Abstract
We examined the evolutionary origins of the ether-à-go-go (EAG) family of voltage-gated K+ channels, which have a strong influence on the excitability of neurons. The bilaterian EAG family comprises three gene subfamilies (Eag, Erg and Elk) distinguished by sequence conservation and functional properties. Searches of genome sequence indicate that EAG channels are metazoan specific, appearing first in ctenophores. However, phylogenetic analysis including two EAG family channels from the ctenophore Mnemiopsis leidyi indicates that the diversification of the Eag, Erg and Elk gene subfamilies occurred in a cnidarian/bilaterian ancestor after divergence from ctenophores. Erg channel function is highly conserved between cnidarians and mammals. Here we show that Eag and Elk channels from the sea anemone Nematostella vectensis (NvEag and NvElk) also share high functional conservation with mammalian channels. NvEag, like bilaterian Eag channels, has rapid kinetics, whereas NvElk activates at extremely hyperpolarized voltages, which is characteristic of Elk channels. Potent inhibition of voltage activation by extracellular protons is conserved between mammalian and Nematostella EAG channels. However, characteristic inhibition of voltage activation by Mg2+ in Eag channels and Ca2+ in Erg channels is reduced in Nematostella because of mutation of a highly conserved aspartate residue in the voltage sensor. This mutation may preserve sub-threshold activation of Nematostella Eag and Erg channels in a high divalent cation environment. mRNA in situ hybridization of EAG channels in Nematostella suggests that they are differentially expressed in distinct cell types. Most notable is the expression of NvEag in cnidocytes, a cnidarian-specific stinging cell thought to be a neuronal subtype.
KEY WORDS: Ether-à-go-go, Potassium channel, Nematostella, Mnemiopsis
INTRODUCTION
Sub-threshold voltage-gated K+ channels have a strong influence on neuronal excitability because they prevent small stimuli from eliciting action potentials. Inward currents produced by excitatory stimuli must overcome outward K+ flux through these channels to effectively excite a neuron. Therefore, the responsiveness of a neuron to incoming stimuli can be tuned by regulation of sub-threshold K+ conductance. Ether-à-go-go (EAG) family K+ channels make significant contributions to sub-threshold K+ currents in mammalian neurons, but their specific physiological roles are not yet fully understood. Genetic deletion or pharmacological block of various EAG family channels demonstrates that they have significant influence on excitation threshold. For instance, knockout of the EAG family K+ channel Kv12.2 in mice reduces sub-threshold K+ current and lowers action potential threshold in hippocampal pyramidal neurons (Zhang et al., 2010). Both knockout of Kv12.2 (Elk2) in mice and a gain-of-function mutation in Kv10.2 (Eag2) in humans cause seizures (Yang et al., 2013; Zhang et al., 2010), and knockout of Eag1 in mouse causes generalized hyperexcitability (Ufartes et al., 2013). Similarly, pharmacologic block of Erg (Eag-related gene) channels enhances the excitability of neurons in several brain regions (Hardman and Forsythe, 2009; Hirdes et al., 2009; Hirdes et al., 2005; Ji et al., 2012; Niculescu et al., 2013). A role for Eag and Erg channels in the regulation of neuromuscular excitation appears conserved in Drosophila and Caenorhabditis elegans (Collins and Koelle, 2013; Garcia and Sternberg, 2003; LeBoeuf and Garcia, 2012; Srinivasan et al., 2012; Titus et al., 1997), but the function of Elk (Eag-like K+ channel) has not yet been examined in invertebrate model systems.
EAG family channels have a unique subunit structure consisting of a cytoplasmic eag domain with Per–Arnt–Sim domain homology (PAS) at the N-terminus, a typical six-transmembrane domain voltage-gated K+ channel core, followed by a cytoplasmic gating domain homologous to cyclic-nucleotide binding domains (CNBHDs) (Ganetzky et al., 1999; Morais Cabral et al., 1998). The eag domain and CNBHD both regulate channel kinetics (Gianulis et al., 2013; Gustina and Trudeau, 2013; Haitin et al., 2013; London et al., 1997; Morais Cabral et al., 1998), but the mechanisms are not fully understood. For instance, cyclic nucleotides are not ligands for the CNBHD of EAG channels (Brelidze et al., 2009; Brelidze et al., 2012). The EAG gene family is comprised of three separate subfamilies classified by high intra-family sequence conservation: Eag, Erg and Elk (Ganetzky et al., 1999; Jegla et al., 2009). Each subfamily is present in vertebrates and protostome invertebrates, and members of each family have been identified in the genome of the starlet sea anemone Nematostella vectensis (Jegla et al., 2009; Martinson et al., 2014; Putnam et al., 2007), pointing to an origin prior to the cnidarian–bilaterian divergence.
Each subfamily has distinctive functional properties, suggesting that they evolved to serve separate physiological roles. Evidence that the Eag, Elk and Erg subfamilies are functionally independent includes the finding that although voltage-gated K+ channels are tetrameric, co-assembly of subunits from distinct subfamilies does not occur (Wimmers et al., 2001; Zou et al., 2003). We have previously expressed Nematostella (sea anemone) Erg channel paralogs and demonstrated that an inactivating IKr-like phenotype of mammalian Erg channels is the likely functional phenotype of Erg channels in the cnidarian–bilaterian ancestor (Martinson et al., 2014). Erg channels are specialized for delayed repolarization of broad action potentials and/or regulation of excitation threshold (Garcia and Sternberg, 2003; LeBoeuf and Garcia, 2012; Martinson et al., 2014; Sanguinetti and Tristani-Firouzi, 2006; Titus et al., 1997). Mammalian Erg1 plays a critical role in the repolarization of cardiac action potential plateaus and loss-of-function mutations in humans are a significant cause of long QT syndrome, which is characterized by delayed repolarization of cardiac action potential (Sanguinetti and Tristani-Firouzi, 2006).
The function of Eag and Elk subfamily channels has not been examined outside the bilaterian lineage, so the functional evolution of these subfamilies is less well understood. Bilaterian Eag channels display comparatively rapid gating and a pronounced Mg2+ sensitivity of activation kinetics (Terlau et al., 1996). Mg2+ sensitivity of Eag channels is primarily conferred by acidic residues in the voltage sensor (Silverman et al., 2000) that are unique to the EAG family of K+ channels, but are shared with hyperpolarization-gated cation channels (HCNs) and cyclic-nucleotide-gated cation channels (CNGs). Erg channel kinetics are sensitive to both Mg2+ and Ca2+ (Fernandez et al., 2005; Jo et al., 1999), whereas Elk channels are insensitive to these ions, but are inhibited by Zn2+ (Zhang et al., 2009). Protons inhibit voltage-dependent activation of all EAG family channels examined so far through protonation of these same EAG family acidic residues (Kazmierczak et al., 2013) and compete with divalent cations for the binding site (Kazmierczak et al., 2013; Terlau et al., 1996). Only vertebrate Elk channels have been expressed to date; they share a hyperpolarized activation threshold but variable inactivation, making the ancestral gating phenotype less clear (Engeland et al., 1998; Trudeau et al., 1999; Zou et al., 2003).
In this study, we examine the functional evolution of Eag and Elk channels by characterization of the currents produced by cloned and expressed Nematostella Eag and Elk channels. We used the diagnostic subunit structure and sequence conservation patterns to identify and classify EAG family channels in early branching metazoan lineages. Examination of metazoan and choanoflagellate genomes and phylogenetic analysis suggests that the EAG family is metazoan-specific and that the Eag, Elk and Erg channels first evolved in a common ancestor of cnidarians, placozoans and bilaterians, after the split from ctenophores and sponges. We found that the biophysical properties of Eag and Elk currents, like Erg, are conserved between Nematostella and bilaterians suggesting conservation of function. Expression patterns of EAG family members in Nematostella are diverse, indicating that the channels are important to a wide variety of physiological processes. These results suggest that the functional diversification of sub-threshold EAG family K+ channels continued after the evolution of the first nervous systems, but was largely complete before the cnidarian–bilaterian divergence.
