Abstract
CRISPR-Cas systems provide adaptive microbial immunity against invading viruses and plasmids. The cariogenic bacterium Streptococcus mutans UA159 has two CRISPR-Cas systems: CRISPR1 (type II-A) and CRISPR2 (type I-C) with several spacers from both CRISPR cassettes matching sequences of phage M102 or genomic sequences of other S. mutans. The deletion of the cas genes of CRISPR1 (ΔC1S), CRISPR2 (ΔC2E), or both CRISPR1+2 (ΔC1SC2E) or the removal of spacers 2 and 3 (ΔCR1SP13E) in S. mutans UA159 did not affect phage sensitivity when challenged with virulent phage M102. Using plasmid transformation experiments, we demonstrated that the CRISPR1-Cas system inhibits transformation of S. mutans by the plasmids matching the spacers 2 and 3. Functional analysis of the cas deletion mutants revealed that in addition to a role in plasmid targeting, both CRISPR systems also contribute to the regulation of bacterial physiology in S. mutans. Compared to wild-type cells, the ΔC1S strain displayed diminished growth under cell membrane and oxidative stress, enhanced growth under low pH, and had reduced survival under heat shock and DNA-damaging conditions, whereas the ΔC2E strain exhibited increased sensitivity to heat shock. Transcriptional analysis revealed that the two-component signal transduction system VicR/K differentially modulates expression of cas genes within CRISPR-Cas systems, suggesting that VicR/K might coordinate the expression of two CRISPR-Cas systems. Collectively, we provide in vivo evidence that the type II-A CRISPR-Cas system of S. mutans may be targeted to manipulate its stress response and to influence the host to control the uptake and dissemination of antibiotic resistance genes.
INTRODUCTION
CRISPRs (clustered regularly interspaced short palindromic repeats) and their associated cas (CRISPR-associated) genes found in bacteria provide a sequence-based adaptive immunity against mobile genetic elements such as phages, invasive conjugative plasmids, and transposable elements (1–8). CRISPRs consist of short repeats interspersed with nonrepetitive nucleotides of 26 to 72 bp called spacers derived from exogenous genetic elements (9–11). CRISPRs are often associated with a set of cas genes that encode proteins that mediate the defense process. This CRISPR-mediated defense system targets invading DNA in three steps: (i) adaptation via incorporation of foreign genetic element-derived spacers into the CRISPR array, (ii) transcription of CRISPR RNAs containing spacer-repeat units, and (iii) interference with the invasive nucleic acid, leading to their degradation (12). According to the current classification, there are three major types of the CRISPR-Cas systems, i.e., the type I, type II, and type III systems, respectively, that differ by the repertoires of cas genes, the organization of cas operons, and the structure of repeats in the CRISPR array (12). In the type I CRISPR-Cas systems the maturation of the precursor CRISPR RNA (pre-crRNA) is mediated by an endonuclease, namely, Cse3 (type I-E), Csy4 (type I-F), and Cas5d (type I-C), whereas in the type III system Cas6 is responsible for crRNA maturation (13–20). In the type II systems transactivating CRISPR RNA (tracrRNA) binds to the pre-crRNA, forming a dual-RNA that is essential for both crRNA maturation by RNase III and invading DNA cleavage by Cas9 (21–25). Similar to type I, type II CRISPR-Cas systems require a short protospacer adjacent motif (PAM) that is located immediately adjacent to the protospacer on the foreign DNA element (26–29).
Although CRISPR interference was originally defined as a phage resistance mechanism, CRISPR-Cas systems are now known to play a broader role in limiting horizontal gene transfer (30). In Staphylococcus epidermidis and Streptococcus pyogenes the CRISPR-Cas systems were shown to prevent the acquisition of plasmids or prophages by blocking entry in a manner akin to that performed against phage DNA (30, 31). Similar observations have been made in Enterococcus faecalis, Enterococcus faecium, and Campylobacter jejuni (32, 33). However, recently in Streptococcus pneumoniae and Neisseria meningitidis, CRISPR-Cas systems were shown to prevent natural transformation (34, 35). Beyond their now canonical function in foreign nucleic acid defense, CRISPR-Cas systems have also been implicated in various aspects of bacterial physiology, virulence, and gene regulation (12, 36–42).
In Streptococcus mutans, one of the primary pathogens implicated in dental caries, relatively little is known about its virulent phages (43, 44). Only five phages, designated M101, M102AD, M102, e10, and f1, have been shown to have lytic activity against S. mutans strains of serotypes c, e, and f, respectively (44, 45). Except for S. mutans strain OMZ381, all S. mutans serotype c strains, including UA159, are known to be resistant to phage infection by M102; only strain OMZ381 showed sensitivity to phage infection, resulting in cell lysis (45). Despite this knowledge, the mechanisms responsible for resistance to M102 in S. mutans serotype c remain unknown. S. mutans strain UA159 harbors two distinct CRISPR-Cas systems: a type II-A CRISPR1-Cas system and a type I-C CRISPR2-Cas system (45–48). The analysis of CRISPR cassettes in 29 S. mutans strains revealed that CRISPR spacers had high sequence similarity with M102, a virulent siphophage specific for S. mutans, suggesting that phage-derived spacers present in these strains likely resulted from M102-like phage attacks (45). Subsequently, it was shown that M102 adheres to phage-sensitive and phage-resistant S. mutans serotype c strains, indicating that factors besides phage adsorption determine resistance of S. mutans serotype c strains to infection by M102 phage (49). Despite these studies that explored the role of CRISPR-Cas systems in S. mutans in conferring phage immunity, recent transcriptome studies suggest other functions that CRISPR-Cas systems might have in S. mutans are poorly understood (50–55).
Here, we investigated the role of CRISPR-Cas systems in phage defense, natural transformation, and stress resistance of S. mutans by utilizing cas gene deletion mutants in S. mutans UA159. We found that S. mutans CRISPR-Cas systems are not essential for phage resistance against M102. However, we demonstrate that the S. mutans type II-A CRISPR1-Cas system inhibits plasmid transformation. Furthermore, we show that CRISPR-Cas systems are regulated by the VicR/K signaling system to modulate environmental stress tolerance and DNA repair, thereby expanding the role of CRISPR-Cas systems in this pathogen.
MATERIALS AND METHODS
Strains, plasmids, phage, and growth conditions.
Bacterial strains, plasmids, and the phage used in the present study are listed in Table 1. All S. mutans strains were grown in Todd-Hewitt broth supplemented with 0.3% yeast extract (THYE; Becton Dickinson, Sparks, MD) as static cultures or on THYE medium with 1.5% (wt/vol) agar (Bioshop, Burlington, Ontario, Canada) at 37°C in a 5% (vol/vol) CO2 atmosphere. Kanamycin (1 mg/ml), spectinomycin (1 mg/ml), and/or erythromycin (10 μg/ml) were added as needed. Escherichia coli DH5α was used as the host for propagation of plasmids and was routinely cultured in Luria-Bertani medium supplemented (when necessary) with spectinomycin (100 μg/ml) at 37°C with aeration. Phage M102 was propagated in S. mutans strain OMZ381, and the phage titer was determined using a plaque assay as described previously (43). Phage resistance was assayed using both the plaque formation assay as described previously (45) and liquid growth assays. For plaque assays, 100 μl of exponentially growing bacterial cultures was mixed with 100 μl of undiluted (∼108 PFU) and 100-fold serial dilutions of phage M102. After incubation at 37°C for 20 min, 4 ml of THYE soft top agar was added and immediately poured on THYE plates, followed by incubation for 48 h at 37°C. Phage sensitivity was assessed based on the number of discrete plaques. For liquid growth assays, 350 μl of exponentially growing cultures were challenged with 5 μl of undiluted phage M102 (∼108 PFU), and phage sensitivity was monitored by using an automated growth reader (Bioscreen C Labsystems, Finland). Growth kinetic experiments were performed under the following stress conditions: pH 5.5, 0.4 M NaCl, 0.003% H2O2, 0.004% sodium dodecyl sulfate (SDS), or 25 mM paraquat, as previously described (56). No antibiotics were used in growth assays in order to avoid additional stress. For heat shock resistance assays, mid-log-phase cells were incubated at 50°C for 60 min. Samples after heat exposure or incubation at 37°C for 1 h were serially diluted and plated on THYE plates, and the CFU were counted.
TABLE 1.
