly to a temporal pattern of sequential neuronal generation that matches what has been observed during corticogenesis in vivo [33, 34]. Thus, a large body of data collectively support, yet do not prove, the theory that all PNs may be generated from the same multipotent progenitors and that fate distinctions are mostly temporally controlled. This model has been recently challenged with the discovery of a fate restricted progenitor lineage (expressing the transcription factor Cux2), which largely produces callosal PNs of L2/3 [35]. In this study the authors used a Cux2-CreERT2 knock-in line to fate map cortical progenitors of the early VZ and found that a large proportion of these progenitors give rise to upper layer PNs. Cux2-Cre positive progenitors were present in the VZ as early as E10.5 and they mostly divided symmetrically (to replenish themselves) and more rarely asymmetrically (to generate neurons) during the window of time when CFuPNs are being produced. Notably, when forced to differentiate during production of deep layer neurons such progenitors still chose to generate upper layer neurons, suggesting fate commitment. These results challenge the long-held model that establishment of PN diversity relies only on multipotent progenitors able to temporally specify different classes of neurons and indicates that progenitor pre-fated to a specific PN identity may also play a central role.
This concept is exciting, although the data currently stand in apparent contrast to a second study [36] where comparable percentages of the progeny of Cux2+ progenitors (lineage-fated with the same Cux2-CreERT2 reporter line) expressed either the CFuPN deep layer marker CTIP2 or the upper layer marker CUX1, when analyzed at P0. It is difficult to exactly explain this apparent discrepancy of results. Things to consider may be the importance to analyze the class specific identity of fate-mapped neurons later than P0 (when neurons are still migrating and often share overlapping sets of markers), and the need to use retrograde labeling to define, beyond molecular markers, the class-specific identity of the neurons mapped. In addition, some of the canonical CFuPN markers, for example CTIP2, are also expressed at low levels in cortical interneurons, which are also labeled by Cux2. These are early days for molecular fate mapping of PN subtypes in the cortex and it is likely that a more definitive answer will come from integrating results from the use of multiple Cre lines and from labeling experiments that permanently “barcode” single progenitors and their neuronal progeny.
Initial work in this direction has used a transgenic BAC line driving CreERT2 from the Fezf2 locus to determine whether progenitors preferentially fated to a deep layer neuron identity exist in vivo [36]. In their first implementation these experiments appear to suggest that progenitors mapped by this line are multipotent, able to generate both different classes of neurons and glia. However, a cautionary note should accompany the use of BAC lines for this type of complex experiments. BACs often do not reproduce at the single-cell level the temporally and spatially-regulated expression of a given locus in vivo. In addition, variation in copy number and in integration sites within the BAC transgene can be a source of great animal-to-animal variability and influence the behavior of the targeted progenitors, respectively. Extension of this early work to include more driver loci and the use of knock-in Cre lines instead of transgenes should in the near future clarify these initial results.
Progenitors clearly play critical roles in specifying neuronal identities. However, several of the molecular decisions that shape projection neuron diversity occur outside of the germinal zones. Several transcription factors (TFs) important to control acquisition of PN class-specific traits are expressed in distinct classes of PNs postmitotically rather than at the progenitor stage [14–20]. In addition, it is known that reciprocal regulation between these postmitotic TFs is an element of the molecular strategy employed to achieve progressive refinement of neuronal subtype identity during corticogenesis [4, 37]. While a recent review has exhaustively covered the role of postmitotic determinants in PN development [4], here we highlight selected examples that relate to the acquisition of distinct aspects of PN identity.
The precise sequential generation of PN subtypes is critical to generate appropriate cortical architecture and connectivity, which requires multiple levels of regulation. Sox5 is one example of a TF expressed postmitotically in subplate (SP) neurons (the first neurons generated in the cortex) and CFuPNs that is required for their generation in the appropriate temporal order. In the absence of Sox5, SP neurons prematurely acquire ScPN characteristics (normally generated two days later), and CThPNs projections are severely compromised [38, 39].
The acquisition of appropriate PN class-specific identity within defined functional areas is also at least partly regulated by TFs postmitotically. Prime examples are Bhlb5 and Lmo4, which regulate area specific differentiation of CSMNs. In the absence of Bhlhb5, CSMNs from caudal motor cortex are not properly specified and fail to connect to the spinal cord [40] ; while in the absence of Lmo4, CSMNs in the rostral motor cortex lack backward projecting collaterals [12]. Another cardinal example of a TF acting postmitotically in PNs is Ctip2, which was one of the first TFs shown to control the lineage-specific axon extension and fasciculation decisions of ScPNs [14]. Finally, the chromatin remodeling protein Satb2 and its partner Ski [41–43] are also restricted to postmitotic stages of CPN development and are central to the generation of a normal complement of CPNs. In the absence of either Satb2 or Ski the majority of CPN axons fail to cross the corpus callosum and project instead ipsilaterally, to subcortical targets. Several subtype-specific molecular markers of CPNs are also not expressed in the absence of Satb2 [41–43].