RESULTS
Phylogenetic analysis of the EAG family
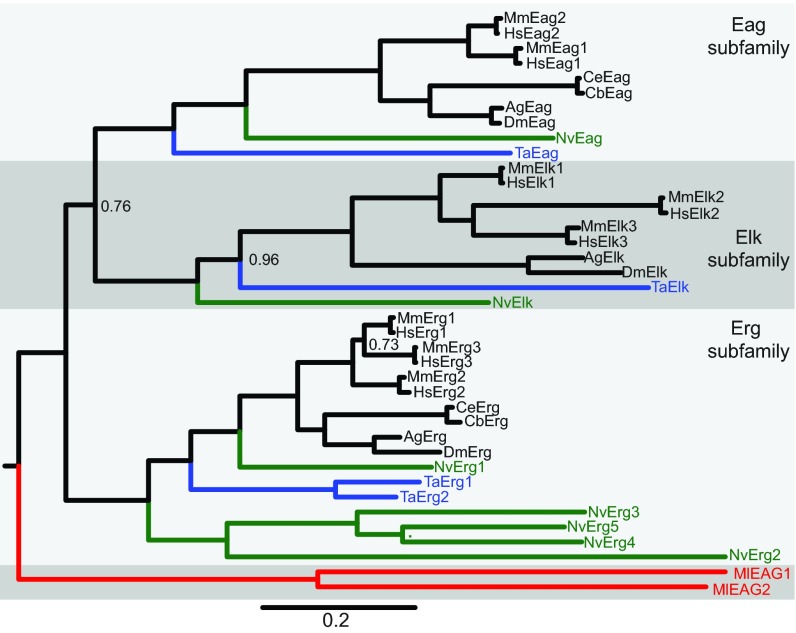
In order to reconstruct the evolution of the EAG family, we sought to identify putative EAG family members in phylogenetically informative extant taxa and use those sequences to build a phylogeny of the EAG gene family. We first used a BLAST search strategy to look for Eag, Elk and Erg subfamily channels in genome drafts and gene annotations from cnidarians (Nematostella vectensis) (Putnam et al., 2007), placozoans (Trichoplax adhaerens) (Srivastava et al., 2008), ctenophores (Mnemiopsis leidyi) (Ryan et al., 2013) and sponges (Amphimedon queenslandica) (Srivastava et al., 2010). We also searched for EAG family members in the choanoflagellates Monosiga brevicollis and Salpingoeca rosetta, because choanoflagellates are believed to be the most closely related outgroup to the metazoans (Fairclough et al., 2013; King et al., 2008). We previously used this strategy to identify five Erg family channels from Nematostella and two Erg family channels from Trichoplax (Martinson et al., 2014). Most EAG family channels were readily identified by the presence of the unique combination of the eag domain, a voltage-gated K+ channel core and CNBHD. Furthermore, EAG family channels from these species had significantly higher sequence similarity to confirmed bilaterian EAG family channels than any other proteins in reciprocal BLAST searches. Some Erg channels in Nematostella have lost the eag domain, but could still be readily identified by specifically high sequence similarity to bilaterian Erg channels (Martinson et al., 2014). We identified two putative divergent EAG channels in the ctenophore Mnemiopsis leidyi and putative Elk and Eag subfamily channels in both Nematostella and Trichoplax, but did not find EAG family channels in sponge or choanoflagellates by subunit structure or sequence homology. We verified the Nematostella and Mnemiopsis channels by cloning of full-length open reading frames from cDNA samples. Coding sequences, amino acid translations and GenBank accession numbers are provided in supplementary material Table S1. We built a Bayesian inference phylogeny of metazoan EAG family proteins, including the sequences from Nematostella, Trichoplax and Mnemiopsis (Fig. 1). We found that our putative Nematostella and Trichoplax Eag and Elk family members group strongly within each of these subfamilies (Fig. 1). The two Mnemiopsis EAG genes form an out-group to the Eag, Elk and Erg subfamilies (Fig. 1, red branch). We next searched an additional ctenophore genome, Pleurobrachia bachei (Moroz et al., 2014), and five diverse ctenophore transcriptomes, including Mnemiopsis and Pleurobrachia (Moroz et al., 2014; Ryan et al., 2013) with bilaterian, cnidarian and Mnemiopsis EAG family channels. Each contained fragments of highly conserved specific orthologs of the two Mnemiopsis EAG channels, but we found no evidence for the existence of other EAG family channels (data not shown). Furthermore, we found no evidence for EAG family genes in identical BLAST searches of eight diverse sponge transcriptomes (Riesgo et al., 2014). Because we found no EAG genes encoded in the choanoflagellate genome and the EAG genes that appear to predate the Eag, Erg and Elk subfamilies in the ctenophores, we suggest that the EAG family evolved specifically in the metazoans prior to divergence of ctenophores. Our data also strongly suggest that diversification of the Eag, Erg and Elk subfamilies occurred after the emergence of sponges and ctenophores, but prior to the divergence of placozoans, cnidarians and bilaterians.
Fig. 1.
Phylogeny of the metazoan EAG K+ channel family. A Bayesian inference phylogeny of the EAG K+ channel family including sequences from bilaterians (black), cnidarians (green), placozoans (blue) and ctenophores (red). The phylogeny is midpoint rooted for display and the Eag, Elk and Erg family branches are labeled and differentially shaded. Ctenophore EAG channels branch separately from the Eag, Elk and Erg subfamilies, whereas cnidarian and placozoan EAG channels are placed within the subfamilies. The scale bar indicates substitutions/site and node posterior probabilities of less than 1 are indicated. Most nodes had a posterior probability of 1, indicating a strong support for the phylogeny. All sequences used in the phylogeny are provided in supplementary material Table S2, with database links to sequence resources. Species prefixes are as follows: Ag, Anopheles gambiae (mosquito); Cb, Caenorhabditis briggsae (nematode); Ce, Caenorhabditis elegans (nematode); Dm, Drosophila melanogaster (fruit fly); Hs, Homo sapiens (human); Ml, Mnemiopsis leidyi (ctenophore); Mm, Mus musculus (mouse); Nv, Nematostella vectensis (sea anemone); Ta, Trichoplax adhaerens (placozoan).
Analyzing mRNA expression patterns for EAG genes in Nematostella vectensis
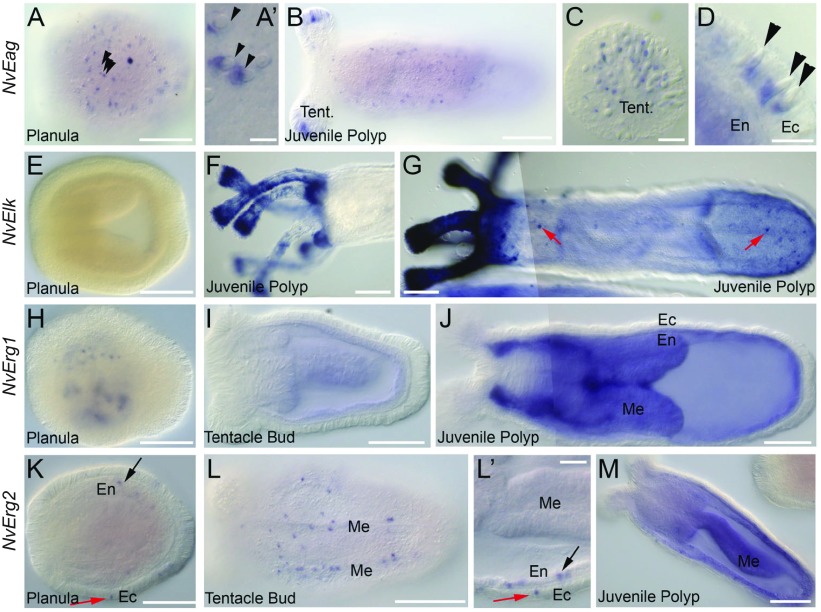
To determine which cell types might express EAG family members in non-bilaterian animals we performed mRNA in situ hybridization against the Nematostella EAG family genes NvEag, NvElk, NvErg1 and NvErg2 (Fig. 2). NvEag expression was first detected in a salt and pepper pattern within the ectoderm of the planula larva (Fig. 2A, arrowheads). Ectodermal salt and pepper patterns in Nematostella larva probably represent expression in neurons, gland cells or cnidocytes (cnidarian-specific ‘stinging’ cells). The cnidocysts (the harpoon-containing capsules of cnidocytes) are readily visible under DIC optics. We always observed NvEag expression in a pattern that is in direct contact with the cnidocyte capsule (Fig. 2A′, arrowheads) suggesting that NvEag is expressed specifically in cnidocytes. Expression adjacent to cnidocysts was found throughout larval and polyp stages (Fig. 2A–D). We detected NvEag in the cnidocyte-rich tentacle tips (Fig. 2B,C) in the body column (Fig. 2B) and adjacent to cnidocytes in ectoderm at the aboral end of juvenile polyps (Fig. 2D, arrowheads). Not all cnidocysts were associated with NvEag expression, suggesting the channel may not be expressed in all three types of cnidocytes identified in Nematostella (Zenkert et al., 2011), that NvEag expression in some cnidocytes is too low to detect by mRNA in situ hybridization or that NvEag is most strongly expressed in a distinct temporal window of cnidocyte development, as described for other cnidocyte-specific genes (Zenkert el al., 2011).
Fig. 2.
In situ hybridization analysis of Nematostella EAG family channel expression patterns. (A–D) NvEag expression is expressed in an ectodermal salt-and-pepper pattern in planula larva (A). Staining is adjacent to the cnidocyst capsules of the cnidocyte stinging cells (arrowheads, A′). NvEag expression is detected into juvenile polyps along the entire body axis, but is highly concentrated in the tentacle tips (B). Magnification of the tentacle tips reveals that NvEag staining in tentacles is also associated with cnidocyst capsules (C). Magnification of the polyp body wall shows that cnidocyst-associated expression is ectodermal (D, arrowheads). (E–G) NvElk expression is absent in planulae and during the early stages of polyp formation (E), but is elevated to high levels in polyp tentacles (F,G) and around the oral region (G). Some cells in the polyp body wall ectoderm also express NvElk (red arrows, G). (H–J) NvErg1 expression begins as punctae in the endoderm of planulae (H) and transitions to diffuse endodermal staining in polyps. (I,J) NvErg2 is expressed in a salt-and-pepper pattern in the ectoderm (red arrow) and endoderm (black arrow) of planula larva (K). Salt-and-pepper expression persists in endoderm (black arrow) and ectoderm (red arrow) through tentacle bud stage (L,L′). Expression appears slightly more concentrated around the mesentery structures (L). At juvenile polyp stages, punctate NvErg2 remains in the ectoderm, whereas endodermal expression becomes ubiquitous with individual cells enriched for NvErg2 (M). All images are lateral views with oral to the left and aboral to the right. Structures indicated are: tentacles (Tent.), endoderm (En), ectoderm (Ec) and mesenteries (Me). Scale bars: 100 μm (A,B,E–L,M); 10 μm (A′,C,D,L′).
Expression of NvElk was not observed until polyp stages (Fig. 2E–G). Expression was highly concentrated in the tentacles, tentacle tips and head regions (Fig. 2F,G). If development of the mRNA probe was allowed to continue, it became clear that individual ectodermal cells in the body column express NvElk (Fig. 2G, red arrows). These cells are found distributed more or less equally along the oral aboral axis and we did not observe any obvious correlation between cnidocyte capsules and NvElk expression. Currently, we are not able to definitively identify NvElk-positive cell types and a more detailed examination will be required to determine the relationship between NvElk expression and cell types within the tentacles. We conclude that NvElk is expressed in a subset of ectodermal cells throughout the body column of Nematostella polyps and a subset of cells in both the ectoderm and endoderm of the polyp tentacles.