Bacterial strains, plasmids, and phage used in this study
| Strain, phage, or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| UA159 | Wild-type strain; Erms Kans Spcs | 75 |
| OMZ381 | Wild-type strain; Erms Kans Spcs | 45 |
| SmuvicK | VicK-deficient mutant derived from UA159; Ermr | 66 |
| CRISPR deletion mutants derived from UA159 | ||
| ΔC1S | Lacking cas9 to csn2; Spcr | This study |
| ΔC1K | Lacking cas9 to csn2; Kanr | This study |
| ΔC2E | Lacking cas3 to cas2; Ermr | This study |
| ΔC1SC2E | Lacking all cas genes; Ermr Spcr | This study |
| ΔC1KC2E | Lacking all cas genes; Ermr Kanr | This study |
| ΔCR1SP13E | Lacking four repeats and spacers 1 to 3; Ermr | This study |
| Phage M102 | 43 | |
| Plasmids | ||
| pCG1 | Streptococcus-E. coli shuttle vector; Spcr | 58 |
| pCG1SP | pCG1 plasmid containing irrelevant spacer; Spcr | This study |
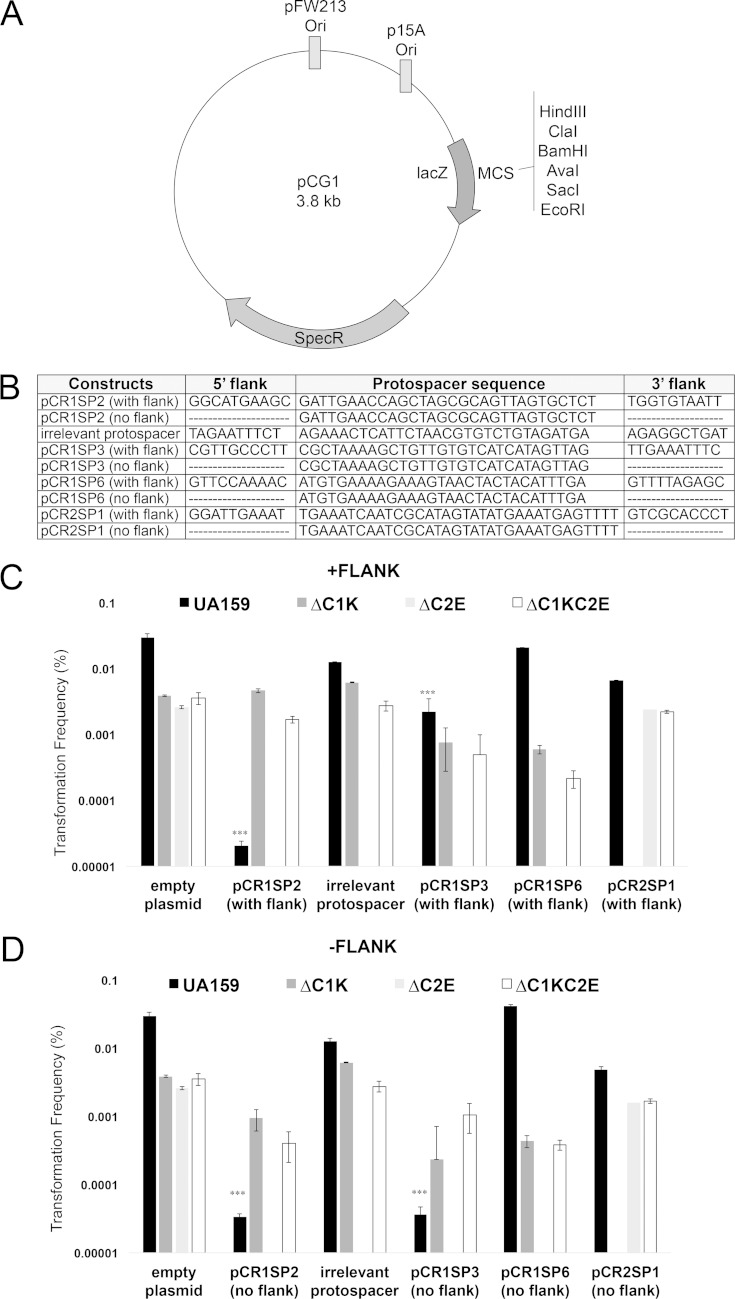
| pCR1SP2 (with flank) | pCG1 plasmid containing spacer 2 with potential PAM; Spcr | This study |
| pCR1SP2 (no flank) | pCG1 plasmid containing spacer 2 with no potential PAM; Spcr | This study |
| pCR1SP3 (with flank) | pCG1 plasmid containing spacer 3 with potential PAM; Spcr | This study |
| pCR1SP3 (no flank) | pCG1 plasmid containing spacer 3 with no potential PAM; Spcr | This study |
| pCR1SP6 (with flank) | pCG1 plasmid containing spacer 6 with potential PAM; Spcr | This study |
| pCR1SP6 (no flank) | pCG1 plasmid containing spacer 6 with no potential PAM; Spcr | This study |
| pCR2SP1 (with flank) | pCG1 plasmid containing spacer 1 with potential PAM; Spcr | This study |
| pCR2SP1(no flank) | pCG1 plasmid containing spacer 1 with no potential PAM; Spcr | This study |
Erm, erythromycin; Spc, spectinomycin; Kan, kanamycin. Superscripts: r, resistant; s, sensitive.
Construction of mutants in S. mutans.
PCR ligation mutagenesis (57) with the primers listed in Table S1 in the supplemental material was utilized to construct nonpolar deletion mutants in cas genes or CRISPR spacers in S. mutans UA159 wild-type strain: (i) two deletion mutants in the cas9-csn2 operon within the CRISPR1-Cas system (strain ΔC1S and ΔC1K), (ii) one deletion mutant in the cas3-cas2 operon within the CRISPR2-Cas system (strain ΔC2E), (iii) two deletion mutants in all cas genes within the CRISPR1-Cas system and CRISPR2-Cas system (strain ΔC1SC2E and strain ΔC1KC2E), and (iv) a deletion mutant lacking four repeats and three spacers within the CRISPR1 array, including the spacer identical to M102 (strain ΔCR1SP13E). Successful mutagenesis was validated using nucleotide sequence analysis and quantitative reverse transcription-PCR (qRT-PCR).
Plasmid construction for transformation studies.
Shuttle vector pCG1 that replicates in both E. coli and S. mutans was used to clone predicted protospacers for the purposes of plasmid transformation assays (58). This plasmid has a β-galactosidase gene that can be disrupted by inserting the spacer sequence and can be quickly screened (blue/white on X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]) for successful cloning. Oligonucleotides (50 to 54 nucleotides [nt]) corresponding to protospacer candidates (matching spacers 2, 3, and 6 of the CRISPR1 locus and spacer 1 of the CRISPR2 locus), along with or without 10-nt upstream and downstream sequences were obtained from ACGT Toronto, Ontario, Canada (see Table S2 in the supplemental material). Protospacer candidates were selected as containing a sequence with >85% similarity to the S. mutans UA159 spacer sequences and originating from virulent phage M102 or genomic DNA from closely related species (S. mutans GS-5 and LJ23 genomes). Extra bases corresponding to SacI restriction sites were added onto the synthesized oligonucleotides so that sticky ends were created when annealed oligonucleotides were digested. After digestion, these protospacers were ligated to pCG1 digested with SacI and dephosphorylated with alkaline phosphatase (New England BioLabs). All constructs were transformed and propagated in E. coli DH5α prior to transformation of S. mutans UA159. Successful spacer cloning was validated using nucleotide sequence analysis.
Competence assays.
In the present study, natural transformation of planktonic-cell suspensions of UA159 and ΔC1K, ΔC2E, and ΔC1KC2E mutants was assessed by using streptococcal plasmid pCG1 constructs (Table 1; see also Fig. 2 and associated text for further details) and compared to an empty vector control. Overnight cultures of UA159 and its mutant strains were diluted 20-fold in THYE and incubated at 37°C until an optical density at 600 nm (OD600) of ∼0.1 was reached, and transformation frequency (TF) assays were conducted as described previously (56).
FIG 2.
The CRISPR1-Cas system of S. mutans UA159 provides immunity against plasmid transformation. (A) Schematic representation of cloning vector pCG1 used for the construction of plasmids for the transformation interference assay. Plasmids for interference assays were produced by inserting a protospacer and 10 nt on both sides of the protospacers into pCG1 plasmid. (B) pCG1 constructs containing potential targets for different S. mutans UA159 spacers (spacers 2, 3, and 6 within CRISPR1 array and spacer 1 within CRISPR2 array). pCG1derivatives in the presence (C) or absence (D) of flank sequences were tested by natural transformation assays using the wild-type S. mutans UA159, ΔC1K, ΔC2E, and ΔC1SC2E strains. The transformation frequency was calculated as the transformant CFU divided by the total number of viable cells. The results shown are representative of at least two independent experiments. ***, constructs showing targeting phenotype.
In vivo assay for DNA damage.