Do selector genes for individual projection neuron classes exist in the mammalian cortex?
Elegant work in C. elegans and Drosophila has defined key transcription factors and decoded part of the molecular grammar that establishes and maintains neuronal diversity in the invertebrate nervous system (reviewed in [44]). In C. elegans, the establishment of neuronal diversity relies on a plethora of TFs that alone or in combination act as master selector genes. Are these rules and principles directly applicable to the mammalian CNS? Does the extreme neuronal diversity of the mammalian cerebral cortex rely on the use of selector genes for individual neuronal classes?
The logic governing the coordinated regulation of genes defining an individual neuronal class of the neocortex is not known; however at least one of such powerful TFs has been recently defined as a selector gene for CSMNs (and ScPNs more broadly): the TF Fezf2 (forebrain embryonic zinc finger 2). Fezf2 is necessary for the fate specification of CSMNs [45–47]. In the absence of Fezf2 subcerebral projection neurons, including all CSMNs, fail to generate. In agreement, CSMN-specific genes are not expressed in L5b of the Fezf2 mutant cortex, a deficiency accompanied by changes in dendritic morphology and a lack of axonal projections to the spinal cord [45–47]. Conversely, Fezf2 alone can cell-autonomously instruct the acquisition of CSMN-specific features when expressed in diverse, permissive cellular contexts, in vivo [21, 48–50].
Recent insight into the mechanisms of action of Fezf2 demonstrates that this gene embodies key properties of selector genes described in invertebrates. Fezf2 is sufficient to activate and repress a broad program of neuron subtype-specific genes, specifically promoting the expression of CSMN signature genes and repressing genes of an alternative neuronal fate (i.e. CPNs of L2/3 identity). Importantly, this occurs by direct binding to the proximal promoters of target genes followed by transcriptional regulation, and it includes control over expression of functionally relevant “effector” genes, able to orchestrate the acquisition of CSMN defining features. Both class-specific and pan-projection neuron genes necessary to “build” CSMNs are controlled by Fezf2. For example, Fezf2 directly instruct the expression of EphB1, a neuronal subtype-specific axon guidance receptor expressed in CSMNs, which in turn executes critical ipsilateral axon guidance decisions of the corticospinal tract [21]. This also indicates that the same transcription factor that instructs most other aspects of neuronal subclass identity of an individual PN type in the neocortex also directly controls the expression of class-specific axon guidance receptors necessary to wire the neurons to the correct long distance targets, without secondary activation of intermediate regulatory genes.
In invertebrates, selector genes have extensively been studied with regards to their ability to instruct and maintain terminal neuronal features, like class-specific neurotransmitter identity [44]. In mammals, these studies are more limited but evidence exists especially with regards to the acquisition of specific monoaminergic features. For example, in mouse midbrain dopaminergic (DA) neurons the transcription factor Nurr1 is necessary for the expression and maintenance of the genes for tyrosine hydroxylase (TH), the dopamine transporter (DAT) and vesicular monoamine transporter (VMAT2), and thus controls dopaminergic identity [51, 52]. Similarly, the homeodomain protein Lhx7 is necessary for the expression of choline acetyltransferase (and other class-specific genes) in cholinergic interneurons of the striatum, which are re-specified into Lhx6-positive, GABAergic interneurons when Lhx7 is ablated from young postmitotic neurons of the cholinergic lineage [53]. Not much is known regarding selection of terminal features for cortical neurons. In this regard it is interesting that Fezf2 induces the glutamatergic identity of CSMNs via direct activation of Vglut1 (Slc17a7) and other genes involved in the synthesis and signaling of glutamate, and inhibits a GABAergic fate by directly repressing transcription of Gad1. This can occur in vitro [21], but it is most notably true in vivo, where overexpression of Fezf2 in progenitors of GABAergic medium spiny neurons of the developing striatum results in a switch to a glutamatergic identity [48]. The data collectively indicates that orchestrated gene expression directly downstream of a common selector gene is one component of the regulatory logic responsible for the establishment of CSMN identity.