We next analyzed mRNA expression of the two Nematostella erg genes. NvErg1 staining is first detected in a punctate pattern in the planula (Fig. 2H). The number of cells and distribution of cells beginning to express NvErg1 varies between individuals during planula stages. However, by juvenile polyp stages, the expression appears ubiquitous in the endodermal tissue (Fig. 2I,J). Thus, it is unclear whether all, or just the majority, of endodermal cells express NvErg1. Expression of NvErg2 mRNA in the planula also is detected in a punctate pattern, but cells can be observed in both the endoderm and ectoderm (Fig. 2K, black and red arrows, respectively). The punctate salt-and-pepper expression persists in both endodermal and ectodermal tissues throughout the tentacle bud and juvenile polyp stages (Fig. 2L,M). We noticed that expression was enriched around the mesentery structures, where myoepithelial cells and neurons are concentrated, but we cannot confirm the identity of NvErg2-positive cells at this time. Many animals had ubiquitous endodermal expression in juvenile polyps, with a punctate distribution of cells enriched for NvErg2 in the endoderm, as well as maintained expression of NvErg2 in individual cells in the ectoderm (Fig. 2M). The overlap in expression between NvErg1 and NvErg2 raises the possibility that some cells express heteromeric Erg channels. These initial studies show diverse expression patterns for EAG family channels in Nematostella, but more detailed studies will be needed to determine cell-type-specific expression.
Functional analysis of NvEag and NvElk
We functionally expressed NvEag and NvElk channels in Xenopus oocytes in order to better understand the functional evolution of the Eag and Elk gene subfamilies. Outward K+ currents from voltage-clamped oocytes expressing NvEag or mouse Eag1 (MmEag1, Kv10.1) are compared in Fig. 3A. The currents have similar fast activation and deactivation kinetics, lack inactivation and have almost superimposable voltage activation (G–V) curves which show substantial channel activity at typical sub-threshold voltage ranges (Fig. 3B). V50 and slope values for the single Boltzmann distribution fits of the voltage-activation data shown in Fig. 3B are reported in Table 1. These results suggest that the channels could play similar physiological roles in vivo, and extend our previous finding that Erg channel function is highly conserved between cnidarians and bilaterians (Martinson et al., 2014).
Fig. 3.
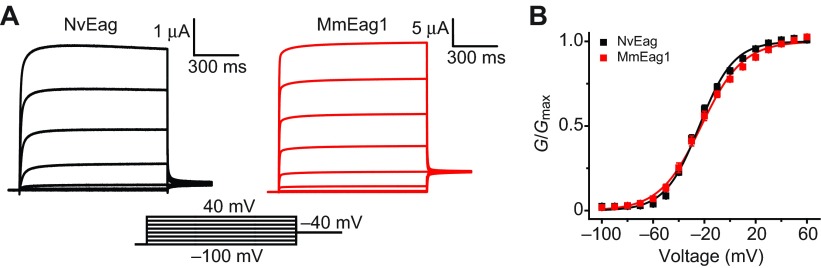
Comparison of NvEag and MmEag1 voltage-activation properties. (A) NvEag and MmEag1 current traces at pH 7 elicited by 1 s voltage steps from −100 mV to 40 mV (in 20 mV increments) from a holding voltage of −100 mV. (B) Normalized G–V curves for NvEag and MmEag1 at pH 7. Error bars show s.e.m. (n=6–11) and smooth curves show fits with a single Boltzmann distribution (parameters in Table 1).
Table 1.
Boltzmann fit parameters from voltage-activation curves of EAG channels
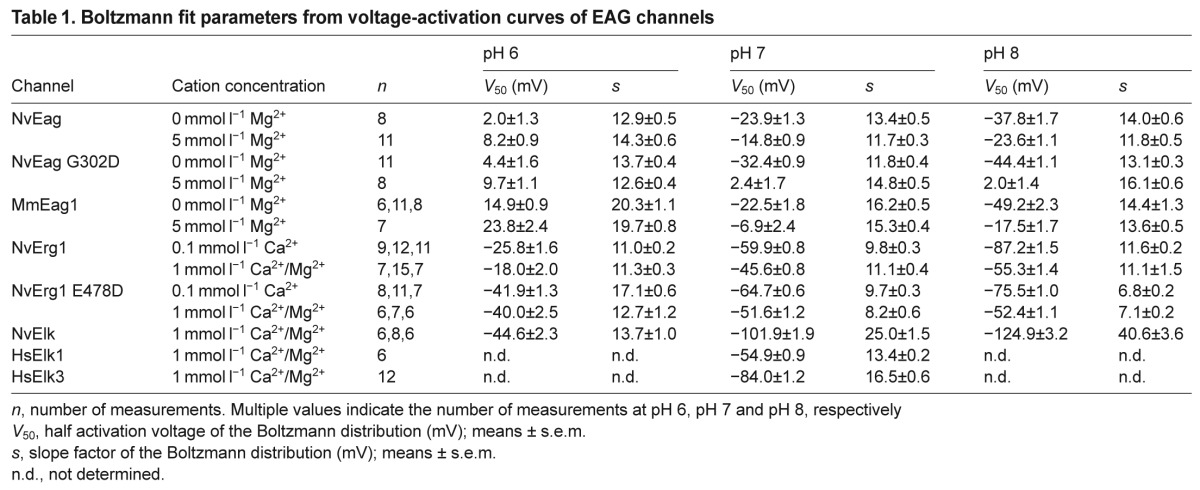
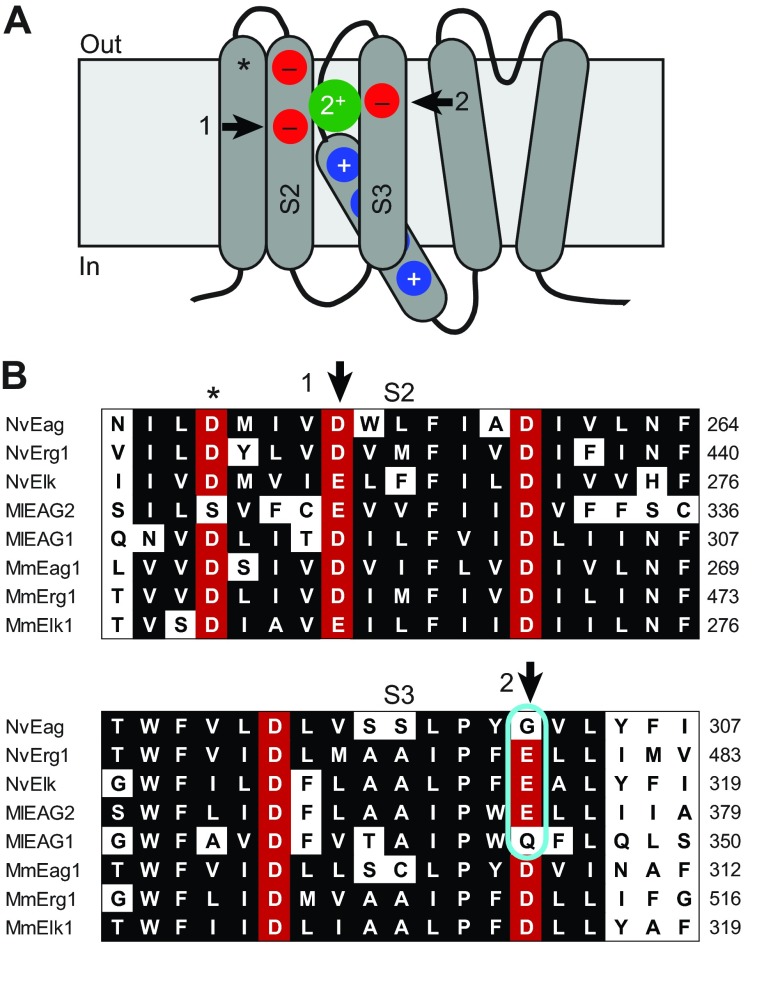
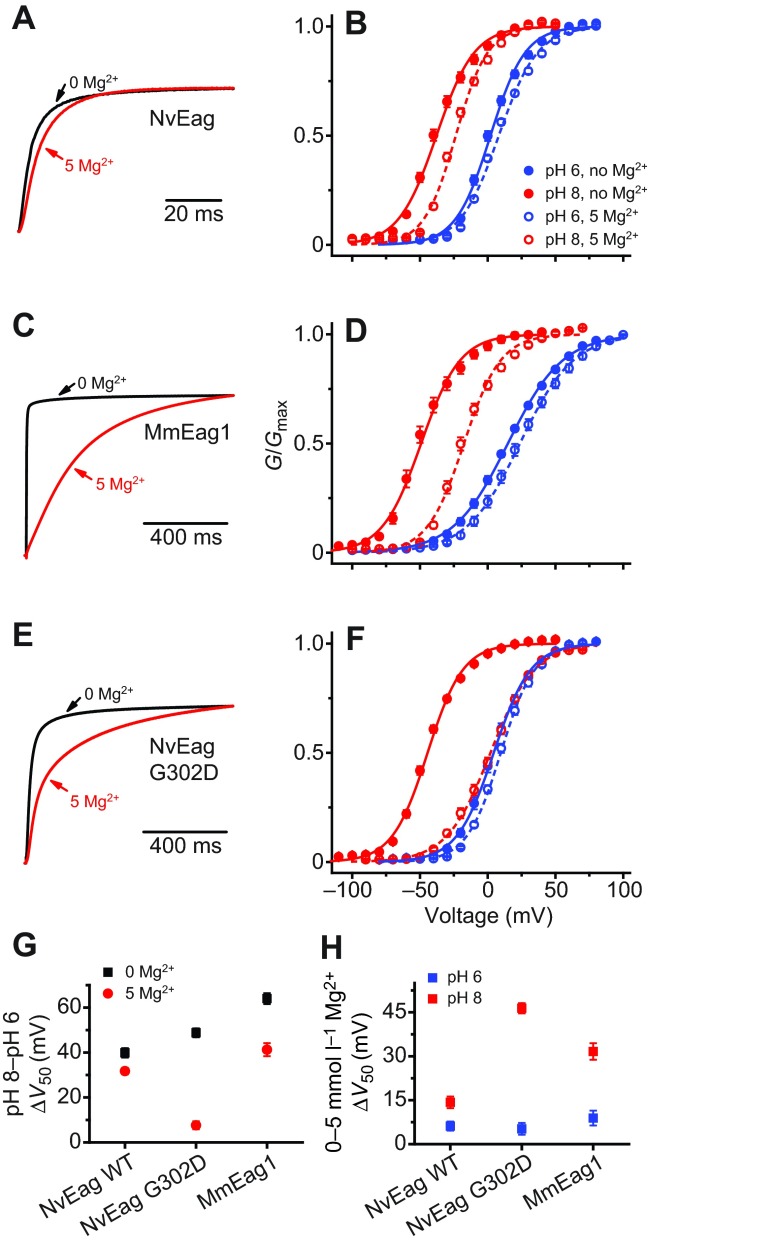
We next examined the Mg2+ dependence of voltage activation because activation of bilaterian Eag channels is significantly slowed by extracellular Mg2+ (Terlau et al., 1996), which binds to a site formed in part by the two solvent-accessible EAG family acidic residues in the outer voltage sensor (Fig. 4A,B) (Silverman et al., 2000). We reasoned that if NvEag was similarly inhibited by low concentrations of Mg2+, it might not be able to activate at sub-threshold potentials in osmoconformers, such as cnidarians living in sea water. However, we observed that NvEag has a Gly residue instead of the bilaterian consensus Asp residue at the S3 EAG charge position (Fig. 4B). Neutralization of this charge in Drosophila Eag eliminates Mg2+ sensitivity (Silverman et al., 2000) and the activation rate of NvEag was much less sensitive to 5 mmol l−1 Mg2+ than was the activation rate of MmEag1 (Fig. 5A,C). We quantified the effect of Mg2+ on voltage activation by measuring the shift in V50 of a single Boltzmann distribution fit to G–V data, as shown in Fig. 5B,D. Boltzmann fit parameters for all data in Fig. 5 are reported in Table 1. Because protons compete with divalent cations at the acidic residue binding site and also inhibit the voltage activation of EAG superfamily channels (Kazmierczak et al., 2013; Terlau et al., 1996), we examined the shift in V50 caused by 5 mmol l−1 Mg2+ at pH 8 to maximize Mg2+ sensitivity, and compared it with the V50 shift caused by 5 mmol l−1 Mg2+ at pH 6, where site-specific Mg2+ sensitivity should be greatly reduced (Fig. 5H). At pH 8, 5 mmol l−1 Mg2+ caused a 31.6±2.8 mV shift in the G–V curve of MmEag1, and the shift was significantly reduced (P<0.001) to 8.9±2.5 mV at pH 6. For NvEag, the V50 shift caused by 5 mmol l−1 Mg2+ was significantly reduced more than twofold with respect to MmEag1 to 14.3±2.0 mV at pH 8 (P<0.001), but was not significantly different from the MmEag1 shift at pH 6 (6.2±1.6 mV). These results indicate that NvEag has reduced Mg2+ sensitivity, which could facilitate activation of the channel at physiologically relevant membrane potentials in the brackish water in which Nematostella resides. We did not examine higher Mg2+ concentrations in order to avoid non-specific effects of surface charge on our measurements.