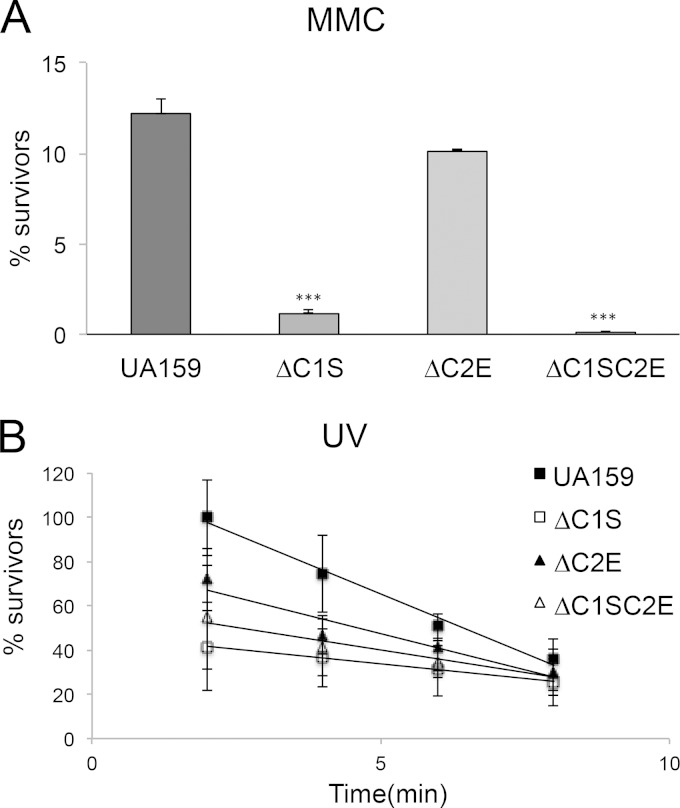
Cells in mid-exponential phase were exposed to UV light (at an intensity of ∼125 μW/cm2) for 0, 2, 4, 6, 8, and 10 min, and then serially diluted cultures were spotted on THYE agar plates, followed by incubation in the dark at 37°C for 48 h. The CFU were then counted. For mitomycin C (MMC) sensitivity assays, exponentially growing strains in THYE broth were harvested by centrifugation, resuspended in THYE in the presence or absence (control) of 0.05 μg of MMC/ml, and incubated at 37°C for 90 min. Sensitivity was quantitatively assessed by plating cells after incubation.
Gene cloning and protein purification.
Cas5d (SMU.1763c) was cloned using genomic DNA from S. mutans UA159 and primers in Table S1 in the supplemental material into the modified pET15b plasmid as previously described (59). For enzymatic assays, Cas5d protein was overexpressed as a fusion with an N-terminal His6 tag in E. coli BL21(DE3) strain (Novagen) which contains no cas genes (2). The protein was purified to >95% homogeneity using metal-chelate affinity chromatography on a nickel affinity resin and subsequent ion-exchange chromatography on a MonoQ column as previously described (59).
RNase assays.
RNA1 (39 nt; 5′-AAAUACGUUUUCUCCAUUGUCAUAUUGCGCAUAAGUUGA) and RNA2 (40 nt; 5′-UUUCAAUUCCUUUUAGGAUUAAUCUUGAAGAUAGAGUUAA) substrates were obtained from Integrated DNA Technologies. Labeling of RNA substrates was conducted using [γ-32P]ATP and T4 polynucleotide kinase according to the manufacturer's instructions (Fermentas). Reaction conditions were as follows: 50 mM Tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.1 nM substrate, and enzyme for 1 h at 37°C. Samples were run on 15% PAGE gels containing 8 M urea, and reaction products were visualized by autoradiography. When RNA from UA159 was used as the substrate, total RNA was extracted from cells grown to mid-log phase (OD600 ≈ 0.4) in THYE medium. One microgram of total RNA was incubated with 1 μg of Cas5d protein in transcription buffer (50 mM Tris-HCl [pH 8.5], 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT]) for 1 h at 37°C. RNA samples were purified by using an RNeasy kit (Qiagen) and then subjected to microarray analysis as described below (51).
Cell preparation for gene expression and microarray analysis.
For quantitative real-time PCR (qRT-PCR), cells from S. mutans UA159 and SmuvicK were grown to mid-log phase (OD600 of ∼ 0.4) under regular or acidic conditions (pH 7.5 versus pH 5.5). To study cas gene expression under regular conditions, overnight cultures were diluted 1:20 in sterile prewarmed THYE and grown to mid-log phase. Cells were harvested by centrifugation, snap-frozen in liquid nitrogen, and stored at −80°C until required. To measure cas gene expression under acidic conditions, overnight cells were diluted 20-fold in sterile prewarmed TYE supplemented with 0.5% glucose (pH 7.5) and grown to mid-log phase (OD600 = 0.4 to 0.5). Cultures were then divided into two aliquots, and cells were collected by centrifugation. Pellets were resuspended in 0.5% glucose supplemented TYE adjusted to pH 7.5 or pH 5.5, and cultures were incubated for 1 h at 37°C with 5% CO2. The cells were then harvested by centrifugation, snap-frozen in liquid nitrogen, and stored at −80°C until RNA isolation. For microarray analysis, cells from S. mutans UA159 at mid-log phase were harvested by centrifugation and utilized for RNA isolation. For qRT-PCR to study nonpolar effects on the downstream genes, cells from UA159 and cas deletion mutants were grown in THYE until an OD600 of ∼0.4 was reached, harvested by centrifugation, snap-frozen in liquid nitrogen, and stored until used for cDNA synthesis. For Northern blot analysis, cells from S. mutans UA159 were grown to early log phase (OD600 of ∼0.1), mid-log phase (OD600 of ∼0.4), and early stationary phase (OD600 of ∼1.0) in THYE at 37°C. For competence-stimulating peptide (CSP) treatment, cells from UA159 grown to early-log phase (OD600 of ∼0.1) were supplemented with 0.2 μM CSP and incubated for 2 h at 37°C until a final OD600 of ∼0.4 was reached. For heat shock and oxidative stress analyses, cells that reached an OD600 of ∼0.4 were pelleted, and the cells exposed to (0.003%) H2O2 and 50°C were incubated for 20 min. The cells were then pelleted, snap-frozen in liquid nitrogen, and stored at −80°C until used.
Gene expression analysis using qRT-PCR.
DNase treatment, cDNA synthesis, qRT-PCR and expression analysis were carried out as previously described (56). Primers used for qRT-PCR are listed in Table S1 in the supplemental material. Expression was normalized against internal standards, gyrA, 16S rRNA, and gtfB. Changes in gene expression were determined using the Pfaffl method (60).
Global transcriptome analysis.
RNAs from mid-exponential-phase cells of S. mutans UA159 were incubated in the presence (experimental) or absence (control) of Cas5d (SMU.1763c) protein for 60 min at 37°C. The RNA samples were transcribed into cDNA using a First-Strand synthesis kit (Invitrogen) as specified in the manufacturer's protocol. Control and experimental cDNAs were used for microarrays (51). A class comparison analysis was performed to identify statistically significant genes. The statistical algorithm used was the two-sample t test (with random variance model) with the parametric P value cutoff set to P < 0.05. Selected genes that showed significant differential expression under experimental conditions were validated utilizing qRT-PCR.
In vitro transcription analysis.
RNA transcripts from DNA templates of SMU.995 and SMU.1502c were obtained by in vitro transcription using MAXIscript kit (Ambion). Transcription reactions (20 μl) containing 10 mM (each) ATP, CTP, GTP, and UTP, 1 μg of PCR product for each transcript, 2 μl of transcription buffer, and 2 μl of T7 phage RNA polymerase was incubated for 1 h at 37°C. Turbo DNase (1 μl) was added to the reaction to remove the template DNA. The transcripts were purified using 3 M sodium acetate and 100% ethanol and then used for RNase activity assay as described above.
Northern blot analysis.
Total RNA was isolated from UA159 cultures incubated under different growth conditions, as indicated above, using Direct-zol RNA MiniPrep (Zymo Research). Five micrograms of total RNA was loaded by lane and resolved on a 8% (wt/vol) polyacrylamide denaturing gel containing 7 M urea. Size-fractionated RNA was transferred to a positively charged nylon membrane (Fermentas) using a Bio-Rad Mini Trans-Blot cell and subjected to UV cross-linking for 5 min. Membranes were prehybridized using DIG Easy Hyb (Roche) for 30 min at 42°C and followed by hybridization with DIG High-Prime DNA probes (25 ng/ml) in DIG Easy Hyb hybridization buffer (Roche) at 42°C overnight. The probes for tracrRNA were PCR amplified from UA159 gDNA using tracrRNA-For and tracrRNA-Rev primers (see Table S1 in the supplemental material) and labeled using the digoxigenin (DIG) High Prime Labeling kit (Roche) according to the supplier's instructions. The probed membrane was incubated with CSPD (Roche), and the chemiluminescent signal was visualized using a chemiluminescent detector (Bio-Rad) and photographed. Densitometry was used to determine the transcript expression levels within the detected bands, and these levels were quantified using the ImageJ64 program (National Institutes of Health, Bethesda, MD) (61). 5S rRNA served as a loading control.
RESULTS
The S. mutans UA159 CRISPR-Cas systems.
