Abstract
The synthesis, computer modeling, and biological activity of an octawalled molecular umbrella short interfacing RNA (siRNA) conjugate is described. This molecular umbrella–siRNA conjugate exhibited mRNA knockdown activity in vitro in the absence of a transfection reagent. Evaluation of this molecular umbrella conjugate in vivo, using the rat eye via intravitreal injection, resulted in sequence specific mRNA knockdown in the retina with no obvious signs of toxicity, as judged by ophthalmic examination.
siRNA mediated gene silencing continues to hold great potential as a therapeutic. Despite impressive advances in this field, new methodologies for the cytosolic delivery of siRNA are still needed.1 The central challenge that remains is the need to translocate either the cellular or the endolysosomal membrane in a safe and efficient manner. One attractive approach is the use of “siRNA–conjugates”, i.e., conjugation of a well-defined single molecule to siRNA in which the conjugated fragment has properties that overcome this delivery challenge. Such designs include lipid (cholesterol, stearyl) and peptide conjugates (R9, Tat).1 In general, such conjugates improve either membrane solubility or solubility in the hydrophilic environment of the intra/extracellular compartments. However, these conjugates do not possess properties that facilitate the interaction with both hydrophilic and hydrophobic domains. Consequently, most conjugates are “trapped” in either the lipophilic or aqueous compartment of the cell thereby limiting their transfection efficiency.2 An improved design would rely upon a conjugate having significant affinity toward both polar extremes.
Molecular umbrella conjugates are a unique class of amphiphilic molecules that have potential for the in vivo delivery of biologically active agents in general.3 These molecules are composed of two or more facial amphiphiles (i.e., “walls”) that are attached to a central scaffold. When immersed in a hydrophilic environment, molecular umbrellas create a hydrophilic exterior. Conversely, when immersed in a hydrophobic environment, these same molecules create a hydrophobic exterior. Such “amphomorphism” behavior is illustrated in Figure 1A. For convenience, only a diwalled molecular umbrella is shown.
Figure 1.
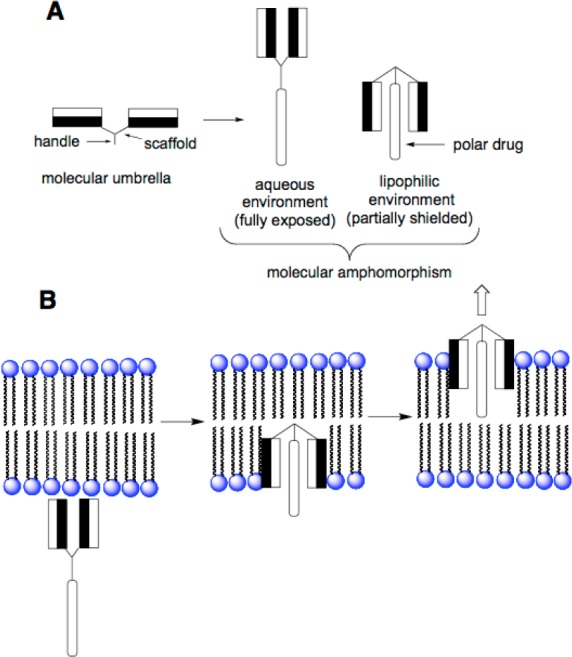
Stylized illustrations showing (A) a diwalled molecular umbrella exhibiting molecular amphomorphism and a molecular umbrella–polar drug conjugate in an exposed and shielded conformation in an aqueous and lipophilic environment, respectively, and (B) a hypothetical “needle and thread” transport mechanism. In these illustrations, the filled and unfilled rectangles that are directly attached to each other are the hydrophobic and hydrophilic faces of an amphiphilic wall, respectively. For this work, the polar drug will be a siRNA.