Fig. 4.
A distinct pattern of acidic side chains confers divalent cation sensitivity to the EAG superfamily voltage sensors. (A) Schematic diagram of an EAG channel family subunit with transmembrane domains shown as rounded rectangles; cytoplasmic gating domains have been omitted. The voltage sensor (four transmembrane domains on left) is depicted with the S4 helix, which contains basic gating charges (blue circles) in a down, or closed, position. Conserved acidic residues in the S2 and S3 transmembrane domains that are exposed to the extracellular solution in the closed state are depicted with red circles. An acidic charge found in the majority of voltage-gated ion channels is marked with an asterisk, and two charges found only in EAG channels and their close relatives are marked with arrows. The green circle indicates the approximate position of a closed state binding site for divalent cations, which can inhibit activation of the voltage sensor. (B) Amino acid alignment of the S2 and S3 transmembrane domains sea anemone (Nv), ctenophore (Ml) and select mouse (Mm) EAG family channels. Gene name is given at the left margin and amino acid position is given at the right margin. The external acidic side chain positions are marked with an asterisk or arrows as in A, and positions conserved in more than half of the sequences are shaded in black or red (acidic side chains). Nematostella and Mnemiopsis sequences do not have the consensus Asp at the EAG S3 acidic site (cyan outline), a residue that is important for divalent cation binding.
Fig. 5.
Mg2+ sensitivity in voltage activation is greatly reduced in NvEag. (A) Normalized current traces recorded in response to voltage steps to +20 mV (from a holding potential of −100 mV) for NvEag in 0 mmol l−1 Mg2+ (black) or 5 mmol l−1 Mg2+ (red). (B) NvEag voltage-activation (G–V) curves at pH 8 and pH 6 for NvEag in 0 mmol l−1 Mg2+ or 5 mmol l−1 Mg2+. Curves show fits with a single Boltzmann distribution (parameters in Table 1). Similar analyses are shown for MmEag1 (C,D) and NvEag G302D (E,F). Scale bars are given for time course in A,C and F. (G) Comparison of pH 8 to pH 6 V50 shifts for NvEag, MmEag1 and NvEag G302D with and without 5 mmol l−1 Mg2+. Shifts were calculated as the differences in average V50 values at pH 8 and pH 6. (H) Comparison of 5 mmol l−1 Mg2+-induced V50 shifts for NvEag, MmEag1 and NvEag G302D at pH 8 and pH 6 calculated from comparison of V50 values determined at 0 mmol l−1 Mg2+ and 5 mmol l−1 Mg2+. Data points in B,D,F–H are means ± s.e.m. (n=6–11).
Restoration of the S3 EAG-specific acidic residue in NvEag (G302D) greatly enhanced Mg2+ sensitivity (Fig. 5E,F); the V50 shift caused by 5 mmol l−1 Mg2+ at pH 8 increased significantly (P<0.001) to 46.4±1.8 mV, whereas it remained essentially unchanged at pH 6 at 5.2±2.0 mV (Fig. 5H). NvEag retains high pH-sensitivity of voltage activation, which is characteristic of EAG family channels, despite the loss of the S3 EAG acidic residues that contributes to the relevant proton binding site (Kazmierczak et al., 2013). However, the shift in V50 from pH 8 to pH 6 at 0 mmol l−1 Mg2+ (39.9±2.1 mV) was significantly smaller (P<0.001) than that observed for MmEag1 (64.0±2.4 mV; Fig. 5G), and the shift had relatively low Mg2+ sensitivity: 5 mmol l−1 Mg2+ only reduced the pH 8–pH 6 V50 shift to 31.7±1.4 mV. In NvEag G302D, the pH 8–pH 6 V50 shift in 0 mmol l−1 Mg2+ increased to 48.8±2.0 mV (P<0.01) and became highly sensitive to Mg2+, indicating enhanced competition between Mg2+ and protons for the voltage sensor binding site. The pH 8–pH 6 V50 shift was reduced to just 7.6±1.8 mV in 5 mmol l−1 Mg2+ (Fig. 5G). These results suggest that neutralization of the EAG-specific acidic residue in S3 is largely responsible for the reduced Mg2+ sensitivity of NvEag. The neutralization does reduce inhibition of activation by extracellular protons, but pH sensitivity nevertheless remains high.
We examined the pH and divalent cation sensitivity of NvErg1 activation because voltage activation of human Erg1 is also inhibited by binding of protons and Ca2+ and Mg2+ to the EAG-specific voltage sensor site (Fernandez et al., 2005; Jo et al., 1999; Kazmierczak et al., 2013). The pH sensitivity of human Erg is extremely low at physiological concentrations of Ca2+, but increases dramatically if Ca2+ is reduced to micromolar levels (Kazmierczak et al., 2013). NvErg1 has a charge preserving Asp to Glu substitution at the EAG S3 acidic site (Fig. 4B). We reasoned that this could alter divalent cation sensitivity because charge-preserving mutations at the EAG-specific acidic residues can alter the sensitivity of Eag and Elk channels for divalent cations (Silverman et al., 2000; Zhang et al., 2009). G–V curves for NvErg1 are shown for pH 6 and 8 in Fig. 6A at both 1 mmol l−1 Ca2+/1 mmol l−1 Mg2+ and 100 μmol l−1 Ca2+. Human Erg1 shows a pH 8–pH 6 V50 shift of over 40 mV at 100 μmol l−1 Ca2+, but less than 15 mV at 1 mmol l−1 Ca2+, demonstrating high-affinity block of voltage sensor proton binding site by Ca2+ (Kazmierczak et al., 2013). NvErg1 has a pH 8–pH 6 V50 shift of 61.4±2.2 mV in 100 μmol l−1 Ca2+, but maintains a relatively large 37.2±2.4 mV shift in 1 mmol l−1 Ca2+/1 mmol l−1 Mg2+, suggesting a reduced divalent cation sensitivity (Fig. 6C). Restoration of the EAG-specific S3 Asp residue in NvErg1 E478D significantly reduces the pH 8–pH 6 V50 shift in 1 mmol l−1 Ca2+/1 mmol l−1 Mg2+ to 12.4±2.7 mV (P<0.001), suggesting an increase in divalent cation sensitivity (Fig. 6B,C). At 100 μM Ca2+, the E478D shift increases to 33.6±1.6 mV, indicating that pH sensitivity is retained in the mutant. Because we did not test lower Ca2+ concentrations, we were not able to determine whether NvErg1 E478D has reduced pH sensitivity relative to the wild type (WT), or whether the proton binding site is still significantly occupied by Ca2+ at 100 μM. We also did not separately examine sensitivity to Ca2+ and Mg2+. These results indicate that replacement of the consensus Asp at the S3 Eag-specific charge position in both NvErg1 and NvEag serves to reduce divalent cation inhibition of voltage gating and may therefore play an important role in maintaining sub-threshold activation of these channels in true osmoconformers such as Nematostella. Glu is also substituted for Asp in NvErg2–NvErg4 (Martinson et al., 2014), and Eag and Erg orthologs from Acropora digitifera (Shinzato et al., 2011) contain the same substitutions as Nematostella. The EAG family channels from multiple ctenophores (Moroz et al., 2014; Ryan et al., 2013), typified by MlEAG1 and MlEAG2 presented here, also substitute Gln or Glu for the Asp (Fig. 4B). Trichoplax adhaerens Erg and EAG channels also contain the Glu substitution (supplementary material Table S2). Thus, the typical bilaterian Asp/Asp S2/S3 pattern of Eag charges in EAG and Erg channels is absent in many simple osmoconforming metazoans that may benefit from reduced Mg2+ and/or Ca2+ inhibition of voltage gating.