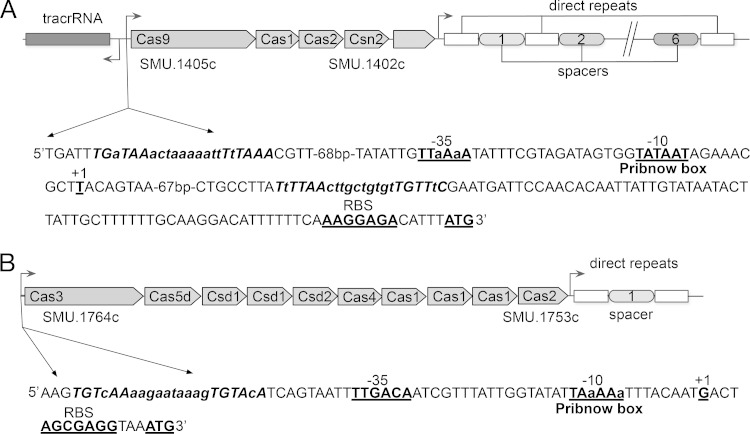
The S. mutans UA159 genome contains two CRISPR-Cas systems: the CRISPR1-Cas system (Fig. 1A) and the CRISPR2-Cas system (Fig. 1B). The CRISPR1-Cas system of subtype II-A spans ∼5.8 kb and has four cas genes organized in an operon: the genes for Cas1 (SMU.1404c, 288 amino acids [aa]) and Cas2 (SMU.1403c, 109 aa) encoding core proteins predicted to be implicated, together with Csn2 (SMU.1402c, 190 aa) in spacer acquisition, and the gene for Cas9 (also known as Csn1) (SMU.1405c, 1,345 aa), which encodes the hallmark protein of type II systems associated with the interference step. A CRISPR array (located between open reading frames [ORFs] SMU.1400 and SMU.1398) consists of seven repeats (36 bp) interspaced by six spacers (30 bp). Spacer 3 shared 100% nucleotide identity with a 30-bp sequence from S. mutans phage M102, and it was previously hypothesized to confer resistance against M102-like phage infection (45). Our sequence similarity searches using the NCBI database for the other spacer sequences within the CRISPR1 locus revealed at least one potential target. For simplicity, we considered only candidate protospacers that matched the CRISPR spacers without or with a few mismatches within the CRISPR spacers. Spacer 2 partially matched (26 bp of 30 bp with 100% sequence identity) to phage M102 and spacer 6 had 100% sequence identity to a segment in the S. mutans GS-5 genome, suggesting that the CRISPR1-Cas system of S. mutans UA159 might target not only M102 phages but also incoming DNA from other S. mutans genomes (see Table S4 in the supplemental material). This possibility is in agreement with the recent studies that examined in vivo expression of the CRISPR1 locus in S. mutans UA159, suggesting that the CRISPR1 locus is transcriptionally active (21, 22). Our deduced protein sequence homology search using BLASTP revealed that the cas genes associated with CRISPR1 encoded hypothetical proteins that are predicted to have nuclease activity involved in defense mechanisms or DNA repair (see Table S3 in the supplemental material) (46, 62). The CRISPR2-Cas system of subtype I-C spans ∼8.0 kb and consists of 10 cas genes organized in an operon: the genes for three Cas1 subunits, i.e., SMU.1757c (94 aa), SMU.1755c (199 aa), and SMU.1754c (131 aa), and Cas2 (SMU.1753c, 97 aa), predicted to act in the adaptation step; genes encoding proteins that are predicted to form Cascade-like complexes involved in the interference stage, i.e., Cas3 (SMU.1764c, 131 aa), Cas4 (SMU.1758c, 214 aa), and Cas5d (SMU.1763c, 249 aa); and three Cas genes representing the CRISPR subtype group Dvulg, i.e., SMU.1760c (291 aa), SMU.1761c (469 aa), and SMU.1762c (187 aa). An array of two 32-bp repeats interspaced by one 34-bp spacer was present downstream of its associated cas genes. Spacer 1 within CRISPR2 array matched 100% to a genomic nucleotide sequence of S. mutans LJ23 (see Table S4 in the supplemental material). Using in silico analysis of CRISPR2 array, we found a leader sequence upstream of the first repeat possibly acting as a promoter for the transcription of the array CRISPR (data not shown). However, in accordance with previous findings (45, 47), we observed that Cas1, required for spacer acquisition, is very unusual. Unlike most other Cas1 proteins that typically are encoded by one gene (forming asymmetrical homodimers) (36, 63, 64), Cas1 of the type I-C CRISPR2-Cas system has three apparent ORFs in UA159. Since both SMU.1757c (94 aa) and SMU.1754c (133 aa) appear to be too short to encode a functional assembly of Cas1, they might represent truncated regions that prevent the genes from functioning properly. Alternatively, they might result from annotation errors. Since in this locus only one spacer was identified, it is possible that the CRISPR2 locus has lost its ability to incorporate novel CRISPR2 spacers as hypothesized previously (45, 47). However, the in vivo activity of the CRISPR2 array remains to be elucidated. Similar to cas genes from CRISPR1, our BLASTP analysis of Cas proteins revealed high sequence similarity to nucleases, helicases and DNA repair proteins (see Table S3 in the supplemental material) (46, 62). Promoter analysis of CRISPR1 cas genes and CRISPR2 cas genes revealed classical elements, including a putative Pribnow box (−10 box; TATAAT and TAaAAaT, respectively) and the −35 element (TTaAaA and TTGACA, respectively), suggesting that cas genes within the same cluster are likely to be cotranscribed. We detected a putative binding site (TGTWAHNNNNNTGTWAH) (65) for the VicR response regulator protein, which is the regulatory component of the VicR/K signaling system for CRISPR1 cas genes regulation (located at positions −118 to −139 and positions +84 to +105 from the putative transcriptional start site) and for CRISPR2 cas regulation (located at positions −47 to −68 from the putative transcriptional start site). This finding is consistent with our published (51) and unpublished work. It is likely that VicR may bind these target sequences to activate or repress the expression of cas genes (65, 66) (Fig. 1).
FIG 1.
Gene maps of the CRISPR-Cas systems in S. mutans UA159. (A) CRISPR1-Cas system. (B) CRISPR2-Cas system. Analysis of the promoter regions of cas genes identified the putative −10 box, −35 box, transcriptional start site (TSS), and ribosome binding site (RBS) (all underlined in boldface), as well as the putative VicR binding consensus sequence (TGTWAH-6/10 bp-TGTWAH) for cas gene regulation.
Loss of the M102-specific CRISPR spacers or cas genes of both CRISPR-Cas systems have no effect on the phage resistance phenotype of S. mutans UA159.
Since spacers 2 and 3 within the CRISPR1 array matched sequences of the M102 genome, we hypothesized that their presence might facilitate phage defense in S. mutans UA159. To test whether the M102-targeting spacers and CRISPR-Cas systems confer immunity against M102 phage infection, S. mutans UA159, OMZ381 (a phage-sensitive strain) and CRISPR-Cas-deficient strains were assessed for phage resistance by challenging with the virulent phage M102 in both liquid growth assays and plaque formation assays. Deletion of cas genes of CRISPR1 and/or CRISPR2 or removal of spacers 2 and 3 within the CRISPR1 array in S. mutans UA159 did not affect phage sensitivity of UA159, since none of these strains were lysed by M102 in plate or liquid-based lytic assays (data not shown). However, in accordance with previous findings by Van der Ploeg (45), the control OMZ381 strain displayed sensitivity to phage, as judged by its complete lysis in the presence of M102 (see Fig. S1A and B in the supplemental material). Since all mutant strains remained resistant to infection by phage M102, we concluded that an as-yet-unidentified and CRISPR-independent mechanism(s) is responsible for the M102-resistant phenotype displayed by UA159.
The type II-A CRISPR-Cas system prevents natural transformation by plasmids in S. mutans UA159.