Is this principle true for other classes of projection neurons? It is hard to imagine a scenario where individual selector genes exist for each of the many classes of projections neurons that populate the mammalian cortex, although it is possible that a small number of these master transcription factors do exist and await discovery. It is likely that other regulatory mechanisms are in place to integrate selector gene functions and guarantee development, evolution and maintenance of this outstanding diversity of neuronal subtypes.
Reprogramming neuronal identity postnatally: a new route to enhance brain plasticity?
All neurons of the mammalian cerebral cortex are generated only during embryonic development, after which time neuronal class-specific, distinguishing traits remain unchanged for the life of the organism. When put in practical terms, this signifies that human neurons are capable of maintaining their class specific identity for a hundred years. How is this incredible task achieved? Are there permanent, irreversible changes that take place as neurons mature that preclude a change in identity imposed postnatally? Or, rather, is neuronal identity actively maintained and thus amenable to change?
Understanding of the mechanisms that maintain neuronal identity in mammalian neurons is in its infancy. Once again, work in invertebrates indicates that expression of key developmental transcription factors need to be maintained into adulthood in order for neurons to keep class-specific properties [54, 55]. It has been shown that sustained expression of such terminal transcription factors is achieved via direct autoregulation, a common strategy by which postmitotic neurons “lock-in” their subtype identity [55]. Much less is known about how neurons preserve their identity in the mammalian CNS. Examples mostly come from the monoaminergic system [54] and the retina [56]. In serotonergic (5-HT) neurons, postmitotic removal of TFs required for the acquisition of serotonergic fate during development (e.g. Lmx1b, Gata-3 or Pet-1) compromises the expression of genes essential to retain aspects of neurotransmitter identity [57, 58]. Similarly, the transcription factor Nurr1 is necessary for midbrain dopaminergic neurons to maintain terminal features such as dopaminergic identity and for expression of some class specific genes [51, 52]. In the retina, it is notable the role of the TF Nrl, a gene critical for the developmental specification of rods over cones [59, 60]. In this case, conditional removal of Nrl in the adult rod photoreceptors not only results in the loss of identity, but it is sufficient to instruct reprogramming of rods into cones [56]. This suggests a dual role of Nrl in the maintenance of rod identity, simultaneously promoting rod traits and suppressing the alternative cone fate. Therefore terminal neuronal identity in mammalian neurons may at least partly be maintained via ‘active’ mechanisms of transcriptional regulation, like in invertebrates. Whether similar conclusions can be applied to neurons of other regions of the mammalian brain, most notably the cerebral cortex, is currently unknown.
Some evidence that no irreversible genetic or epigenetic changes preclude reprogramming of neuronal identity also came from experiments where the nuclei of some neuronal classes (i.e. neurons from the olfactory epithelium) could support the development of an entire mouse upon somatic cell nuclear transfer into enucleated eggs [61]. Intriguingly though, the same reversion to pluripotency has been much harder to achieve when starting from cortical neurons [62], possibly reflecting different plasticity by different neurons.
Do neurons of the cortex loose the ability to convert from one class into another once fate specified? Are they different (i.e. less plastic) than the plethora of other differentiated cell types that could be successfully reprogrammed into other cell classes by potent transcription factor cocktails (reviewed in [63])? We still do not know whether neurons of the adult cerebral cortex (and for that matter from any region of the mammalian CNS) can be directly reprogrammed from one class into another. However, recent evidence demonstrates that differentiated projection neurons are more plastic than previously thought. Ectopic overexpression of Fezf2, able to select directly multiple features of identity of CSMNs [21, 48] when expressed in a plastic cellular context, was also sufficient to directly reprogram postmitotic callosal projection neurons of L2/3 and stellate glutamatergic interneurons of L4 [49, 50] into CFuPNs, in vivo. This shows that the postmitotic nature of neurons does not per se preclude reprogramming. However, neuronal nuclear plasticity progressively declines over the first postnatal weeks, and reprogramming capabilities in response to Fezf2 have exhausted by P21 [50]. This progressive loss of ability to reprogram parallels what was observed in the retina where the ability of rods to reprogram into cones decreases sharply with age [56], suggesting that additional levels of regulation take place later during neuronal maturation, presumably for the ultimate safeguarding of specific circuit function.