Fig. 6.
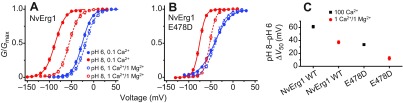
NvErg1 exhibits reduced divalent cation sensitivity. Normalized G–V curves for NvErg1 WT (A) and NvErg1 E478D (B) at pH 8 (red) and pH 6 (blue) with either 100 μmol l−1 Ca2+ (solid lines and symbols) or 1 mmol l−1 Ca2+/1 mmol l−1 Mg2+ (dashed lines and open symbols). (C) Comparison of pH 8 to pH 6 V50 shifts for NvErg1 WT and NvErg1 E478D at 100 μmol l−1 Ca2+ and 1 mmol l−1 Ca2+/1 mmol l−1 Mg2+. Shifts were calculated as the differences in average V50 values at pH 8 and pH 6. Data points are means ± s.e.m. (n=6–11) and smooth curves show fits with a single Boltzmann distribution (parameters in Table 1).
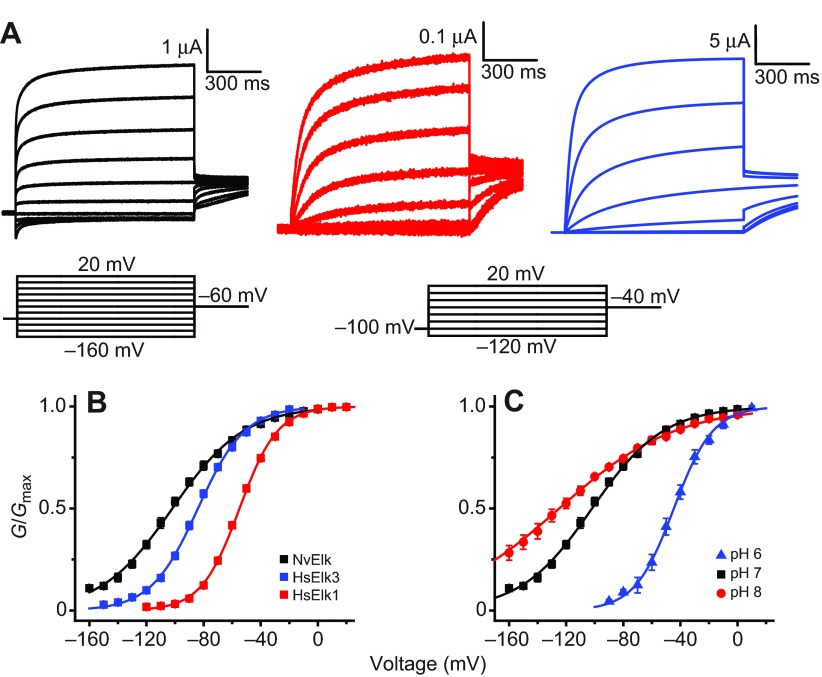
We next functionally expressed NvElk in order to determine which functional properties of Elk channels are broadly conserved in metazoans. A hyperpolarized voltage-activation range is a defining feature of mammalian Elk channels, and these channels contribute significantly to resting K+ conductance in neurons (Zhang et al., 2010). NvElk currents recorded from voltage-clamped Xenopus oocytes are compared with human Elk1 (HsElk1) and Elk3 currents in Fig. 7A, and G–V curves are shown for pH 7 in Fig. 7B. The signature feature of NvElk is its extremely hyperpolarized voltage-activation range. We determined a −101.9±1.9 mV V50 for NvElk from Boltzmann fits of voltage-activation data. NvElk currents show a similar kinetic profile to HsElk3, the mammalian Elk ortholog with the most left-shifted G–V curve (V50 at pH 7 of −84.0±1.2 mV). These results suggest that hyperpolarized voltage activation of Elk channels has been conserved throughout metazoan evolution and demonstrate that both Nematostella and mammals have Elk channels that are almost fully active at typically resting potentials. Like mammalian Elk channels (Kazmierczak et al., 2013), voltage activation of NvElk is highly sensitive to extracellular pH, with a pH 8–pH 6 V50 shift of 80.3±4.0 mV in 1 mmol l−1 Ca2+/1 mmol l−1 Mg2+ (Fig. 7C). We did not examine the divalent cation sensitivity of NvElk because Elk channels are insensitive to Mg2+ and Ca2+ because of a conserved substitution of the S2 EAG-specific Asp with Glu (Zhang et al., 2009). An analogous substitution in Drosophila Eag eliminates Mg2+ sensitivity (Silverman et al., 2000). Inactivation, which is present only in the vertebrate Elk2 ortholog (Engeland et al., 1998; Trudeau et al., 1999; Zhang et al., 2010), is absent in NvElk, suggesting that, in contrast to Erg, inactivation may not be an ancestral feature of Elk channels.
Fig. 7.
Comparison of NvElk and mammalian Elk currents. (A) Current traces recorded from Xenopus oocytes expressing NvElk (black), HsElk3 (red) and HsElk1 (blue) at pH 7 in response to a series of 1 s voltage steps from a holding potential of −100 mV. The voltage steps range from −160 mV to 20 mV for NvElk and from −120 mV to 20 mV for both HsElk3 and HsElk1 in 20 mV increments. Tail currents were taken at −60 mV for NvElk and at −40 mV for HsElk3 and HsElk1. (B) Normalized G–V curves for NvElk, HsElk3 and HsElk1 at pH 7. Data were measured from isochronal tail currents following 1 s steps to the indicated voltages. (C) Normalized G–V curves for NvElk at pH 6, 7 and 8. Data points in B and C show mean ± s.e.m. (n=6–12) and smooth curves represent a single Boltzmann distribution fit (parameters reported in Table 1).
DISCUSSION
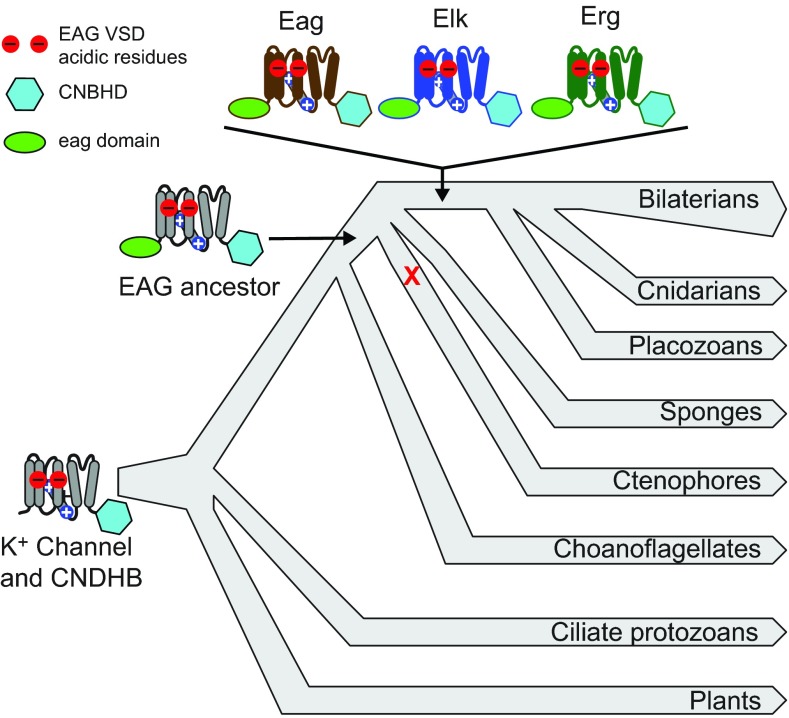
Our results indicate that the unique subunit structure of EAG family K+ channels, consisting of an eag domain, K+ channel core and CNBHD, first appeared in a common ancestor of ctenophore lineage and cnidarian–placozoan–bilaterian lineage. However, the functionally distinct Eag, Elk and Erg subfamilies appeared only later in this lineage after the divergence from ctenophores. This conclusion is based on the presence of ctenophore-specific EAG family genes and the absence of Eag, Elk or Erg genes in two ctenophore genomes and five ctenophore transcriptomes. Therefore, incomplete coverage of ctenophore sequence or species is unlikely to account for this result. A summary of our current view of EAG K+ channel evolution is presented in Fig. 8. Because ctenophores are currently believed to be the most basal branching metazoan lineage (Dunn et al., 2008; Hejnol et al., 2009; Moroz et al., 2014; Ryan et al., 2013), the ancestral EAG family channel probably first appeared in a common ancestor of all extant metazoans. If so, the EAG family seems to have been lost in the sponge lineage – a group of animals devoid of distinct neurons.
Fig. 8.
Summary of EAG K+ channel family evolution. Important events in the evolution of EAG family channels are overlaid on a schematic eukaryotic phylogeny. Channel subunits diagrams are used to illustrate the presence of three diagnostic EAG channel features: (1) the EAG family voltage sensor acidic residues (red circles); (2) the CNBHD (cyan hexagon); and (3) the N-terminal eag domain (green oval). A K+ channel with the EAG voltage sensor acidic residues and the CNBHD was inherited from prokaryotes (base of phylogeny) and is widely present in eukaryotic lineages, including plants, ciliates and metazoans. The eag domain was appended in an ancestral metazoan before divergence of ctenophores, and may have been lost in the sponge lineage (red X). The Eag, Elk and Erg subfamilies originated in an ancestor of placozoans, cnidarians and bilaterians and are highly conserved on a functional level between cnidarians and bilaterians. The functional phenotypes of ctenophore EAG family channels have not yet been determined, but it is likely that the original metazoans had a reduced diversity of sub-threshold K+ channels for control of excitation. The branching order of metazoan lineages is based on recent findings from ctenophore genome sequencing (Moroz et al., 2014; Ryan et al., 2013).