In addition to conferring phage immunity, CRISPR-Cas systems were shown to constitute an effective barrier against artificial means of transformation (e.g., electroporation) in several bacteria (22, 30, 67, 68). Further, it was shown that the introduction of the engineered Streptococcus pyogenes CRISPR-Cas system reduced the transformation efficiency in a heterologous host, Streptococcus pneumoniae (34). Zhang et al. (35) found that the native meningococcal CRISPR-Cas system was able to prevent natural transformation of spacer-matching sequences, suggesting that it can limit the horizontal spread of virulence genes. These studies raised the question whether naturally transformable S. mutans employs CRISPR-Cas systems to form an effective barrier to limit foreign DNA acquisition by transformation. The S. mutans spacer sequences have potential matches to either phage M102 or other bacterial species present in the dental plaque (data not shown); however, for simplicity we selected only candidate protospacers that fully matched spacer sequences from UA159 or had only a few mismatches, namely, CR1SP2, CR1SP3, and CR1SP6 within the CRISPR1 locus and CR2SP1 within the CRISPR2 locus of S. mutans UA159 (Fig. 2). Previously, it was shown that in type II systems, the PAM sequence is located at the protospacer 3′ end, whereas for the type I systems it is located at the 5′ end of the protospacer (23, 68, 69). To deduce putative PAM motifs for S. mutans UA159, the identified 10-nt sequences located directly downstream and upstream of the protospacer sequences were aligned using WebLogo (data not shown). Sequence logos revealed that all potential natural targets except for the one matching spacer 2, with 3′-PAM(5′-TGGTGTAATT-3′) downstream of the protospacer 2, have flanking sequences that deviate significantly from the PAM consensus identified in other S. mutans strains (see Table S4 in the supplemental material) where 5′-NGG-3′ is located at 3′ end for the type II-A system and 5′TTC-3′ at the 5′ end for the type I-C system (45, 70). Since we were not able to identify the most common nucleotides that could represent the PAM sequence, we designed our plasmid constructs containing the protospacers matching CR1SP2, CR1SP3, and CR1SP6 within the CRISPR1 locus and CR2SP1 within the CRISPR2 locus of S. mutans UA159 and included 10 nt on both sides of the protospacer. For comparison, we also cloned protospacers lacking flanking sequences (Fig. 2B). The resulting pCG1vectors were used in transformation assays into wild-type UA159 and its cas deletion mutant strains. The transformation frequency of each plasmid carrying a protospacer was compared to that of an empty vector (Fig. 2C and D). Empty pCG1 consistently exhibited percent transformation frequencies (%TF) of (2 to 4) × 10−2, an observation consistent with previous work (data not shown). Protospacers with identity to spacers 2 and 3 within CRISPR1 had dramatically decreased %TF values compared to the native plasmid, suggesting that the type II-A CRISPR-Cas system is functional against the plasmids carrying the protospacers matching CR1SP2 and CR1SP3. In contrast, plasmids carrying targets for spacer 6 within CRISPR1 and spacer 1 within CRISPR2 (both matching sequences from other S. mutans genomes) exhibited transformation frequencies comparable to those of the empty vector, suggesting that they were recognized as self after introduction by transformation and were not targeted by CRISPR machinery (Fig. 2C and D). Similar to previous work (71), we found that the degree of inhibition of transformation varied widely depending on the protospacer tested. For example, the protospacer from M102 phage with 86% identity to the CR1SP2 spacer caused an ∼1,000-fold inhibition of transformation while the protospacer from M102 phage with 100% identity to CR1SP3 resulted in only ∼10-fold inhibition. Intriguingly, despite the previously demonstrated importance of the PAM sequence in interference in type II-A systems (35, 68, 69, 72, 73), protospacers matching CRISPR spacers 2 and 3 cloned without any flanking sequences, consistently failed to yield transformants, indicating that they likely elicited CRISPR interference (Fig. 2D). Transformation frequency of ΔC1K and ΔC1KC2E in the presence of plasmids targeting CR1SP2 was restored to that of empty vector, as well as in the presence of CR1SP3, though to a lesser extent, indicating that the CRISPR1-Cas function was abolished by deletion of cas genes.
Purified recombinant SMU.1763c (Cas5d) has RNase activity.
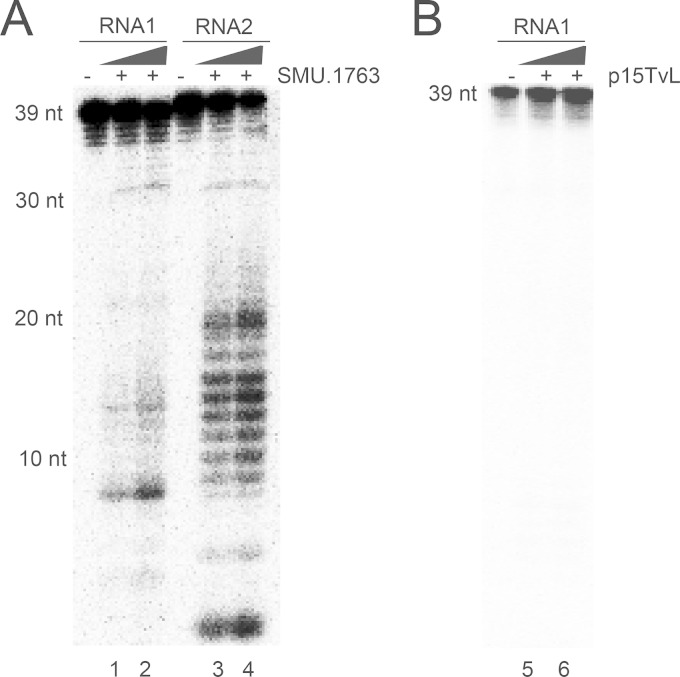
Cas5d protein belongs to the subtype I-C/Dvulg CRISPR-Cas system, and recent work provided evidence that pre-crRNA processing, which is the key molecular event that initiates the CRISPR interference, is mediated by the Cas5d protein which, after the maturation process, assembles with crRNA, Csd1, and Csd2 proteins to form an interference complex (18). To investigate whether SMU.1763c (Cas5d) in S. mutans UA159 is capable of cleaving the RNA, the protein was overexpressed in E. coli, and purified protein was assayed for nuclease activity in a dose-dependent manner using the 32P-labeled single-stranded (ss) synthetic oligoribonucleotides as the substrates. As shown in Fig. 3A, purified SMU.1763c cleaved ssRNA substrates into small fragments. A control protein fraction purified from E. coli cells containing the empty plasmid was used to confirm that the observed RNase activity was associated with SMU.1763c and not with contaminating E. coli RNases (Fig. 3B). Although the purified SMU.1763c was >95% pure (see Fig. S2 in the supplemental material), at this stage we cannot exclude copurification of contaminant proteins, and we will address this in future work using site-directed mutagenesis. Similar results were obtained when the DNase-treated RNA from UA159 was used as a substrate (data not shown). These results indicate that SMU.1763c protein is a RNase with activity against ssRNA.
FIG 3.
Cleavage of the synthetic ssRNA substrates by the Cas5d protein from S. mutans UA159. 5′-32P-labeled RNA1 or RNA2 (0.05 μm) was incubated in the absence or in the presence of 100 ng (lanes 1 and 3) or 200 ng of Cas5d (SMU.1763c) (lanes 2 and 4) or in the presence of 100 or 200 ng of purification product obtained from E. coli cells transformed with an empty p15TvL vector (lanes 5 and 6) at 37°C for 30 min in the presence of 50 mM Tris-HCl (pH 7.0), 5 mm MnCl2, 100 mM KCl, and 1 mM DTT. Reaction products were separated on a 15% PAGE–8 M urea gel and visualized by phosphorimaging. 39-nt RNA1 and 40-nt RNA2 were prepared by using the oligonucleotides 5′-AAAUACGUUUUCUCCAUUGUCAUAUUGCGCAUAAGUUGA and 5′-UUUCAAUUCCUUUUAGGAUUAAUCUUGAAGAUAGAGUUAA, respectively.
SMU.1763c cleaves cellular RNAs.
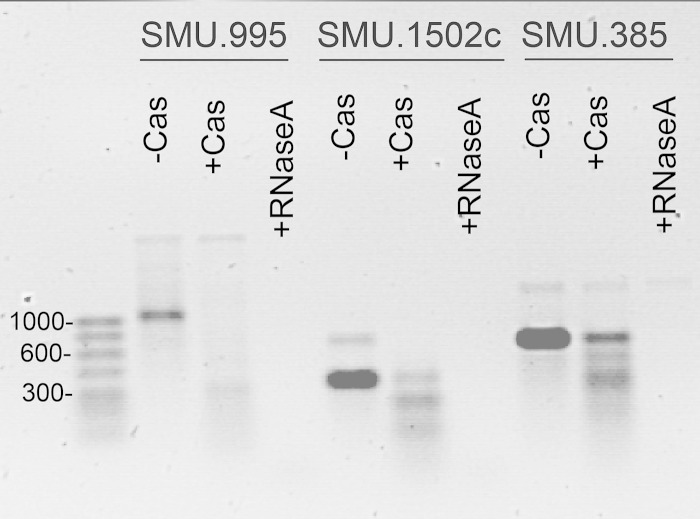
To identify potential RNA substrates targeted and cleaved by the SMU.1763c protein, we used RNAs from mid-exponential-phase cells of S. mutans UA159 in the presence (experimental) or absence (control) of this protein. DNase-treated RNA samples were converted to cDNA and used for global microarray analysis. Five transcripts were downregulated >1.8-fold by the addition of Cas5d protein (P < 0.05) (see Fig. S3 in the supplemental material), and four were confirmed as significantly reduced by qRT-PCR (P < 0.05), including a putative ABC transporter (SMU.995), a putative cell envelope protein (SMU.246c), and two hypothetical proteins (SMU.1502c and SMU.2075c). To confirm the ability of SMU.1763c to specifically target these substrates, we performed “in vitro” transcription analysis using full-length DNAs of SMU.995, SMU.1502c, and SMU.385 (a random substrate from UA159) and T7 phage RNA polymerase. SMU.1763c cleaved all targets, including the control SMU.385, suggesting that Cas5d did not exhibit sequence specificity in its RNA cleavage activity as anticipated from the microarray experiment (Fig. 4). It is possible that RNAs that appear as differentially regulated in the presence of Cas5d were more accessible to RNase activity due to their abundance or their location on the genome.
FIG 4.
Cas5d (SMU.1763c) cleavage of RNA transcripts of SMU.995 and SMU.1502c generated by in vitro transcription.
Cas proteins are involved in sensitivity to DNA damage.