Previously, we have demonstrated that molecular umbrellas are capable of crossing liposomal membranes, even when they are relatively large in size and high in hydrophilicity. Remarkably, passive transport rates were found to increase on going from diwalled to tetrawalled to octawalled molecular umbrellas.4 We have also shown that certain molecular umbrellas can transport hydrophilic agents across liposomal membranes despite the fact that they are capable of shielding only a small segment of the agent. For example, a small diwalled molecular umbrella was found capable of transporting a 16-mer oligonucleotide (S-dT16) across liposomal membranes.5 We and others have also demonstrated that molecular umbrellas can enter cells, in vitro, and that passive transport appears to play a significant role.6,7 Most recently, we have found that the covalent attachment of octaarginine to the “handle” of a molecular umbrella can be used to deliver siRNA to HeLa cells, as judged by the knockdown of enhanced green fluorescent protein expression.8 Although the precise mechanism by which a molecular umbrella transports partially shielded hydrophilic agents across lipid membranes remains to be established, one plausible model involves a “needle and thread” pathway, that is, where the umbrella molecule (i.e., the “needle”) inserts into the bilayer with partial coverage of the attached agent (i.e., the thread), allowing both to cross the membrane (Figure 1B).5
Encouraged by these positive results, we were strongly motivated to test the feasibility of using a molecular umbrella–siRNA conjugate for the in vivo transport of siRNA. In the work that is reported herein, we have focused on local delivery by intravitreal injection in the eye.9−12 In addition to therapeutic opportunities, we chose to focus on ocular delivery because it minimizes many of the hurdles associated with systemic delivery including serum nuclease exposure, biodistribution, and specific tissue targeting. We also chose to use a large octawalled umbrella to maximize coverage of an attached siRNA and because our past studies have shown that larger molecular umbrellas can cross lipid bilayers by passive transport faster than smaller analogs due to greater partitioning into the membrane.4
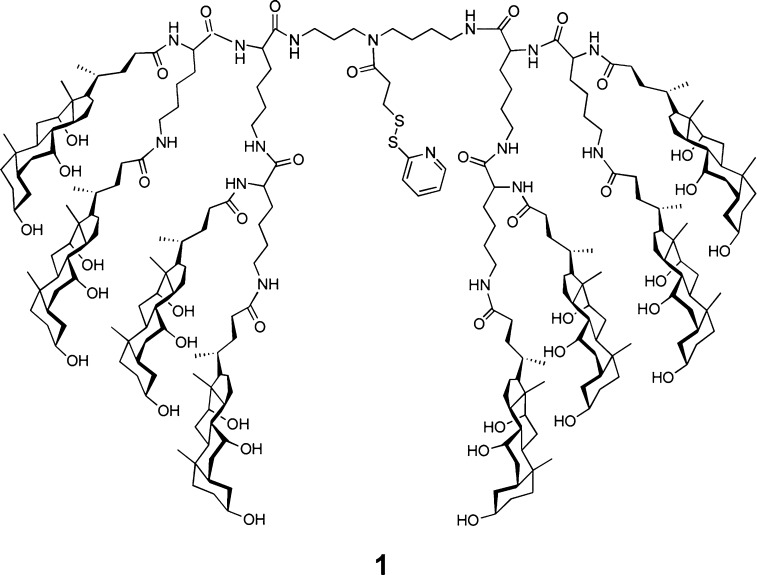
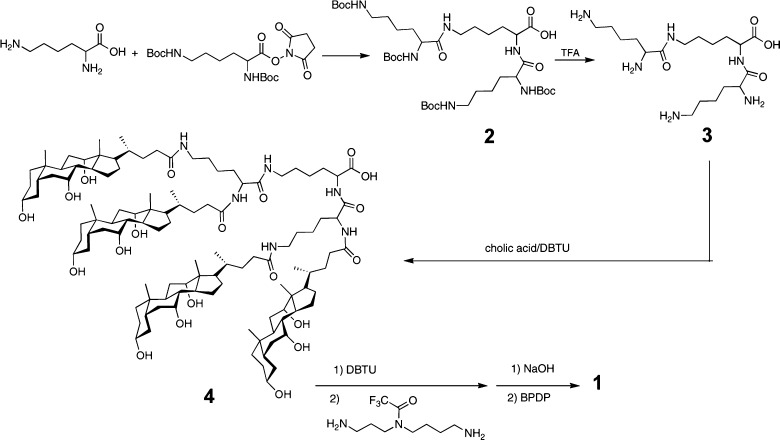
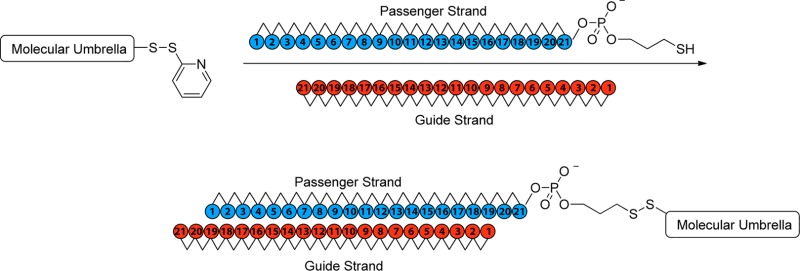
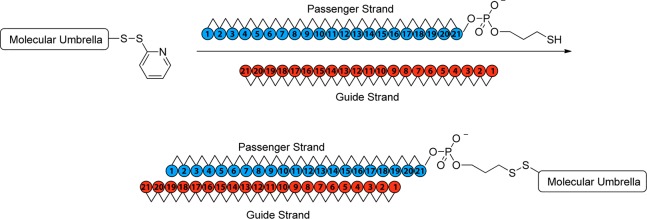