The EAG ancestor appears to have evolved from an ancient channel lineage dating back to prokaryotes. Prokaryotes have K+ channels with a voltage sensor and a CNBD (Brams et al., 2014; Nimigean et al., 2004). The voltage sensor of these prokaryotic channels already contained the EAG family acidic charges. K+ channels based on this motif can also be found in plants (Sentenac et al., 1992) and ciliate protozoans such as Paramecium (Jegla and Salkoff, 1995), indicating that such a channel might have been present in the first eukaryotes. CNGs and hyperpolarization-gated channels appear to be the closest metazoan relatives of EAG channels (Yu and Catterall, 2004) and may have evolved from the same ancestor because they also have the cyclic nucleotide-binding domain and the additional EAG family acidic charges in the voltage sensor. The key step in EAG channel evolution in metazoans was the addition of the PAS-based eag domain (Morais Cabral et al., 1998) to the N-terminus of the ancestral channel. This domain regulates channel function by stabilizing an open state and contributes to the characteristically slow deactivation of many EAG channels (Morais Cabral et al., 1998; Wang et al., 1998). The domain has been lost independently several times within the Erg lineage, producing an alternate rapidly deactivating Erg channel phenotype (Martinson et al., 2014). EAG channels have retained the inherited cyclic nucleotide binding domain, but we refer to it as a CNBHD because extant EAG channels do not bind cyclic nucleotides (Brelidze et al., 2009). The CNBHD nevertheless appears to play an important role in channel gating in part through interaction with the eag domain (Gianulis et al., 2013; Haitin et al., 2013). We were not able to functionally express the ctenophore EAG channels MlEAG1 and MlEAG2 in Xenopus oocytes, so we were not able to determine with more precision when cyclic-nucleotide sensitivity might have been lost during EAG family evolution.
Our functional analyses of Nematostella Eag, Elk and Erg channels show that many of the distinctive characteristic features of these subfamilies were already in place before the cnidarian–bilaterian divergence. For instance, NvEag is highly similar in terms of basic kinetic profile and voltage dependence to bilaterian Eag channels such as MmEAG1 (as shown here) or Drosophila Eag (Robertson et al., 1996). Eag channels deactivate more rapidly than typical Elk and Erg channels, and the results presented here suggest that this rapid gating is ancestral. The hallmark of bilaterian Elk channels is a hyperpolarized G–V curve that overlaps with typical neuronal resting potentials (Engeland et al., 1998; Trudeau et al., 1999; Zou et al., 2003). In mice, Elk channels contribute to basal K+ conductance and sub-threshold excitability of neurons (Zhang et al., 2010). This property is conserved and taken to an extreme in NvElk, which appears to be almost completely activated at typical resting potentials. The ancestral Erg channel phenotype is specialized for delayed repolarization by a combination of rapid inactivation and recovery (Martinson et al., 2014). In the human heart, Erg1 plays a key role in repolarizing cardiac action potential plateaus (Sanguinetti and Tristani-Firouzi, 2006). It has a functional phenotype that is highly similar to NvErg1, suggesting a potential ancestral role for Erg channels in repolarization of broad action potentials. However, Erg channels also activate at sub-threshold potentials and could therefore play a role in the regulation of sub-threshold excitability. Some Erg channels have lost inactivation but retained sub-threshold activation (Martinson et al., 2014), and the channels have been implicated in the regulation of excitation threshold in Caenorhabditis, Drosophila and mouse (Collins and Koelle, 2013; Hardman and Forsythe, 2009; Hirdes et al., 2009; LeBoeuf and Garcia, 2012; Titus et al., 1997). Sub-threshold activation seems to be a unifying functional feature of all EAG family channels that have been functionally expressed.
This sub-threshold activation appears to be conferred in part by the EAG family acidic residues in the voltage sensor. Mutagenic neutralization of these charges in mammalian Eag, Elk and Erg channels depolarizes the voltage-activation curve (Fernandez et al., 2005; Silverman et al., 2000; Zhang et al., 2009). Furthermore, voltage activation of EAG channels is universally depolarized by extracellular protons through neutralization of the EAG acidic charges (Kazmierczak et al., 2013). In mammalian EAG channels, pH sensitivity may have physiological relevance, because native K+ channels inhibited by similar concentrations of protons have been observed in a variety of neurons and remain unexplained at the molecular level (González et al., 2009; Mulkey et al., 2007). It has not yet been established whether EAG family K+ channels contribute to such currents. The role of pH sensitivity in Nematostella EAG family channels is less clear because the pH of the extracellular environment has not been characterized. However, seawater has a pH in excess of 8, suggesting that extracellular pH could be comparatively high. All Nematostella EAG family channels expressed so far show significant sub-threshold activation at pH 7 and 8, and this would support the idea that regulation of sub-threshold activity is an important role of EAG family channels. The typical acidic charge pattern of the EAG voltage sensor that supports both divalent cation and proton sensitivity is altered in Nematostella Eag and Erg channels to reduce Mg2+ and Ca2+ sensitivity. Both types of channel substitute the characteristic EAG Asp in S3 with either Gly (NvEag) or Glu (NvErg1–NvErg5). These mutations reduce divalent cation block and therefore may enhance sub-threshold activation in sea water, but they do not eliminate pH sensitivity. Because it is possible to greatly reduce or eliminate pH sensitivity by substitution of the S3 Asp (Kazmierczak et al., 2013), this suggests there could be selective pressure to maintain pH sensitivity. However, it is also possible that there is simply an overlap at this site between mutations that preserve sub-threshold activation with those that preserve pH sensitivity.
We show here that the biophysical properties of Elk and Eag channels are highly conserved between cnidarians and bilaterians, similar to what we have previously found for Erg channels (Martinson et al., 2014). There is also a pattern of high conservation for other voltage-gated K+ channels, including the Shaker, Shal and Shaw families, which encode A-currents and delayed rectifiers and probably play important roles in action potential repolarization and patterning (Bouchard et al., 2006; Jegla and Salkoff, 1997; Jegla et al., 1995; Jegla et al., 2012; Sand et al., 2011). Therefore, the high molecular conservation of channel genes between cnidarians and bilaterians extends to functional conservation, at least at the level of channel biophysics. Molecular genetic analysis, which is now becoming possible in model systems such as Nematostella, will be needed to determine what roles these channels play in physiology, and thus why there has been such strong selective pressure to maintain specific biophysical properties over such a vast evolutionary distance.
The in situ results on mRNA expression presented here give some initial clues to the roles of some EAG family channels in Nematostella. NvEag appears to be expressed in a subset of cnidocytes and we therefore suggest that it could play a role in regulating discharge. The in vivo physiology of bilaterian Eag channels is still poorly understood, but they are highly expressed in neurons, and contribute to K+ currents and excitability control in Drosophila motor neurons (Srinivasan et al., 2012) and neuromuscular excitability in C. elegans (LeBoeuf and Garcia, 2012). Both mammalian Eag orthologs have been genetically associated with neuronal excitability control (Ufartes et al., 2013; Yang et al., 2013), but the cellular mechanisms have not been explored. As cnidocytes are a cnidarian-specific cell type believed to be derived from a neuronal cell type (Pantin, 1942), it is tempting to speculate that ancestral Eag subfamily channels regulated sub-threshold excitability of neurons. NvErg2 and NvElk are also present in a few cells scattered around the body wall that could potentially represent neurons but remain undefined. Elk channels regulate resting properties of neurons and sub-threshold excitability in bilaterians (Zhang et al., 2010), and our functional analysis opens the possibility that regulation of neuronal excitation is an ancestral role of the Elk family. Additionally, it should be noted that all of the Eag genes in Nematostella are expressed at relatively low levels and the in situ results require multiple days to development. This is consistent with numerous expression studies in a variety of species that indicate that ion channels are low-abundance proteins. Thus, it is possible that additional cells express these EAG genes, but at levels below the sensitivity threshold of mRNA in situ hybridization. Ultimately, genetic manipulation of NvEag channels will be required to provide further evidence for putative neuronal roles for NvEag and NvElk.
Erg channels play a dual role in bilaterians, regulating excitation threshold of neurons and muscle (Collins and Koelle, 2013; Hardman and Forsythe, 2009; Ji et al., 2012; LeBoeuf and Garcia, 2012; Niculescu et al., 2013) and regulating repolarization of broad action potentials in the vertebrate heart (Sanguinetti and Tristani-Firouzi, 2006). We previously found that NvErg1 and NvErg4 show sub-threshold activation and showed that the inactivating IKr phenotype of cardiac Erg channels in vertebrates that enables repolarization of broad action potentials is present in NvErg1 and is probably the ancestral Erg channel phenotype (Martinson et al., 2014). Here, we find that NvErg1 and NvErg2 are broadly expressed in endoderm, and that NvErg2 is also expressed in a punctate pattern that we speculate is consistent with a neuronal distribution. Although cnidarian myoepithelial cells are endodermal, we were not able to conclusively determine whether Erg channels are expressed primarily in Nematostella myoepithelium. The functional properties of NvErg2 have not yet been examined and we did not look at the mRNA distribution of NvErg3–NvErg5.
Our results show that the molecular and functional diversity of EAG family K+ channels had already been established before the divergence of cnidarians and bilaterians, providing a rich palette of sub-threshold K+ conductances for regulation of excitability. The reduced molecular diversity of EAG family channels in ctenophores suggests that new mechanisms for the regulation of excitability continued to evolve after establishment of the first nervous systems in basal metazoans. However, the functional diversity of EAG channels and voltage-gated K+ channels as a whole remains to be determined in ctenophores. Our data adds to a growing body of evidence that ion channel function is highly conserved between cnidarians and bilaterians at the gene family level. Interestingly, no new families of voltage-gated ion channel appear to have evolved in either the cnidarian or bilaterian lineages after their divergence. Together, these observations suggest that the major mechanisms for controlling neuronal excitability predate the cnidarian–bilaterian divergence. These cnidarian–bilaterian channels provide a diverse, adaptable set of conductances that enable a broad range of neuronal activity patterns and physiological responses.