Cas1 and Cas2 proteins have been predicted to be involved in DNA repair (74). Recent work in E. coli demonstrated that a mutant deficient in Cas1 had a DNA repair-deficient phenotype (36). Since proteins encoded by SMU.1403c, SMU.1404c, SMU.1754c, SMU.1753c, SMU.1755c, and SMU.1757c within CRISPR-Cas systems display sequence similarity to the Cas1 and Cas2 family proteins (see Table S3 in the supplemental material), we tested cas deletion mutants under DNA-damaging conditions to evaluate their putative roles in DNA repair. Hence, we examined the survival of S. mutans UA159 and mutant strains under DNA-damaging conditions induced by 0.05 μg mitomycin C (MMC)/ml or UV irradiation. The survival of ΔC1S, and ΔC1SC2E cells was drastically altered when exposed to MMC (P < 0.005) or UV irradiation (P < 0.005) relative to the wild-type strain, suggesting a role in DNA repair (Fig. 5). Further, to validate that our phenotypic changes were caused only by the lack of cas genes, we carried out qRT-PCRs on their downstream genes. Deletion of cas genes within CRISPR1 (ΔC1S strain), CRISPR2 (ΔC2E strain), and CRISPR1+2 (ΔC1SC2E) had no polar effects on the downstream genes, as judged by expression analysis using UA159 and mutant strains (see Fig. S4 in the supplemental material). Together, our results suggested a role for these proteins in conferring protection to DNA damaging agents.
FIG 5.
Effects of MMC or UV irradiation on viability of S. mutans UA159 and mutant strains. (A) S. mutans UA159, as well as ΔC1S, ΔC2E, and ΔC1SC2E strains, was exposed to 0.050 μg of MMC/ml for 1 h. The results shown are representative of at least two independent experiments. Statistical analyses were performed using the Student t test (***, P < 0.005). (B) Actively growing cells of UA159 and its mutant strains were exposed to UV irradiation for 2, 4, 6, and 8 min. The results here represent the average of two independent experiments ± the standard errors. The differences were statistically significant (P < 0.005; P < 0.05 [Student test]).
ΔC1S responds to oxidative, SDS, acid, and high-temperature stressors.
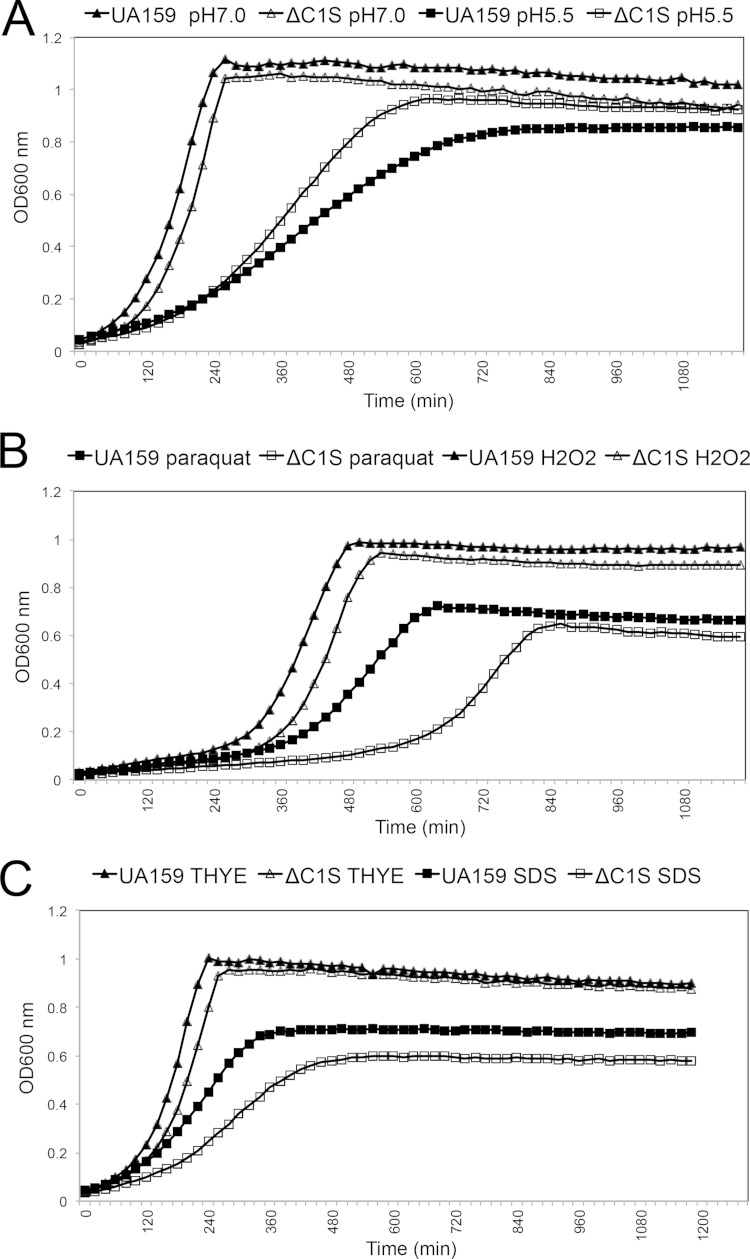
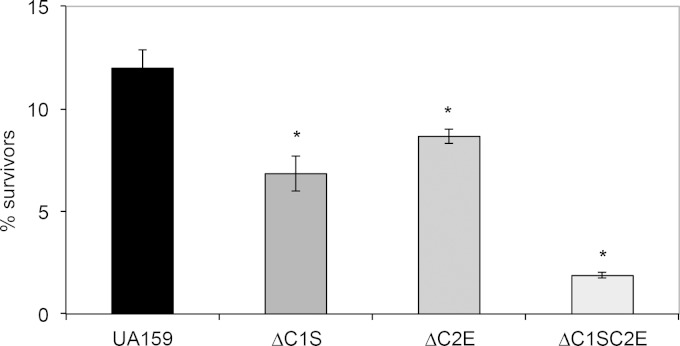
Since previous transcriptome studies in S. mutans linked CRISPR-Cas systems with environmental stress tolerance (50–55), we monitored growth kinetics of UA159 and cas deficient mutants under low pH (5.5), H2O2 (0.003%), SDS (0.004%), paraquat (25 mM), NaCl (0.4 M), and ethanol (2%), using an automated growth reader. ΔC1S strain grew faster under low pH and had a higher yield compared to the wild-type strain, suggesting that cas genes associated with the CRISPR1 locus have a role in the acid tolerance of S. mutans (Fig. 6A). The ΔC1S strain was impaired in its ability to tolerate stresses induced by paraquat, H2O2 and SDS, suggesting the ΔC1S played a role in responding to intracellular oxidative stress (paraquat), extracellular oxidative stress (H2O2), and cell membrane stress (SDS) (Fig. 6B and C). In the presence of other stresses induced by NaCl and ethanol, the ΔC1S mutant grew similarly to UA159 (data not shown). Although the ΔC2E mutant did not reveal drastically altered growth rates compared to wild-type UA159 strain under any of the environmental stressors tested (see Fig. S5 in the supplemental material), ΔC1SC2E mutant displayed growth phenotypes similar to those of the ΔC1S strain (data not shown). Under high-temperature stress, the survival of all mutant strains was impaired (P ≤ 0.05) compared to the wild type, suggesting that S. mutans cas genes have a role in temperature stress tolerance (Fig. 7). Since the double mutant displayed sensitivity higher than either of the single mutants, it is possible that both CRISPR-Cas systems work cooperatively or sequentially to combat temperature stress.
FIG 6.
Growth kinetics of S. mutans UA159 and ΔC1S under various stressors: pH 7.0 or 5.5 (A), 25 mM paraquat or 0.003% H2O2 (B), and THYE or 0.004% SDS (C). Each point represents the average of four independent optical density values per sample. These results shown are representative of two independent experiments conducted with the mutant and UA159 parent strain.
FIG 7.
Survival of S. mutans UA159 and mutant strains after exposure to 50°C temperature stress for 1 h. The results represent mean CFU counts ± the standard deviations. The differences were statistically significant (P ≤ 0.05, Student t test). These results shown are representative of two independent experiments conducted with the mutants and UA159 parent strain.
The S. mutans cas genes are transcriptionally regulated by the two-component regulatory system VicR/VicK.