The molecular design strategy that we have employed is illustrated in Figure 2. Thus, an oligonucleotide that can form a duplex with an siRNA is covalently attached to the handle of an activated molecular umbrella via thiolate–disulfide exchange. The umbrella-bound oligonucleotide is then used as a “passenger” strand to anneal with the complementary “guide” strand to give the biologically active siRNA. The specific activated molecular umbrella that we have used in this work is shown in Figure 3. In Figure 4 is shown the method that was used to prepare 1. Thus, acylation of l-lysine with N1,N2-diBoc-l-lysine-N-(O-succinimidyl ester) to produce 2, followed by deprotection to give 3, and acylation with an activated form of cholic acid afforded 4. Finally, activation of 4, followed by condensation with spermidine-N2-trifluoroacetamide, deprotection and acylation with N-[O-1,2,3-benzotriazin-4(3H)one-yl]-3,(2-pyridyldithio)propionate (BPDP) afforded 1. Covalent attachment of a given passenger strand to a molecular umbrella framework, illustrated in Figure 5 and detailed in the Supporting Information, was then accomplished via thiolate disulfide exchange by reacting 1 with the thiol form of an oligonucleotide at room temperature. The latter was generated in situ by reducing a disulfide protected form of the oligonucleotide with dithiothreitol in aqueous PBS (pH 7.4). All molecular umbrella–siRNA conjugates that were investigated in vitro and in vivo were purified by reverse phase HPLC. The purified single strand molecular umbrella conjugates were then duplexed to the corresponding guide strand to yield 1b and 5 (Figure 5). Analytical purity and confirmation of duplex formation, as detailed in the Supporting Information, were performed with high resolution mass spectrometry and capillary electrophoresis.13 For these studies, siRNA against Sjögren syndrome type B antigen (SSB) was used to prepare conjugate 1b, and a control sequence was used to prepare conjugate 5. The specific siRNA duplex oligonucleotide sequences used are described in the Supporting Information (S7). The gene target SSB is expressed in all cell types allowing for a general understanding of mRNA knockdown. The control sequence, which does not target SSB mRNA, gives confidence that the effects observed with SSB siRNA are not due to off-target effects. Additional information on SSB si RNA can be found in ref (13).
Figure 2.
Stylized illustration of an activated molecular umbrellas being covalently coupled to a passenger strand, which is then used to form a duplex with the complementary guide strand of siRNA, that is, the active agent for mRNA knockdown.
Figure 3.
Structure of an activated molecular umbrella used for coupling to a guide strand.
Figure 4.
Synthetic scheme used to prepare 1.
Figure 5.
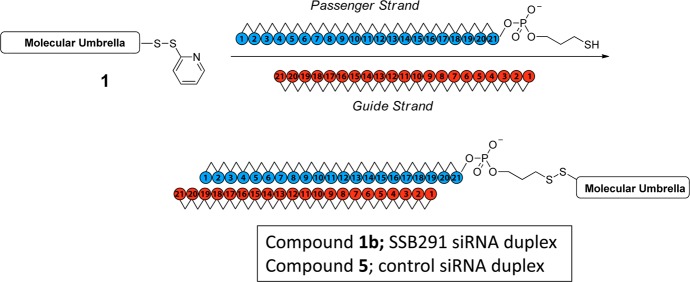
Attachment of molecular umbrella 1 to the siRNA passenger strand and subsequent duplex formation to duplexes 1b and 5. Here, the 1b duplex contains SSB siRNA and duplex 5 contains a control siRNA sequence that does not have sequence homology with SSB mRNA.