MATERIALS AND METHODS
Identification and cloning of EAG family genes
EAG family genes were identified in Nematostella vectensis, Trichoplax adhaerens and Mnemiopsis leidyi using TBLASTN (Altschul et al., 1997) searches of gene predictions and genome drafts (Putnam et al., 2007; Ryan et al., 2013; Srivastava et al., 2008) with a panel of bilaterian Eag, Elk and Erg sequences. Sequence from candidate EAG family genes was reciprocally searched against mouse and Drosophila RefSeq databases (Pruitt et al., 2012) to confirm their identity. EAG family genes from Nematostella, Trichoplax and Mnemiopsis had clear best matches to mouse and Drosophila genes in these reciprocal searches. The strategy was used successfully in a previous study to identify Nematostella Erg genes (Martinson et al., 2014). Protein sequence predictions were adjusted manually as necessary based on homology, and standard PCR cloning strategies were used to amplify expression clones for NvEag, NvElk and MlEAG1 and MlEAG2 from oligo dT-primed cDNA samples prepared from total RNA from whole animals. RACE PCR was used to identify and confirm the 3′ end of all ORFs and the 5′ ends of MlEAG1 and MlEAG2. Full-length clones of each gene were assembled using ligation and/or overlap PCR, inserted into the pOX expression vector (Jegla and Salkoff, 1997) for expression in Xenopus oocytes and sequence was confirmed. Clones matching the coding sequences reported in supplementary material Fig. S1 were used in expression studies.
Phylogenetic analysis
Amino acid sequence alignments for phylogenetic analysis were generated using ClustalW as implemented in MEGA5 (Tamura et al., 2011), and were hand adjusted as necessary to produce optimal alignments. Analysis was limited to the core voltage-gated K+ channel and CNBHD motifs. The eag domain was eliminated from analysis because it has been lost in several Erg lineages. Mr. Bayes 3 (Ronquist and Huelsenbeck, 2003) was used to generate a Bayesian inference phylogeny under a mixed model. Two independent replicates of four chains were run for 1 million generations. Trees were sampled every 1000 generations and the first 25% were discarded. A consensus phylogeny is reported in Fig. 1.
In situ hybridization
Animals were raised to appropriate stages in ×1/3 sea water at 25°C and then fixed as described (Wolenski et al., 2013). Nematostella EAG family sequences used for in situ hybridization are given in supplementary material Table S2 and were cloned into pGEM-T. Riboprobe templates were generated using SP6 and T7 primers to amplify each gene out of the plasmid background. Probes were generated and mRNA in situ hybridizations were performed as described (Wolenski et al., 2013). Images were obtained using a Zeiss M2 in conjunction with a Zeiss Axiocam HRc and the Zen software (Zeiss, Jena, Germany).
Functional expression in Xenopus oocytes
Expression plasmids were linearized and capped run-off cRNA transcripts were generated using the T3 mMessage mMachine kit (Life Technologies, Carlsbad, CA, USA). Transcripts were precipitated by LiCl and rinsed with ice-cold 70% ethanol. Dried pellets were dissolved in nuclease-free water supplemented with RNase inhibitor (SUPERase-In; Life Technologies) before storage and use. Mature Xenopus oocytes were injected with 1–5 ng diluted cRNA in a 50 nl volume and incubated for 1–3 days at 18°C in ND96 culture solution before recording. Xenopus laevis ovaries were purchased from Nasco (Fort Atkinson, WI, USA) and Xenopus I (Dexter, MI, USA) and oocytes were enzymatically defolliculated with 1 mg ml−1 type II collagenase (Sigma-Aldrich, St Louis, MO, USA) in Ca2+-free ND98 solution (98 mmol l−1 NaCl, 2 mmol l−1 KCl, 1 mmol l−1 MgCl2, 5 mmol l−1 HEPES, pH 7.2). Following digestion, oocytes were rinsed extensively and then maintained in ND98 culture solution (98 mmol l−1 NaCl, 2 mmol l−1 KCl, 1.8 mmol l−1 CaCl2, 1 mmol l−1 MgCl2 and 5 mmol l−1 HEPES, pH 7.2) supplemented with 2.5 mmol l−1 sodium pyruvate, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (Life Technologies, Carlsbad, CA, USA).
Base recording solution consisted of 98 mmol l−1 NaOH, 2 mmol l−1 KCl, 5 mmol l−1 HEPES and was adjusted to the desired pH value with methanesulfonic acid. CaCl2 and MgCl2 were added to the base solution to the desired concentration. For MmEag1 G–V measurements, K+ concentration was increased to 50 mmol l−1, and Na+ concentration was reduced to 50 mmol l−1. Two-electrode voltage clamp (TEVC) recordings were carried out using a CA-1B amplifier (Dagan, Minneapolis, MN, USA) at room temperature (22–24°C). Glass capillary electrodes were filled with 3 M KCl (0.4–1 MΩ), and bath clamp circuitry was connected by a 1 mol l−1 NaCl–agarose bridge. Data collection and analysis were performed with the pClamp 10 acquisition suite (Molecular Devices, Sunnyvale, CA, USA). Currents were sampled at 10 KHz and filtered at 2 KHz. G–V curves were measured from isochronal tail currents recorded at −40 mV for NvEag, HsElk1 and HsElk3, −60 mV for NvElk and −70 mV for NvErg1. For MmEag1, the reversal potential was empirically determined and used to compute the electrochemical driving force at each voltage step. G–V curves were derived by normalizing steady state current at the end of the 1 s step to the driving force. Data were fit in Origin 8.1 (OriginLab, Northampton, MA, USA) with a single Boltzmann distribution: f(V)=(A1−A2)/(1+e(V−V50)/s)+A2, where V50 is the half-maximal activation voltage, s is the slope factor, and A1 and A2 are the lower and upper bounds, respectively. Reported V50 and slope factor values are the means ± s.e.m. of individual fits. Data points from individual cells were normalized before averaging, and Boltzmann fits shown were generated with arithmetic means of V50 and s values of measurements from individual cells. Statistical comparisons between datasets were made using a two-tailed t-test.
Supplementary Material
Acknowledgements
Parts of this work were presented at the ‘Evolution of the First Nervous Systems II’ meeting, which was supported by the National Science Foundation.
FOOTNOTES
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the Department of Biology and Huck Institutes for the Life Sciences at Penn State University, and by the National Institutes of Health [NS069842 to T.J.]. A.S.M. and F.H.D. were supported by undergraduate research from the Eberly College of Science at Penn State, and A.P.S. and X.L. were supported by the Huck Institutes. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://jeb.biologists.org/lookup/suppl/doi:10.1242/jeb.110080/-/DC1
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., Price R. B., Moneypenny C. G., Thompson L. F., Zillhardt M., Stalheim L., Anderson P. A. (2006). Cloning and functional expression of voltage-gated ion channel subunits from cnidocytes of the Portuguese Man O'War Physalia physalis. J. Exp. Biol. 209, 2979-2989. [DOI] [PubMed] [Google Scholar]
- Brams M., Kusch J., Spurny R., Benndorf K., Ulens C. (2014). Family of prokaryote cyclic nucleotide-modulated ion channels. Proc. Natl. Acad. Sci. USA 111, 7855-7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T. I., Carlson A. E., Zagotta W. N. (2009). Absence of direct cyclic nucleotide modulation of mEAG1 and hErg1 channels revealed with fluorescence and electrophysiological methods. J. Biol. Chem. 284, 27989-27997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T. I., Carlson A. E., Sankaran B., Zagotta W. N. (2012). Structure of the carboxy-terminal region of a KCNH channel. Nature 481, 530-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. M., Koelle M. R. (2013). Postsynaptic Erg potassium channels limit muscle excitability to allow distinct egg-laying behavior states in Caenorhabditis elegans. J. Neurosci. 33, 761-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn C. W., Hejnol A., Matus D. Q., Pang K., Browne W. E., Smith S. A., Seaver E., Rouse G. W., Obst M., Edgecombe G. D., et al. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745-749. [DOI] [PubMed] [Google Scholar]
- Engeland B., Neu A., Ludwig J., Roeper J., Pongs O. (1998). Cloning and functional expression of rat ether-à-go-go-like K+ channel genes. J. Physiol. 513, 647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough S. R., Chen Z., Kramer E., Zeng Q., Young S., Robertson H. M., Begovic E., Richter D. J., Russ C., Westbrook M. J., et al. (2013). Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 14, R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D., Ghanta A., Kinard K. I., Sanguinetti M. C. (2005). Molecular mapping of a site for Cd2+-induced modification of human ether-à-go-go-related gene (hErg) channel activation. J. Physiol. 567, 737-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B., Robertson G. A., Wilson G. F., Trudeau M. C., Titus S. A. (1999). The eag family of K+ channels in Drosophila and mammals. Ann. N. Y. Acad. Sci. 868, 356-369. [DOI] [PubMed] [Google Scholar]
- Garcia L. R., Sternberg P. W. (2003). Caenorhabditis elegans UNC-103 Erg-like potassium channel regulates contractile behaviors of sex muscles in males before and during mating. J. Neurosci. 23, 2696-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianulis E. C., Liu Q., Trudeau M. C. (2013). Direct interaction of eag domains and cyclic nucleotide-binding homology domains regulate deactivation gating in hErg channels. J. Gen. Physiol. 142, 351-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J. A., Jensen L. T., Doyle S. E., Miranda-Anaya M., Menaker M., Fugger L., Bayliss D. A., Burdakov D. (2009). Deletion of TASK1 and TASK3 channels disrupts intrinsic excitability but does not abolish glucose or pH responses of orexin/hypocretin neurons. Eur. J. Neurosci. 30, 57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustina A. S., Trudeau M. C. (2013). The eag domain regulates hErg channel inactivation gating via a direct interaction. J. Gen. Physiol. 141, 229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitin Y., Carlson A. E., Zagotta W. N. (2013). The structural mechanism of KCNH-channel regulation by the eag domain. Nature 501, 444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman R. M., Forsythe I. D. (2009). Ether-à-go-go-related gene K+ channels contribute to threshold excitability of mouse auditory brainstem neurons. J. Physiol. 587, 2487-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A., Obst M., Stamatakis A., Ott M., Rouse G. W., Edgecombe G. D., Martinez P., Baguñà J., Bailly X., Jondelius U., et al. (2009). Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. Biol. Sci. 276, 4261-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirdes W., Schweizer M., Schuricht K. S., Guddat S. S., Wulfsen I., Bauer C. K., Schwarz J. R. (2005). Fast erg K+ currents in rat embryonic serotonergic neurones. J. Physiol. 564, 33-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirdes W., Napp N., Wulfsen I., Schweizer M., Schwarz J. R., Bauer C. K. (2009). Erg K+ currents modulate excitability in mouse mitral/tufted neurons. Pflugers Arch. 459, 55-70. [DOI] [PubMed] [Google Scholar]