Impact of pyramidal neuron diversity on the behavior of cortical neurons and glia
Emerging data seem to point at a central role for distinct projection neuron classes in affecting the behavior of other cell types in the cerebral cortex. Functional maps have shown that distinct projection neurons choose highly-selective synaptic connectivity within the same local circuits [64]. The pattern of connectivity shown by different classes of neighboring PNs reflects the identity of both the pre- and post-synaptic cell types, as demonstrated by simultaneous whole-cell recording of multiple projection neuron types within L5 across different cortical areas [64, 65]. In the visual cortex, for example, cortico-cortical neurons show significantly higher preference to connect with their neighboring corticotectal neurons than with each other [64]. Similar results were obtained in the frontal cortex with paired recordings of retrogradely labeled corticopontine neurons, where these neurons make more numerous excitatory inputs onto cells that share the same long-range axonal target than onto those that project ipsilaterally [65]. Together these results support a model by which the specific identity of PNs influences the nature of the local excitatory subnetworks.
Recent studies on inhibitory cortical networks have also shown that both the choice of the postsynaptic target of inhibitory interneurons and the properties of their synaptic connections, at least in some areas of the cortex, depend on the identity of their projection neuron partners. In the prefrontal cortex, fast-spiking parvalbumin-positive interneurons preferentially inhibit ScPNs over the adjacent CPNs within layer 5 [66]. Similarly, in the medial entorhinal cortex (MEC), inhibitory basket cells (CCK-positive) selectively innervate a specific class of PNs (projecting to the contralateral EC), while avoiding neighboring neurons projecting to the ipsilateral dentate gyrus [67]. In addition to the choice of synaptic partners, the strength of inhibitory networks is also influenced by the identities of the PN partners. Callosal PNs receive a significantly greater number of inhibitory inputs onto their initial axonal segment by chandelier cell than corticothalamic neurons [68, 69]. The mounting evidence that PN diversity imparts a certain level of specificity to the wiring of the local inhibitory network is in agreement with the finding that PN subtypes affect the radial distribution into layers of cortical interneurons during development, and that this effect is a function of the class-specific identity of the projection neurons involved [70, 71].
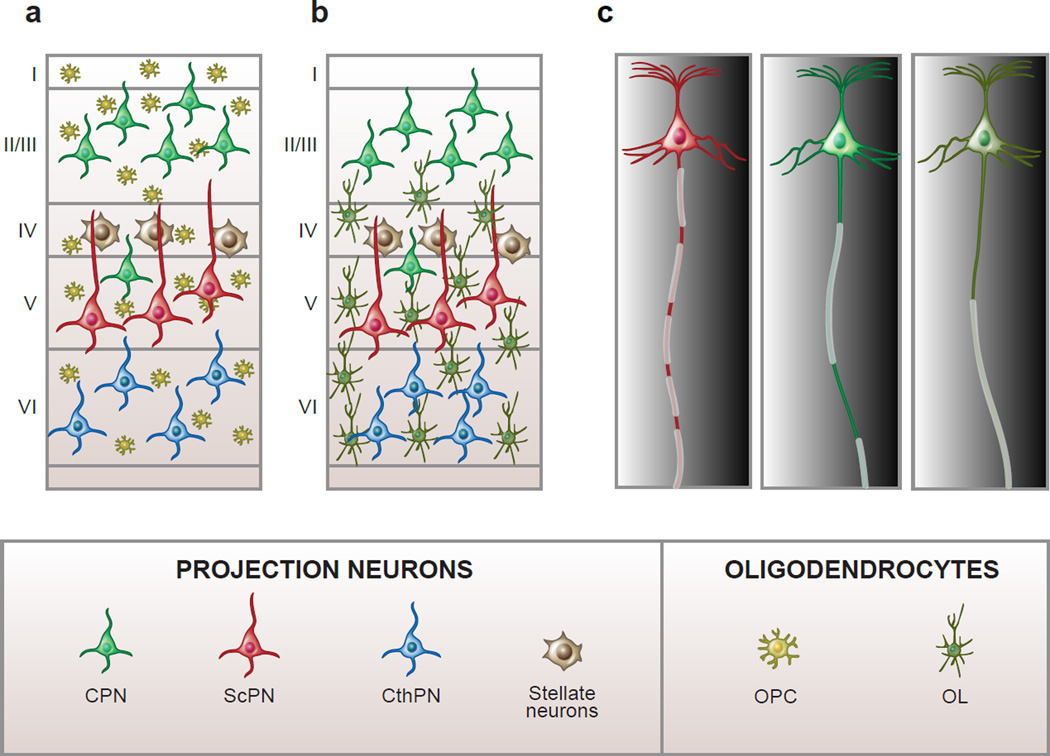
Do projection neurons also influence the behavior of non-neuronal cell types in the cortex? Some data point at such an effect. It is very recent the discovery that PNs in different layers display distinct profiles of myelin distribution along their axons, suggesting an effect of projection neuron identity on the behavior of oligodendrocytes [72] (Figure 2). A novel pattern of myelination termed “intermittent myelin” was found only in L2/3 projection neurons, which display an alternation of myelinated and unmyelinated tracts of variable lengths. In contrast, CFuPNs in L5 and L6 predominantly showed classic profiles of uninterrupted longitudinal myelin segments separated only by small nodes of Ranvier. These results indicate that longitudinal myelin deposition is a defining feature of each neuron and suggests that its establishment reflects idiosyncratic interactions between projection neurons and oligodendrocytes.