To date, molecular modeling on siRNA conjugates has been limited. Furthermore, conjugates 1b and 5 represent the first examples of molecular umbrella–siRNA conjugates. Thus, we saw this as an opportunity to gain insight into the interactions between the umbrella, siRNA, and an aqueous environment. To better understand the intramolecular interactions, classical molecular dynamics (MD) simulations were performed. The AMBER force field was selected for approximating the atomic interactions.14 This force field is particularly appropriate for modeling interactions between nucleic acids and small molecules especially where covalent bonds are involved.15 For this hybrid system we also employed the AM1 bond charge corrected version for which we have extensive experience and confidence.16 Sodium chloride (NaCl) was added at a physiciological concentration of 150 mM. In addition sodium ions (Na+) were added to ensure charge neutrality. The initial conformation of the double strand nucleic acids was placed in the A-form, while the umbrella assumed a more extended form to minimize any preference for the intramolecular umbrella–siRNA interaction. This initial configuration of the umbrella–siRNA conjugate, explicit water, and ions was minimized before slowly heating to ∼310 K. After reaching constant pressure equilibrium, a 40 ns MD simulation was performed using a 0.001 ps time step. Figure 6 shows a snapshot of the final conformation of the conjugate resulting from these calculations. As predicted, the hydrophobic faces of six of the cholic acids condense upon themselves and the hydroxyl moieties interact with the aqueous environment. Encouragingly, two of the cholic acid hydrophobic faces interact with the hydrophobic wedge of the siRNA, lending support to the amphiphilic nature of the umbrella conjugate.
Figure 6.
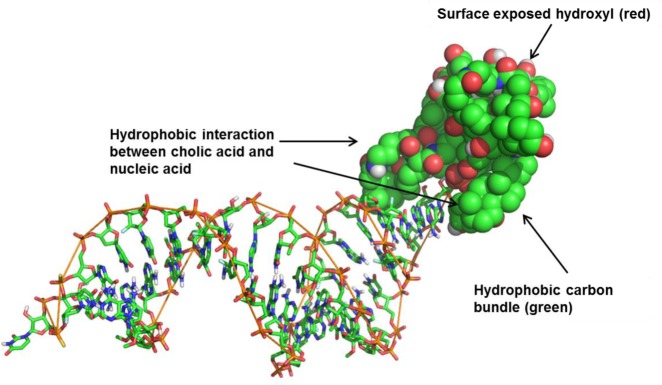
Molecular dynamic simulation of conjugate 1b. Simulation details and additional graphical representations can be found in the Supporting Information.
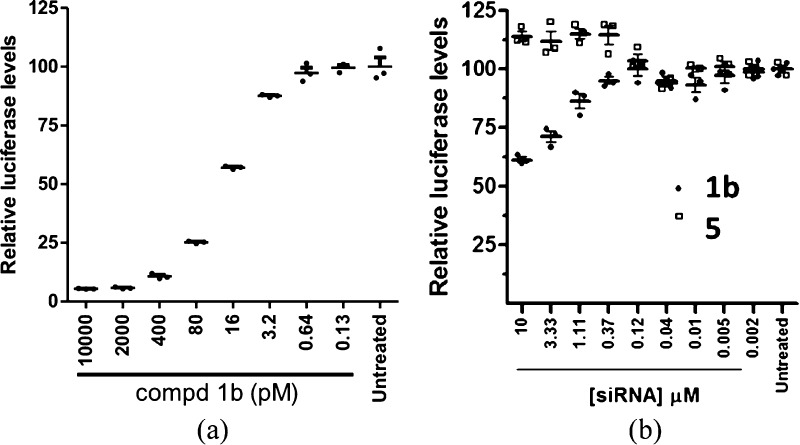
For our in vitro and in vivo experiments, we opted to focus resources on conjugate 1b and the conjugate containing a control sequence (5) as an initial test of this approach. To confirm that siRNA conjugation to the umbrella does not negatively impact RNAi activity, the conjugate 1b was transfected in HEK293 cells using lipofectamine. As shown in Figure 7a, robust silencing activity with an IC50 of ∼20 pM was observed. After confirming the intrinsic potency, we next tested if the conjugates have cell uptake properties that results in silencing in vitro in the absence of lipofectamine. As shown in Figure 7b, conjugate 1b displays dose-dependent target knockdown while the control siRNA conjugate 5 does not show activity. Although 1b exhibited substantially lower activity than that found in the presence of lipofectamine, its ability to knockdown mRNA in vitro renders such duplexes worthy of in vivo testing.