- Jegla T., Salkoff L. (1995). A multigene family of novel K+ channels from Paramecium tetraurelia. Receptors Channels 3, 51-60. [PubMed] [Google Scholar]
- Jegla T., Salkoff L. (1997). A novel subunit for shal K+ channels radically alters activation and inactivation. J. Neurosci. 17, 32-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegla T., Grigoriev N., Gallin W. J., Salkoff L., Spencer A. N. (1995). Multiple Shaker potassium channels in a primitive metazoan. J. Neurosci. 15, 7989-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegla T. J., Zmasek C. M., Batalov S., Nayak S. K. (2009). Evolution of the human ion channel set. Comb. Chem. High Throughput Screen 12, 2-23. [DOI] [PubMed] [Google Scholar]
- Jegla T., Marlow H. Q., Chen B., Simmons D. K., Jacobo S. M., Martindale M. Q. (2012). Expanded functional diversity of shaker K(+) channels in cnidarians is driven by gene expansion. PLoS ONE 7, e51366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Tucker K. R., Putzier I., Huertas M. A., Horn J. P., Canavier C. C., Levitan E. S., Shepard P. D. (2012). Functional characterization of ether-à-go-go-related gene potassium channels in midbrain dopamine neurons – implications for a role in depolarization block. Eur. J. Neurosci. 36, 2906-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S. H., Youm J. B., Kim I., Lee C. O., Earm Y. E., Ho W. K. (1999). Blockade of HErg channels expressed in Xenopus oocytes by external H+. Pflugers Arch. 438, 23-29. [DOI] [PubMed] [Google Scholar]
- Kazmierczak M., Zhang X., Chen B., Mulkey D. K., Shi Y., Wagner P. G., Pivaroff-Ward K., Sassic J. K., Bayliss D. A., Jegla T. (2013). External pH modulates EAG superfamily K+ channels through EAG-specific acidic residues in the voltage sensor. J. Gen. Physiol. 141, 721-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N., Westbrook M. J., Young S. L., Kuo A., Abedin M., Chapman J., Fairclough S., Hellsten U., Isogai Y., Letunic I., et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf B., Garcia L. R. (2012). Cell excitability necessary for male mating behavior in Caenorhabditis elegans is coordinated by interactions between big current and ether-a-go-go family K(+) channels. Genetics 190, 1025-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London B., Trudeau M. C., Newton K. P., Beyer A. K., Copeland N. G., Gilbert D. J., Jenkins N. A., Satler C. A., Robertson G. A. (1997). Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ. Res. 81, 870-878. [DOI] [PubMed] [Google Scholar]
- Martinson A. S., van Rossum D. B., Diatta F. H., Layden M. J., Rhodes S. A., Martindale M. Q., Jegla T. (2014). Functional evolution of Erg potassium channel gating reveals an ancient origin for IKr. Proc. Natl. Acad. Sci. USA 111, 5712-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais Cabral J. H., Lee A., Cohen S. L., Chait B. T., Li M., Mackinnon R. (1998). Crystal structure and functional analysis of the HErg potassium channel N terminus: a eukaryotic PAS domain. Cell 95, 649-655. [DOI] [PubMed] [Google Scholar]
- Moroz L. L., Kocot K. M., Citarella M. R., Dosung S., Norekian T. P., Povolotskaya I. S., Grigorenko A. P., Dailey C., Berezikov E., Buckley K. M., et al. (2014). The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkey D. K., Talley E. M., Stornetta R. L., Siegel A. R., West G. H., Chen X., Sen N., Mistry A. M., Guyenet P. G., Bayliss D. A. (2007). TASK channels determine pH sensitivity in select respiratory neurons but do not contribute to central respiratory chemosensitivity. J. Neurosci. 27, 14049-14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu D., Hirdes W., Hornig S., Pongs O., Schwarz J. R. (2013). Erg potassium currents of neonatal mouse Purkinje cells exhibit fast gating kinetics and are inhibited by mGluR1 activation. J. Neurosci. 33, 16729-16740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimigean C. M., Shane T., Miller C. (2004). A cyclic nucleotide modulated prokaryotic K+ channel. J. Gen. Physiol. 124, 203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin C. F. A. (1942). The excitation of nematocysts. J. Exp. Biol. 19, 294-310. [Google Scholar]
- Pruitt K. D., Tatusova T., Brown G. R., Maglott D. R. (2012). NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130-D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V. V., et al. (2007). Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86-94. [DOI] [PubMed] [Google Scholar]
- Riesgo A., Farrar N., Windsor P. J., Giribet G., Leys S. P. (2014). The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31, 1102-1120. [DOI] [PubMed] [Google Scholar]
- Robertson G. A., Warmke J. M., Ganetzky B. (1996). Potassium currents expressed from Drosophila and mouse eag cDNAs in Xenopus oocytes. Neuropharmacology 35, 841-850. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574. [DOI] [PubMed] [Google Scholar]
- Ryan J. F., Pang K., Schnitzler C. E., Nguyen A. D., Moreland R. T., Simmons D. K., Koch B. J., Francis W. R., Havlak P., Smith S. A., et al. NISC Comparative Sequencing Program (2013). The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand R. M., Atherton D. M., Spencer A. N., Gallin W. J. (2011). jShaw1, a low-threshold, fast-activating K(v)3 from the hydrozoan jellyfish Polyorchis penicillatus. J. Exp. Biol. 214, 3124-3137. [DOI] [PubMed] [Google Scholar]
- Sanguinetti M. C., Tristani-Firouzi M. (2006). hErg potassium channels and cardiac arrhythmia. Nature 440, 463-469. [DOI] [PubMed] [Google Scholar]
- Sentenac H., Bonneaud N., Minet M., Lacroute F., Salmon J. M., Gaymard F., Grignon C. (1992). Cloning and expression in yeast of a plant potassium ion transport system. Science 256, 663-665. [DOI] [PubMed] [Google Scholar]
- Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., Fujie M., Fujiwara M., Koyanagi R., Ikuta T., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320-323. [DOI] [PubMed] [Google Scholar]
- Silverman W. R., Tang C. Y., Mock A. F., Huh K. B., Papazian D. M. (2000). Mg(2+) modulates voltage-dependent activation in ether-à-go-go potassium channels by binding between transmembrane segments S2 and S3. J. Gen. Physiol. 116, 663-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Lance K., Levine R. B. (2012). Contribution of EAG to excitability and potassium currents in Drosophila larval motoneurons. J. Neurophysiol. 107, 2660-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Begovic E., Chapman J., Putnam N. H., Hellsten U., Kawashima T., Kuo A., Mitros T., Salamov A., Carpenter M. L., et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454, 955-960. [DOI] [PubMed] [Google Scholar]
- Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E., Mitros T., Richards G. S., Conaco C., Dacre M., Hellsten U., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H., Ludwig J., Steffan R., Pongs O., Stühmer W., Heinemann S. H. (1996). Extracellular Mg2+ regulates activation of rat eag potassium channel. Pflugers Arch. 432, 301-312. [DOI] [PubMed] [Google Scholar]
- Titus S. A., Warmke J. W., Ganetzky B. (1997). The Drosophila erg K+ channel polypeptide is encoded by the seizure locus. J. Neurosci. 17, 875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau M. C., Titus S. A., Branchaw J. L., Ganetzky B., Robertson G. A. (1999). Functional analysis of a mouse brain Elk-type K+ channel. J. Neurosci. 19, 2906-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufartes R., Schneider T., Mortensen L. S., de Juan Romero C., Hentrich K., Knoetgen H., Beilinson V., Moebius W., Tarabykin V., Alves F., et al. (2013). Behavioural and functional characterization of Kv10.1 (Eag1) knockout mice. Hum. Mol. Genet. 22, 2247-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Trudeau M. C., Zappia A. M., Robertson G. A. (1998). Regulation of deactivation by an amino terminal domain in human ether-à-go-go-related gene potassium channels. J. Gen. Physiol. 112, 637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers S., Wulfsen I., Bauer C. K., Schwarz J. R. (2001). Erg1, erg2 and erg3 K channel subunits are able to form heteromultimers. Pflugers Arch. 441, 450-455. [DOI] [PubMed] [Google Scholar]
- Wolenski F. S., Layden M. J., Martindale M. Q., Gilmore T. D., Finnerty J. R. (2013). Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea anemone, Nematostella vectensis. Nat. Protoc. 8, 900-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Vasylyev D. V., Dib-Hajj F., Veeramah K. R., Hammer M. F., Dib-Hajj S. D., Waxman S. G. (2013). Multistate structural modeling and voltage-clamp analysis of epilepsy/autism mutation Kv10.2-R327H demonstrate the role of this residue in stabilizing the channel closed state. J. Neurosci. 33, 16586-16593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. H., Catterall W. A. (2004). The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci. STKE 2004, re15. [DOI] [PubMed] [Google Scholar]
- Zenkert C., Takahashi T., Diesner M. O., Özbek S. (2011). Morphological and molecular analysis of the Nematostella vectensis cnidom. PLoS ONE 6, e22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bursulaya B., Lee C. C., Chen B., Pivaroff K., Jegla T. (2009). Divalent cations slow activation of EAG family K+ channels through direct binding to S4. Biophys. J. 97, 110-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bertaso F., Yoo J. W., Baumgärtel K., Clancy S. M., Lee V., Cienfuegos C., Wilmot C., Avis J., Hunyh T., et al. (2010). Deletion of the potassium channel Kv12.2 causes hippocampal hyperexcitability and epilepsy. Nat. Neurosci. 13, 1056-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou A., Lin Z., Humble M., Creech C. D., Wagoner P. K., Krafte D., Jegla T. J., Wickenden A. D. (2003). Distribution and functional properties of human KCNH8 (Elk1) potassium channels. Am. J. Physiol. 285, C1356-C1366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.