Figure 2. Projection neurons have distinct profiles of longitudinal myelination.
In the mammalian neocortex oligodendrocyte progenitors (OPCs) are evenly distributed across all layers (a), but oligodendrocytes (OLs) show preferential distribution in the deep layers (b), reflecting higher levels of myelin in L5 and L6. Cortical PNs display diverse myelination patterns along their axons. At least three types of myelination profiles exist in the mouse neocortex (in addition to axons that are not myelinated) (c). Some projection neurons have axons that are myelinated throughout their entire length with short un-myelinated nodes of Ranvier, others display myelinated segments intercalated with myelinated tracts of different lengths (intermittent myelin). Finally, selected neurons have axons with a long unmyelinated tract between the axon hillock and the first internode (c). The two latter patterns of myelin distribution are found preferentially in PNs of the upper layers, suggesting that myelination patterns may be an integral feature of neuronal class-specific identity. Abbreviations: CPN, callosal PN; CthPN, corticothalamic PN; ScPN, subcerebral PN.
The data support an emerging model in which differentiation of projection neuron diversity impacts the behavior of other cells in the cortex (both neurons and glia) to ultimately shape working circuit, allow cortical diversification, and sustain complex behavior.
Concluding Remarks and Perspective
The mammalian cerebral cortex contains an unparalleled diversity of neurons, which has dramatically increased over the course of evolution. The principles and rules that shape this diversity in the embryo, how this process goes wrong in disease, and whether the landscape of developmentally-generated neuronal subtypes can be changed in the adult, are active areas of investigation. Lots of questions still remain unanswered. What strategies are used to generate PN diversity and what is the role of progenitors? Is cortical neuronal diversity in mammals built using similar strategies as in invertebrates? How plastic do cortical neurons remain postnatally and could adult neurons be changed, paving new routes to enhance cortical plasticity? For as difficult as these questions remain, work of the last decade has provided novel molecular substrates to define and push the boundaries of neuronal diversity in the mammalian cerebral cortex, priming the next decade for exciting new answers.
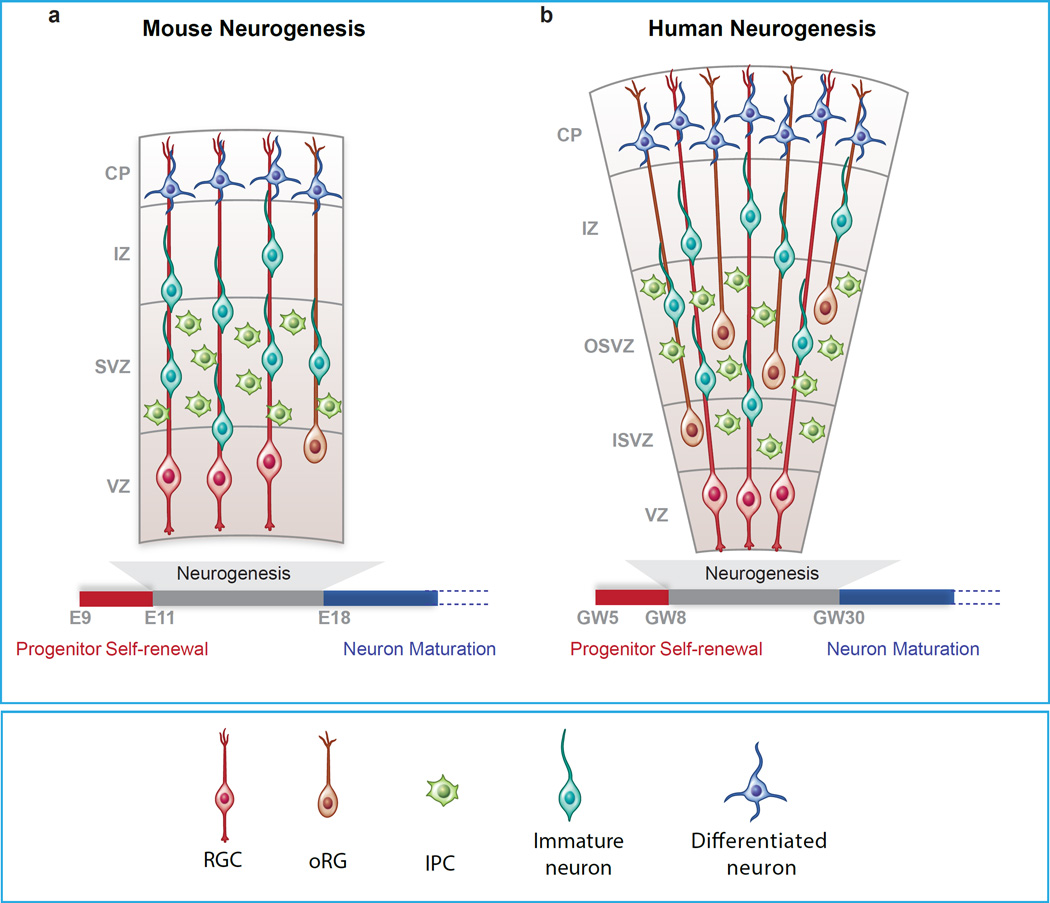
Figure I (Box1). Progenitors of the cerebral cortex in mice and humans.
Different types of progenitors are depicted for mouse (a) and human cortex (b). Time scale of neurogenesis is measured in embryonic days (E) for mice and in gestational week (GW) for humans. Abbreviations: VZ, Ventricular zone; SVZ, Sub-ventricular zone; OSVZ, Outer Sub-ventricular zone; ISVZ, Inner Sub-ventricular zone; IZ, Intermediate zone; CP, Cortical plate; RGC, Radial glial cell; oRG, Outer radial glial cell; IPC, Intermediate precursor cell. The images are not to scale.
BOX 1 Progenitors of the cerebral cortex in mice and humans.