Figure 7.
(a) In vitro RNAi activity of molecular umbrella–siRNA conjugate 1b in the presence of transfection reagent. The HEK293 cells stably transfected with firefly and renilla luciferase containing the siRNA target sequence in the 3′UTR of renilla were used for evaluating knockdown. The cells were treated for 24 h and the luciferase levels were determined to evaluate knockdown. (b) In vitro RNAi activity of molecular umbrella–siRNA conjugates 1b and 5 in the absence of transfection reagent. The HEK293 cells were treated for 72 h and target mRNA levels were analyzed by b-DNA assay.
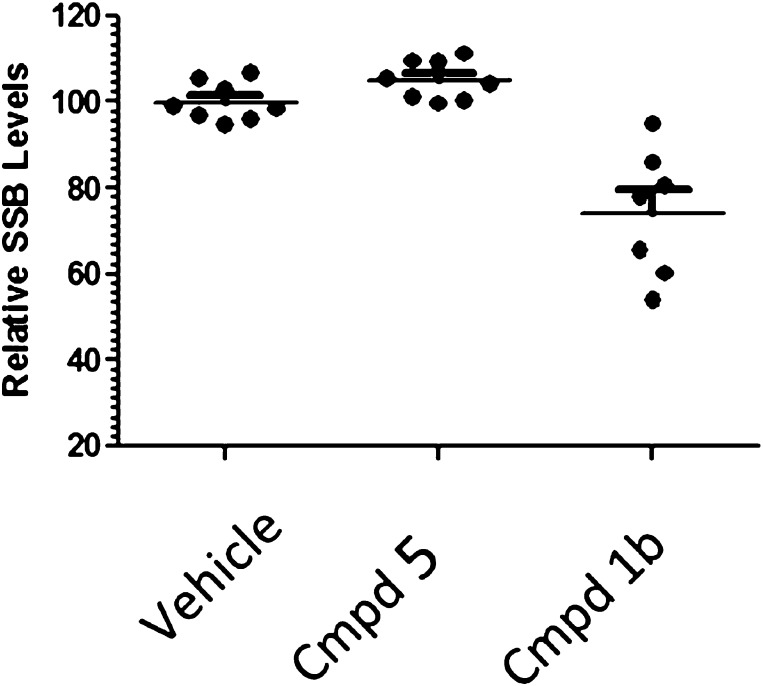
To determine if these results could be extended in vivo, the molecular umbrella conjugates were assessed for mRNA knockdown in rat retina after intravitreal injection. As shown in Figure 8, modest but statistically significant mRNA knockdown (32%) was observed by the umbrella–siRNA conjugate at 100 μg/eye dose. In sharp contrast, no activity was observed with the nontarget umbrella conjugate. It should be noted that, for ocular animal studies, accurate dosing of compounds (5 μL) by intravitreal injection in the relatively small space of the eye is challenging. We attribute the scatter seen in these data to such dosing and, in the case of the conjugate 1b, to modest knockdown levels. For this reason, a larger sample size (n = 8 eyes/treatment) was used for the in vivo studies as compared with triplicate measurements that were made in the cell culture studies. Finally, an ophthalmic examination of eye revealed no signs of obvious toxicity as evidenced by dilation of the pupil and biomicroscopy of the anterior segment and indirect ophthalmicroscopy of the posterior segment. This provides further evidence that mRNA reduction was specific to the siRNA mechanism.
Figure 8.