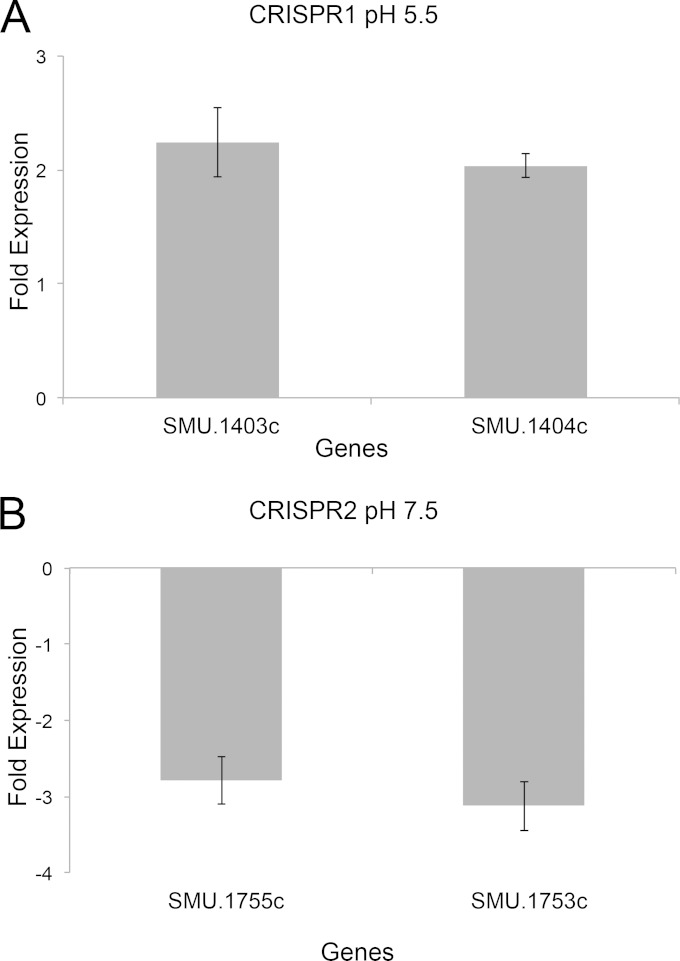
Search of the promoter regions located upstream of the CRISPR1 cas and CRISPR2 cas genes revealed the presence of putative binding sites for the VicR response regulator protein on their expression. To test the regulatory role of the VicR/K system in modulating the activity of cas genes, we performed qRT-PCR using cDNAs isolated from a VicK-deficient mutant (SmuvicK) and UA159 strains and examined the expression of two candidate genes from each CRISPR operon. High expression levels of cas genes from both CRISPR operons were observed in UA159 cells, suggesting that these genes are being expressed under mid-log-growth phase. Loss of VicK caused >2-fold downregulation of SMU.1753c and SMU.1755c expression from the CRISPR2-Cas system, suggesting that VicK mediated a positive regulatory role on their expression (Fig. 8A). Conversely, SMU.1403c and SMU.1404c from the CRISPR1-Cas system were 2-fold upregulated by vicK deletion, suggesting that VicK mediated a negative regulatory role on their expression (Fig. 8B). Hence, it is possible that VicR/K differentially regulates CRISPR systems to prevent or reduce their simultaneous expression.
FIG 8.
Expression of cas genes from the CRISPR1 (A) and CRISPR2 (B) operons. RNA analysis from mid-logarithmic-phase cultures of S. mutans UA159 and SmuvicK grown under regular or acidic conditions. The results are the averages of triplicate samples from three independent experiments ± the standard errors.
DISCUSSION
S. mutans is one of several bacterial species known to be competent for horizontal gene transfer via natural transformation. Only a few phages are known to infect S. mutans and transformation is the key process used by S. mutans to acquire exogenous DNA. Frequent horizontal gene transfer occurs in S. mutans to promote homology-based DNA repair, genetic diversity, or other functions. However, the mechanisms that regulate the transfer, uptake, and recombination of incoming DNA in naturally transformable S. mutans are still poorly understood. Consistent with previous findings (45), we demonstrated that in S. mutans UA159, CRISPR-Cas systems do not play a prominent role in acquired resistance to M102 phage infection. Based on the high variability of the CRISPR spacers (including M102 sequences) between S. mutans serotype c (45), it is unlikely that CRISPR-Cas systems would be so widespread if they were unable to provide adaptive protection to their hosts. Probably, S. mutans has a variety of natural phage resistance mechanisms, including restriction/modification systems and/or CRISPR-Cas systems to target diverse steps of the phage life cycle to prevent M102 phages from attacking these genomes (48, 75).
We also revealed that the native type II-A CRISPR1-Cas system of S. mutans UA159 is important for preventing natural transformation via plasmid DNA. Using transformation assays, pCR1SP2 and pCR1SP3 constructs which contained protospacer sequences matching spacers SP2 (86% sequence identity to M102) and SP3 (100% identity to M102) in the CRISPR1 locus yielded drastically reduced TF compared to that of the empty plasmid. Consistent with previous reports, we found that CRISPR1-Cas machinery can tolerate a few nucleotide mismatches between spacer and protospacer at certain positions (23, 67, 68, 76, 77). As previously observed (23, 71), the degree of inhibition of transformation varied widely depending on the protospacer tested. The UA159 strain could be transformed with the protospacer with no mismatch (pCR1SP3) at higher frequencies than the protospacer with mismatches (pCR1SP2), suggesting that the CRISPR-Cas machinery is more permissive for pCR1SP3. Although it is still unclear, it might reflect a weak or altered interaction between the CRISPR-Cas system and plasmid DNA; however, that has yet to be elucidated. Intriguingly, effective interference was not observed with pCR1SP6 and pCR2SP1 constructs (100% identity to a S. mutans GS-5 spacer and 100% identity to a S. mutans LJ-23 spacer, respectively), possibly suggesting that crRNA transcripts complementary to these targets are only weakly expressed to produce interference. Since UA159 contains only one CRISPR2 spacer, further investigations are warranted to confirm that the observed phenotype is explained by the function of the CRISPR2-Cas machinery and rule out the lack of effective interference activity due to cas gene mutations (45, 47). Surprisingly, the presence or absence of flanking sequences within pCR1SP2 and pCR1SP3 had no effect on the ability to interfere with plasmid transformation. These results contrast to those data obtained using in vitro plasmid cleavage assays, where dual-tracrRNA/crRNA-guided Cas9 from S. mutans could efficiently cleave target DNA in the presence of a NGG sequence (70). Consistent with our findings, it was also observed in Streptococcus thermophilus that plasmids carrying protospacers associated with consensus or with nonconsensus (degenerate) PAMs could not be transformed into the corresponding plasmid-interfering strains, whereas phages carrying the degenerative PAMs could infect the matching phage-insensitive mutants (23, 72). These researchers suggested that the tolerance of PAM degeneracy for CRISPR-Cas function could be due to the lower selective pressure for plasmids compared to phages. Such an activity could theoretically produce a lower level of TF in the presence or absence of flanking sequences observed in our study. Alternatively, the presence of plasmids inside the cell could increase the expression of Cas proteins, reflecting higher interference activity that might not require a PAM site (78).
Using purified recombinant protein, we also demonstrated that SMU.1763c possesses RNase activity against synthetic oligoribonucleotides and total RNA extracted from S. mutans UA159. Further, based on our in vitro transcription and DNA microarray studies, SMU.1763c had no obvious sequence preference in RNA cleavage. Recently, it was also reported that Cas5d ortholog from B. halodurans cleaves pre-crRNA by recognizing both the hairpin structure and the 3′ single-stranded sequence in the CRISPR repeat region (18). Based on these findings and the fact that we were not able to identify sequence specificity in SMU.1763c cleavage, we speculate that RNA secondary structure elements such as stem-loop are required for SMU.1763c to process RNA substrates in a sequence- and site-specific manner.
Consistent with previous studies (36, 79), we found that the CRISPR1 cas-deficient mutant exhibited enhanced sensitivity to killing by MMC (which inhibits growth by causing DNA cross-linkage) (80) or UV irradiation (inhibits growth by causing bulky DNA lesions). In addition, the CRISPR2 cas-deficient mutant was not sensitive to DNA damage, a finding which is in perfect agreement with previous bioinformatic work where Cas1 associated with CRISPR2 appeared to be truncated (45, 47). S. mutans possesses several DNA repair systems to support functions related to DNA protection or repair: RecA (81), apurinic-apyrimidinic endonuclease (82) or a UV repair excinuclease (uvrA) (83). The nucleotide excision repair (NER) has been shown to be the major system for repairing damaged DNA caused by UV light and genotoxic agents such as MMC (83). Although Cas components of the CRISPR1-Cas system possibly act in the NER pathway in response to DNA damage caused by environmental stress, their specific role in this repair pathway remains to be elucidated.
Further, our transcriptional analysis identified that the VicR/K system modulates the expression of S. mutans cas genes. The VicR/K system, one of 14 two-component signal transduction systems (TCSTSs) in S. mutans (66, 75, 84), is comprised of a VicK histidine sensor kinase and an essential VicR response regulator. It was previously shown to be involved in biofilm formation, genetic competence, stress tolerance, bacteriocin production, and cell viability (51, 56, 66, 85). Based on our finding that VicR/K modulates the expression of cas genes, a role of CRISPR-Cas systems in contending with various environmental stressors was not surprising. In S. mutans UA159 it has been proposed that dual crRNA/tracrRNA participates in type II-A CRISPR function (22); therefore, it raised the question as to whether the entire CRISPR1-Cas system is necessary to mediate the stress responses observed for the ΔC1S mutant. Using Northern blot analysis, the expression of the tracrRNA under stress conditions was noted for the wild type (see Fig. S6 in the supplemental material). Although our observations may indicate that tracrRNA possibly with crRNA mediates stress response in vivo, our assumption warrants further investigations. We also noted that ΔC2E did not share the same sensitivity to the tested stressors as that seen with the ΔC1S mutant, suggesting that CRISPR1 cas and CRISPR2 cas genes are differentially regulated to function independently within the environment. In fact, this is the case in S. thermophilus where CRISPR-Cas systems were observed to function independently (86). Furthermore, several transcriptome studies revealed that deletion of virulence or global regulatory genes of S. mutans (including genes involved in stress response) differently affected transcription of cas genes within CRISPR-Cas systems, suggesting different roles for cas genes within the cell (50, 54, 87). As already proven in other systems (88–91), it is possible that different regulatory systems, in addition to VicR/K, interact with S. mutans CRISPR-Cas systems to mediate gene expression in response to cues such as oxidative stress and cell membrane changes or alterations in the internal pH of the cell. The presence of diverse and complex regulatory strategies to modulate the CRISPR-Cas activity might also explain why some phenotypes displayed by ΔC1S and ΔC2E are not compatible with those of a VicK mutant as shown previously (51). Currently, studies are under way to examine whether VicR exerts a direct regulatory role on the transcription of CRISPRs by binding to their respective promoters. Examination of other regulatory systems in S. mutans on CRISPRs transcription and function would be of interest.