In mice, after neural tube closure, neuroepithelial (NE) cells with stem cell-like properties initially divide symmetrically to expand the progenitor pool and later differentiate into more restricted progenitors called radial glial cells (RGCs), which are bipolar cells with radial fibers contacting the apical ventricular zone and the pial surface. RGCs serve as a scaffold for neuronal migration, and they are also multipotent progenitor cells able to generate neurons, astrocytes and oligodendrocytes [73, 74]. At the onset of neurogenesis, the majority of RGCs exhibit asymmetric divisions in the ventricular zone (VZ) to produce an RGC daughter cell and either a neuron or an Intermediate Precursor Cell (IPC) [75]. IPCs then migrate basally to form the Subventricular Zone (SVZ) where they further divide symmetrically to give rise to 2 to 4 neurons [76–78]. The progenitor composition of the human developing cortex is more complex. One key distinction of the SVZ of humans (and that of primates, more broadly) is that in addition to increased numbers of IPCs it contains an expanded new population of progenitor cells named outer Radial Glia (oRG), which lack apical contacts but retain a basal process to pia [79]. Interestingly oRGs are also present in mice but at a very low frequency [80]. A striking difference between oRGs in humans and mice is that murine oRGs directly produce neurons by symmetric division while oRGs in humans divide asymmetrically to self-renew and generate a self-amplifying IPC, which then generate neurons [79–81]. These cells might contribute to the increased number and tangential dispersion of human neurons and to cortical folding (reviewed in [82]). Recent studies in primates have also shown that in addition to IPCs at least 4 different types of oRG cells are present in the SVZ, contributing to increased progenitor diversity [83].
Box 2: Outstanding Questions.
Many pivotal questions remain in the field regarding the principles that define, generate and maintain neuronal diversity in the mammalian cerebral cortex. Among others:
Which criteria should be taken into account to classify neurons in order to understand the true extent of neuronal diversity in the neocortex? Single-cell profiling of large number of cortical cells in high-throughput will soon provide the field with a massive amount of information on the molecular identity of each cell. This will bring about the challenge of mining the data to recognize the existence of new neuronal types as distinct from simply new neuronal states (e.g. a change in molecular composition that reflects a transitory molecular response to stimuli).
What is the relationship between progenitor and neuronal diversity? Lots of questions remain regarding the strategies used by progenitors to generate the large number of cortical projection neurons found in the mammalian cortex. It is likely that experimental strategies involving barcoding of individual progenitors and permanent labeling of their neuronal progeny will contribute to clarify these lineage relationships.