In vivo activity of molecular umbrella conjugate in rat retina delivered by intravitreal injection (dose = 100 μg/eye, based on historical in-house data with different siRNA conjugates that demonstrated activity at this dose). Higher doses were not evaluated due to solubility limitations. Vehicle refers to the dosing solution alone; i.e., only water was administered in this case.
In summary, the first molecular umbrella–siRNA conjugates were prepared and found to exhibit in vitro activity in the absence of lipofectamine and modest ocular in vivo activity. Having demonstrated proof of concept, future work will now focus on elucidating structure–activity relationships, expanding the modeling work to look at conjugate–membrane interactions, tracking metabolic fate, and determining specific cellular localization within the retina to identify therapeutic applicability.
Acknowledgments
This work was funded in part by the Institutes of Health (PHS GM51814 and PHS GM100962).
Supporting Information Available
Experimental procedures, computer modeling details and supplementary data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Stanton M. G.; Colletti S. L. (2010) Medicinal chemistry of siRNA delivery. J. Med. Chem. 53, 7887–7901. [DOI] [PubMed] [Google Scholar]
- Juliano R. L.; Ming X.; Nakagawa O. (2012) Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjugate Chem. 23, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janout V.; Regen S. L. (2009) Bioconjugate-based molecular umbrellas. Bioconjugate Chem. 20, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehiri M.; Chen W.-H.; Janout V.; Regen S. L. (2009) Molecular umbrella transport: exceptions to the classic size/lipophilicity rule. J. Am. Chem. Soc. 131, 1338–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janout V.; Regen S. L. (2005) A needle and thread approach towards bilayer transport: permeation of a molecular umbrella-oligonucleotide conjugate across a phospholipid membrane. J. Am. Chem. Soc. 127, 22–23. [DOI] [PubMed] [Google Scholar]
- Mehiri M.; Jing B.; Ringhoff D.; Janout V.; Cassimeris L.; Regen S. L. (2008) Cellular entry and nuclear targeting by a highly anionic molecular umbrella. Bioconjugate Chem. 19, 1510–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D.; Wu D.; Wang Z.; Shi W.; Wu T.; Zhang A.; Hong S.; Wang J.; Zhang Y.; Ren L. (2009) Cellular uptake mechanism of molecular umbrella. Bioconjugate Chem. 20, 2311–2316. [DOI] [PubMed] [Google Scholar]
- Cline L. L.; Janout V.; Fisher M.; Juliano R. L.; Regen S. L. (2011) A molecular umbrella approach to the intracellular delivery of siRNA. Bioconjugate Chem. 22, 2210–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campochiaro P. A. (2006) Potential applications for RNAi to probe pathogenesis and develop new treatments for ocular disorders. Gene Ther. 13, 559–562. [DOI] [PubMed] [Google Scholar]
- Rajappa M.; Saxena P.; Kaur J. (2010) Potential applications for RNAi to probe pathogenesis and develop new treatments for ocular disorders. Adv. Clin. Chem. 50, 103–121. [PubMed] [Google Scholar]
- Fattal E.; Bochot A. (2008) State of the art and perspectives for the delivery of antisense oligonucleotides and siRNA by polymeric nanocariers. Int. J. Pharm. 364, 237–248. [DOI] [PubMed] [Google Scholar]
- Ng E. W. M.; Shima D. T.; Calias P.; Cunningham E. T.; Guyer D. R.; Adamis A. P. (2006) Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discovery 5, 123–132. [DOI] [PubMed] [Google Scholar]
- Aaronson J. G.; Klein L. J.; Momose A. A.; O’Brien A. M.; Shaw A. W.; Tucker T. J.; Yuan Y.; Tellers D. M. (2011) Rapid HATU-mediated solution phase siRNA conjugation. Bioconjugate Chem. 22, 1723–1728. [DOI] [PubMed] [Google Scholar]
- Cornell W. D.; Cieplak P.; Bayly C. I.; Kollman P. A. (1993) Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 115, 9620. [Google Scholar]
- Cornell W. D.; Cieplak P.; Bayly C. I.; Gould I. R.; Merz K. M.; Ferguson D. M.; Spellmeyer D. C.; Fox T.; Caldwell J. W.; Kollman P. A. (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197. [Google Scholar]
- Jakalian A.; Jack D. B.; Bayly C. I. (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 23, 1623–1641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.