In summary, our data provide the first experimental evidence that the CRISPR1-Cas system of S. mutans UA159 play novel roles in resistance against incoming plasmids that carry matching protospacer sequences and stress response. Given their multiple roles in the cell physiology, the type II-A system may prove to be useful target for therapeutics to diminish the virulence and also to influence S. mutans species to prevent the uptake and dissemination of antibiotic resistance genes.
ACKNOWLEDGMENTS
We thank Richard Mair for assistance with bioinformatic analyses, Deanna Del Re for constructing the ΔC2E mutant, and Greg Brown for cloning cas5d gene.
D.G.C. is a recipient of National Institutes of Health grant R01DE013230-03 and Canadian Institutes of Health Research (CIHR) grant (MT-15431), and M.A.S. is a recipient of a CIHR Strategic Training Fellowship in Cell Signaling in Mucosal Inflammation and Pain and Harron Scholarship. This study was also partially supported by the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405) and the Ontario Research Fund (ORF-GL2-01-004).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02333-14.
REFERENCES
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, Van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 6.Van Der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 9.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 10.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 11.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 12.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garside EL, Schellenberg MJ, Gesner EM, Bonanno JB, Sauder JM, Burley SK, Almo SC, Mehta G, MacMillan AM. 2012. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA 18:2020–2028. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gesner EM, Schellenberg MJ, Garside EL, George MM, MacMillan AM. 2011. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol 18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 17.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence-and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. 2012. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype IC/Dvulg CRISPR-Cas system. Structure 20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sashital DG, Jinek M, Doudna JA. 2011. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 18:680–687. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 20.Wang R, Preamplume G, Terns MP, Terns RM, Li H. 2011. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chylinski K, Le Rhun A, Charpentier E. 2013. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol 10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garneau JE, Dupuis M, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 24.Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. 2013. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol 10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magadán AH, Dupuis M-MÈ Villion M, Moineau S. 2012. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One 7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 27.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mojica F, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 29.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 30.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. 2011. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One 6:e19543. doi: 10.1371/journal.pone.0019543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-Cas. mBio 1:e00227-10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dugar G, Herbig A, Förstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM. 2013. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet 9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. 2013. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol 189:3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sampson TR, Saroj SD, Llewellyn AC, Tzeng Y-L, Weiss DS. 2013. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunderson FF, Cianciotto NP. 2013. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio 4:e00074-13. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Ploeg J. 2007. Genome sequence of Streptococcus mutans bacteriophage M102. FEMS Microbiol Lett 275:130–138. doi: 10.1111/j.1574-6968.2007.00873.x. [DOI] [PubMed] [Google Scholar]
- 44.Delisle AL, Guo M, Chalmers NI, Barcak GJ, Rousseau GM, Moineau S. 2012. Biology and genome sequence of Streptococcus mutans phage M102AD. Appl Environ Microbiol 78:2264–2271. doi: 10.1128/AEM.07726-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Ploeg JR. 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966–1976. doi: 10.1099/mic.0.027508-0. [DOI] [PubMed] [Google Scholar]
- 46.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol 131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 48.Maruyama F, Kobata M, Kurokawa K, Nishida K, Sakurai A, Nakano K, Nomura R, Kawabata S, Ooshima T, Nakai K. 2009. Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. BMC Genomics 10:358. doi: 10.1186/1471-2164-10-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibata Y, Yamashita Y, van der Ploeg JR. 2009. The serotype-specific glucose side chain of rhamnose-glucose polysaccharides is essential for adsorption of bacteriophage M 102 to Streptococcus mutans. FEMS Microbiol Lett 294:68–73. doi: 10.1111/j.1574-6968.2009.01546.x. [DOI] [PubMed] [Google Scholar]
- 50.Chattoraj P, Banerjee A, Biswas S, Biswas I. 2010. ClpP of Streptococcus mutans differentially regulates expression of genomic islands, mutacin production and antibiotic tolerance. J Bacteriol 192:1312–1323. doi: 10.1128/JB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, Cvitkovitch DG. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol 191:6415–6424. doi: 10.1128/JB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Z, Okinaga T, Niu G, Qi F, Merritt J. 2010. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol Microbiol 78:1431–1447. doi: 10.1111/j.1365-2958.2010.07417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192:2546–2556. doi: 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kajfasz JK, Abranches J, Lemos JA. 2011. Transcriptome analysis reveals that ClpXP proteolysis controls key virulence properties of Streptococcus mutans. Microbiology 157:2880–2890. doi: 10.1099/mic.0.052407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J, Wu C, Huang I-H, Merritt J, Qi F. 2011. Differential response of Streptococcus mutans toward friend and foe in mixed-species cultures. Microbiology 157:2433–2444. doi: 10.1099/mic.0.048314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senadheera MD, Lee AWC, Hung DCI, Spatafora GA, Goodman SD, Cvitkovitch DG. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol 189:1451–1458. doi: 10.1128/JB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lau PCY, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–205. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y-YM, Shieh H-R, Lin C-T, Liang S-Y. 2011. Properties and construction of plasmid pFW213, a shuttle vector with the oral Streptococcus origin of replication. Appl Environ Microbiol 77:3967–3974. doi: 10.1128/AEM.02828-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W. 2008. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem 283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–43. [Google Scholar]
- 62.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:1–26. doi: 10.1186/1745-6150-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim T-Y, Shin M, Huynh Thi Yen L, Kim J-S. 2013. Crystal structure of Cas1 from Archaeoglobus fulgidus and characterization of its nucleolytic activity. Biochem Biophys Res Commun 441:720–725. doi: 10.1016/j.bbrc.2013.10.122. [DOI] [PubMed] [Google Scholar]
- 64.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. 2009. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 65.Dubrac S, Msadek T. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Senadheera MD, Guggenheim B, Spatafora GA, Huang Y-CC, Choi J, Hung DCI, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol 187:4064–4076. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lécrivain A-L, Bzdrenga J, Koonin EV, Charpentier E. 2014. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2007. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. 2002. A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res 30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H. 2002. Genome sequence of Streptococcus mutans UA 159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manica A, Zebec Z, Teichmann D, Schleper C. 2011. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 78.Makarova KS, Anantharaman V, Aravind L, Koonin EV. 2012. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct 7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. 2012. Characterization of the CRISPR/Cas subtype IA system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol 194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tomasz M. 1995. Mitomycin C: small, fast, and deadly (but very selective). Chem Biol 2:575–579. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 81.Quivey RG, Faustoferri RC, Clancy KA, Marquis RE. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett 126:257–262. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- 82.Hahn K, Faustoferri R, Quivey R. 1999. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol Microbiol 31:1489–1498. doi: 10.1046/j.1365-2958.1999.01292.x. [DOI] [PubMed] [Google Scholar]
- 83.Hanna MN, Ferguson RJ, Li YH, Cvitkovitch DG. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J Bacteriol 183:5964–5973. doi: 10.1128/JB.183.20.5964-5973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biswas I, Drake L, Erkina D, Biswas S. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J Bacteriol 190:68–77. doi: 10.1128/JB.00990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Senadheera DB, Cordova M, Ayala EA, de Paz LEC, Singh K, Downey JS, Svensäter G, Goodman SD, Cvitkovitch DG. 2012. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carte J, Christopher RT, Smith JT, Olson S, Barrangou R, Moineau S, Glover CV, Graveley BR, Terns RM, Terns MP. 2014. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol Microbiol 93:98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. 2008. Role of RelA of Streptococcus mutans in global control of gene expression. J Bacteriol 190:28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. 2010. Identification and characterization of Escherichia coli CRISPR/Cas promoters and their silencing by H-NS. Mol Microbiol 75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 89.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol 395:270–281. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 90.Medina-Aparicio L, Rebollar-Flores J, Gallego-Hernandez A, Vazquez A, Olvera L, Gutierrez-Rios R, Calva E, Hernandez-Lucas I. 2011. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol 193:2396–2407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shinkai A, Kira S, Nakagawa N, Kashihara A, Kuramitsu S, Yokoyama S. 2007. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J Bacteriol 189:3891–3901. doi: 10.1128/JB.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]