How is neuronal diversity preserved in the adult cerebral cortex? Is there a “unified” molecular strategy or does each neuronal subtype “lock-in” its identity in its own manner? Learning the developmental logic that builds neuronal diversity will certainly inform on mechanisms that may be at play to maintain class-specific traits in the adult. In addition, it will be critical to understand how much environmental factors and experience contribute to preserve neuronal identity unchanged.
To which extent can adult neurons change their identity under the appropriate signals? The next few years will see a surge in experiments aimed at probing the capacity of adult neurons to acquire new traits and functions. In addition, studies from the developmental interactions among different types of neurons and glia points at the exciting prospect of using neuronal reprogramming to enhance neuroplasticity in vivo.
Highlights.
The cerebral cortex contains many sub-types of excitatory and inhibitory neurons
Excitatory (projection) neurons (PNs) send axons to targets within and outside cortex
Several mechanisms responsible for generating PN diversity
Mature PNs retain ability to reprogram class-specific features in vivo
In vivo reprogramming of PN identity could represent novel substrate for plasticity
Acknowledgments
We would like to thank Hsu-Hsin Chen for her insightful comments and critical reading of the manuscript. We are also very grateful to Dennis Sun for his help with the illustrations. This work was supported by grants from the US National Institute of Health (NS062849, MH101268 to P.A.), the New York Stem Cell Foundation, and the Harvard Stem Cell Institute to P.A.; P.A. is a New York Stem Cell Foundation-Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Golgi C. Sulla Fina Anatomia Degli Organi Centrali del Sistema Nervoso. 1886 [Google Scholar]
- 2.Cajal SRy. Histologie du Systéme Nerveaux de l’Homme et des Vertebres. 1911 [Google Scholar]
- 3.No RLd. Cerebral Cortex: Architecture, Intracortical Connections, Motor Projections. Oxford university press; 1949. [Google Scholar]
- 4.Greig LC, et al. Molecular logic of neocortical projection neuron specification, development and diversity. Nature reviews. Neuroscience. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson SA, et al. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 7.DeFelipe J, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nature reviews. Neuroscience. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marin O, Muller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Current opinion in neurobiology. 2014;26:132–141. doi: 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migliore M, Shepherd GM. Opinion: an integrated approach to classifying neuronal phenotypes. Nature reviews. Neuroscience. 2005;6:810–818. doi: 10.1038/nrn1769. [DOI] [PubMed] [Google Scholar]
- 10.Molyneaux BJ, et al. Neuronal subtype specification in the cerebral cortex. Nature reviews. Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 11.Aboitiz F, Montiel J. One hundred million years of interhemispheric communication: the history of the corpus callosum. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas / Sociedade Brasileira de Biofisica … [et al.] 2003;36:409–420. doi: 10.1590/s0100-879x2003000400002. [DOI] [PubMed] [Google Scholar]
- 12.Cederquist GY, et al. Lmo4 establishes rostral motor cortex projection neuron subtype diversity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:6321–6332. doi: 10.1523/JNEUROSCI.5140-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shepherd GM. Corticostriatal connectivity and its role in disease. Nature reviews. Neuroscience. 2013;14:278–291. doi: 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arlotta P, et al. Neuronal Subtype-Specific Genes that Control Corticospinal Motor Neuron Development In Vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molyneaux BJ, et al. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belgard TG, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard A, et al. Transcriptional architecture of the primate neocortex. Neuron. 2012;73:1083–1099. doi: 10.1016/j.neuron.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoerder-Suabedissen A, et al. Expression profiling of mouse subplate reveals a dynamic gene network and disease association with autism and schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3555–3560. doi: 10.1073/pnas.1218510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodato S, et al. Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nature neuroscience. 2014;17:1046–1054. doi: 10.1038/nn.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leid M, et al. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expression Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baslan T, Hicks J. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev. 2014;26C:59–65. doi: 10.1016/j.gde.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 25.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 26.Kohwi M, Doe CQ. Temporal fate specification and neural progenitor competence during development. Nature reviews. Neuroscience. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- 28.Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/s0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 29.Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- 30.Luskin MB, et al. Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron. 1988;1:635–647. doi: 10.1016/0896-6273(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 31.Walsh C, Cepko CL. Clonally related cortical cells show several migration patterns. Science. 1988;241:1342–1345. doi: 10.1126/science.3137660. [DOI] [PubMed] [Google Scholar]
- 32.Shen Q, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nature neuroscience. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 33.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Gaspard N, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 35.Franco SJ, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo C, et al. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80:1167–1174. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srinivasan K, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwan KY, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai T, et al. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 40.Joshi PS, et al. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II–V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 43.Baranek C, et al. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3546–3551. doi: 10.1073/pnas.1108718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nature neuroscience. 2014;17:899–907. doi: 10.1038/nn.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molyneaux BJ, et al. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Chen B, et al. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen JG, et al. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouaux C, Arlotta P. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nature neuroscience. 2010;13:1345–1347. doi: 10.1038/nn.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De la Rossa A, et al. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nature neuroscience. 2013;16:193–200. doi: 10.1038/nn.3299. [DOI] [PubMed] [Google Scholar]
- 50.Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nature cell biology. 2013;15:214–221. doi: 10.1038/ncb2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadkhodaei B, et al. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadkhodaei B, et al. Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2360–2365. doi: 10.1073/pnas.1221077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopes R, et al. Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3119–3124. doi: 10.1073/pnas.1109251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flames N, Hobert O. Transcriptional control of the terminal fate of monoaminergic neurons. Annual review of neuroscience. 2011;34:153–184. doi: 10.1146/annurev-neuro-061010-113824. [DOI] [PubMed] [Google Scholar]
- 55.Hobert O. Regulation of terminal differentiation programs in the nervous system. Annual review of cell and developmental biology. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- 56.Montana CL, et al. Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proceedings of the National Academy of Sciences. 2013;110:1732–1737. doi: 10.1073/pnas.1214387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao ZQ, et al. Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. Journal of Neuroscience. 2006;26:12781–12788. doi: 10.1523/JNEUROSCI.4143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu C, et al. Pet-1 is required across different stages of life to regulate serotonergic function. Nature neuroscience. 2010;13:1190–1198. doi: 10.1038/nn.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mears AJ, et al. Nrl is required for rod photoreceptor development. Nature genetics. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 60.Montana CL, et al. Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. The Journal of biological chemistry. 2011;286:36921–36931. doi: 10.1074/jbc.M111.279026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eggan K, et al. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- 62.Yamazaki Y, et al. Assessment of the developmental totipotency of neural cells in the cerebral cortex of mouse embryo by nuclear transfer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14022–14026. doi: 10.1073/pnas.231489398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amamoto R, Arlotta P. Development-inspired reprogramming of the mammalian central nervous system. Science. 2014;343:1239882. doi: 10.1126/science.1239882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown SP, Hestrin S. Intracortical circuits of pyramidal neurons reflect their long-range axonal targets. Nature. 2009;457:1133–1136. doi: 10.1038/nature07658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morishima M, et al. Highly differentiated projection-specific cortical subnetworks. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:10380–10391. doi: 10.1523/JNEUROSCI.0772-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee AT, et al. Pyramidal neurons in prefrontal cortex receive subtype-specific forms of excitation and inhibition. Neuron. 2014;81:61–68. doi: 10.1016/j.neuron.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varga C, et al. Target-selective GABAergic control of entorhinal cortex output. Nature neuroscience. 2010;13:822–824. doi: 10.1038/nn.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farinas I, DeFelipe J. Patterns of synaptic input on corticocortical and corticothalamic cells in the cat visual cortex. II. The axon initial segment. The Journal of comparative neurology. 1991;304:70–77. doi: 10.1002/cne.903040106. [DOI] [PubMed] [Google Scholar]
- 69.Farinas I, DeFelipe J. Patterns of synaptic input on corticocortical and corticothalamic cells in the cat visual cortex. II. The cell body. The Journal of comparative neurology. 1991;304:53–69. doi: 10.1002/cne.903040105. [DOI] [PubMed] [Google Scholar]
- 70.Hevner RF, et al. Postnatal shifts of interneuron position in the neocortex of normal and reeler mice: evidence for inward radial migration. Neuroscience. 2004;124:605–618. doi: 10.1016/j.neuroscience.2003.11.033. [DOI] [PubMed] [Google Scholar]
- 71.Lodato S, et al. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomassy GS, et al. Distinct profiles of myelin distribution along single axons of pyramidal neurons in the neocortex. Science. 2014;344:319–324. doi: 10.1126/science.1249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malatesta P, et al. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 74.Anthony TE, et al. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 75.Pontious A, et al. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 76.Haubensak W, et al. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 78.Noctor SC, et al. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 79.Hansen DV, et al. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, et al. A new subtype of progenitor cell in the mouse embryonic neocortex. Nature neuroscience. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.LaMonica BE, et al. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nature communications. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nature reviews. Neuroscience. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Betizeau M, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]